Difference between revisions of "BIN-SX-Chimera"
m |
m |
||
| (26 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | <div id=" | + | <div id="ABC"> |
| − | + | <div style="padding:5px; border:4px solid #000000; background-color:#b3dbce; font-size:300%; font-weight:400; color: #000000; width:100%;"> | |
| − | UCSF | + | UCSF ChimeraX: Structure Visualization and Analysis |
| − | + | <div style="padding:5px; margin-top:20px; margin-bottom:10px; background-color:#b3dbce; font-size:30%; font-weight:200; color: #000000; "> | |
| − | + | (UCSF ChimeraX; Structure visualization; Structure analysis) | |
| − | + | </div> | |
| − | |||
| − | <div | ||
| − | |||
| − | UCSF | ||
</div> | </div> | ||
| − | {{ | + | {{Smallvspace}} |
| − | + | <div style="padding:5px; border:1px solid #000000; background-color:#b3dbce33; font-size:85%;"> | |
| − | + | <div style="font-size:118%;"> | |
| − | + | <b>Abstract:</b><br /> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | ||
| − | <div | ||
| − | |||
<section begin=abstract /> | <section begin=abstract /> | ||
| − | + | This unit introduces the molecular viewer UCSF ChimeraX, starts you off on a routine to practice stereo viewing of molecular models, and teaches how to use ChimeraX to solve a number of common visualization and analysis tasks. | |
| − | This unit introduces the molecular viewer UCSF | ||
<section end=abstract /> | <section end=abstract /> | ||
| + | </div> | ||
| + | <!-- ============================ --> | ||
| + | <hr> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td style="padding:10px;"> | ||
| + | <b>Objectives:</b><br /> | ||
| + | This unit will ... | ||
| + | * ... introduce the molecular viewer UCSF ChimeraX; | ||
| + | * ... teach how to use it for a number of structure visualization and analysis tasks; | ||
| + | * ... start you off on learning to view biomolecules with split-screen stereo. | ||
| + | </td> | ||
| + | <td style="padding:10px;"> | ||
| + | <b>Outcomes:</b><br /> | ||
| + | After working through this unit you ... | ||
| + | * ... can competently use ChimeraX for molecular visualization and analysis tasks; | ||
| + | * ... can view structure molecules in stereo; | ||
| + | * ... can create informative images of structures, aiming for publication quality. | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <!-- ============================ --> | ||
| + | <hr> | ||
| + | <b>Deliverables:</b><br /> | ||
| + | <section begin=deliverables /> | ||
| + | <li><b>Time management</b>: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit.</li> | ||
| + | <li><b>Journal</b>: Document your progress in your [[FND-Journal|Course Journal]]. Some tasks may ask you to include specific items in your journal. Don't overlook these.</li> | ||
| + | <li><b>Insights</b>: If you find something particularly noteworthy about this unit, make a note in your [[ABC-Insights|'''insights!''' page]].</li> | ||
| + | <section end=deliverables /> | ||
| + | <!-- ============================ --> | ||
| + | <hr> | ||
| + | <section begin=prerequisites /> | ||
| + | <b>Prerequisites:</b><br /> | ||
| + | This unit builds on material covered in the following prerequisite units:<br /> | ||
| + | *[[BIN-PDB|BIN-PDB (The RCSB-PDB Structure Database)]] | ||
| + | <section end=prerequisites /> | ||
| + | <!-- ============================ --> | ||
| + | </div> | ||
| − | {{ | + | {{Smallvspace}} |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | {{ | + | {{Smallvspace}} |
| − | + | __TOC__ | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| − | === | + | === Evaluation === |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | This learning unit can be evaluated for a maximum of 5 marks. If you want to submit tasks for this unit for credit you have the following options. If you have any questions about these options, discuss on the Discussion Board. | |
| + | <div class="note">Please note that all options for evaluation are to be developed with '''ChimeraX''' only. [https://pymol.org/2/ PyMol], [https://www.cgl.ucsf.edu/chimera/ Chimera], [https://www.ks.uiuc.edu/Research/vmd/ VMD] and [https://en.wikipedia.org/wiki/List_of_molecular_graphics_systems others] are excellent alternatives, but '''ChimeraX''' is the molecular viewer against which examples for this course have been developed.</div> | ||
| − | + | <ol> | |
| − | < | + | <li>Create a new page on the student Wiki as a subpage of your User Page.</li> |
| − | < | + | <li>Put all of your writing to submit on this one page.</li> |
| − | + | <li>When you are done with everything, go to the [https://q.utoronto.ca/courses/180416/assignments Quercus '''Assignments''' page] and open the first Learning Unit that you have not submitted yet. Paste the URL of your Wiki page into the form, and click on '''Submit Assignment'''.</li> | |
| − | < | + | </ol> |
| − | |||
| − | < | ||
| − | |||
| − | + | Your link can be submitted only once and not edited. But you may change your Wiki page at any time. However only the last version before the due date will be marked. All later edits will be silently ignored. | |
| − | + | {{Smallvspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
; Short Report option | ; Short Report option | ||
| Line 87: | Line 88: | ||
::'''A - Helix dipoles: Nucleosome core''' | ::'''A - Helix dipoles: Nucleosome core''' | ||
| − | ::'''A.1''' Open | + | ::'''A.1''' Open ChimeraX and load the structure [http://www.rcsb.org/pdb/explore/explore.do?structureId=5XF3 '''5XF3''']. |
| − | ::'''A.2''' Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings. | + | ::'''A.2''' Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings. |
::'''A.3''' Write up, illustrate, and interpret your findings in a short report. | ::'''A.3''' Write up, illustrate, and interpret your findings in a short report. | ||
::'''B - Helix dipole: Transcription factor''' | ::'''B - Helix dipole: Transcription factor''' | ||
| − | ::'''B.1''' Open | + | ::'''B.1''' Open ChimeraX and load the structure [http://www.rcsb.org/pdb/explore/explore.do?structureId=4Y60 '''4Y60''']. |
::'''B.2''' Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings. | ::'''B.2''' Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings. | ||
::'''B.3''' Write up, illustrate and interpret your findings in a short report. | ::'''B.3''' Write up, illustrate and interpret your findings in a short report. | ||
::'''C - Helix dipole: Transcription factor''' | ::'''C - Helix dipole: Transcription factor''' | ||
| − | ::'''C.1''' Open | + | ::'''C.1''' Open ChimeraX and load the structure [http://www.rcsb.org/pdb/explore/explore.do?structureId=4ZSF '''4ZSF''']. |
::'''C.2''' Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings. | ::'''C.2''' Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings. | ||
::'''C.3''' Write up, illustrate and interpret your findings in a short report. | ::'''C.3''' Write up, illustrate and interpret your findings in a short report. | ||
| + | :'''3.''' When you are done with everything, submit the page via Quercus as described above. | ||
| − | + | <!-- | |
| − | |||
| − | |||
| − | |||
| − | |||
{{Smallvspace}} | {{Smallvspace}} | ||
| − | |||
; Tasks submission option | ; Tasks submission option | ||
:# Create a new page on the student Wiki as a subpage of your User Page. | :# Create a new page on the student Wiki as a subpage of your User Page. | ||
:# There are a number of tasks in which you are explicitly asked you to submit code or other text for credit. Put all of these submission on this one page. | :# There are a number of tasks in which you are explicitly asked you to submit code or other text for credit. Put all of these submission on this one page. | ||
| − | :# When you are done with everything, | + | :# When you are done with everything, submit the page via Quercus as described above. |
| − | |||
| − | |||
--> | --> | ||
| Line 120: | Line 115: | ||
;Option to produce a "protein structure gallery"" entry | ;Option to produce a "protein structure gallery"" entry | ||
| − | :Illustrating a research article well is a crucial skill that can significantly enhance the chances of it being accpted for publication. | + | :Illustrating a research article well is a crucial skill that can significantly enhance the chances of it being accpted for publication. ChimeraX provides great support for all sorts of visual and quantitative analysis, in this deliverable you showcase some of its options to highlight a biological fact, or illustrate the use of the ChimeraX program. Navigate to [http://steipe.biochemistry.utoronto.ca/abc/students/index.php/BIN-SX-Chimera_images the Instructions and Topics page on the Student Wiki] to view an example and choose a topic. |
:* Create a new page on the student Wiki as a subpage of your User Page. Develop your visual analysis there. | :* Create a new page on the student Wiki as a subpage of your User Page. Develop your visual analysis there. | ||
| − | :* When you are done with | + | :* When you are done with everything, submit the page via Quercus as described above. |
| − | |||
| − | |||
| + | {{Smallvspace}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Contents == | == Contents == | ||
| − | |||
| − | ==Molecular graphics: UCSF | + | ==Molecular graphics: UCSF ChimeraX== |
| − | To view molecular structures, we need a tool to visualize the three dimensional relationships of atoms. A ''molecular viewer'' is a program that takes 3D structure data and allows you to display and explore it. For a number of reasons, I use the UCSF | + | To view molecular structures, we need a tool to visualize the three dimensional relationships of atoms. A ''molecular viewer'' is a program that takes 3D structure data and allows you to display and explore it. For a number of reasons, I use the UCSF ChimeraX viewer for this course<ref>Previous versions of the course used [https://www.cgl.ucsf.edu/chimera/ UCSF Chimera] and some of the example code and images might still reflect that - don't be confused.</ref>: |
| − | # | + | # ChimeraX is open, and free for academia; |
# It creates very appealing graphics; | # It creates very appealing graphics; | ||
| − | # It is under ongoing development and is well maintained | + | # It is under ongoing development and is well maintained; |
| − | # It provides an array of useful utilities for structure analysis; | + | # There is a very helpful and professional [https://www.rbvi.ucsf.edu/chimerax/docs/contact.html help-list] available; |
| − | # besides an intuitive, menu driven interface, | + | # It provides an array of useful utilities for structure analysis; |
| + | # besides an intuitive, menu driven interface, ChimeraX can be scripted via its command line, or even programmed via its in-built python interpreter; | ||
| + | # and - a ''remote-control'' option allows to script ChimeraX display and analysis directly from R. | ||
| − | === | + | ===ChimeraX: First steps=== |
| + | |||
| + | {{Smallvspace}} | ||
{{task|1= | {{task|1= | ||
;Installation | ;Installation | ||
| − | # Access the [ | + | # Access the [https://www.rbvi.ucsf.edu/chimerax/ ChimeraX homepage] and navigate to the [https://www.rbvi.ucsf.edu/chimerax/download.html '''Download'''] section. |
| − | # Find the the newest version for your platform in the table and click on the file to download it. | + | # Find the the newest version for your platform in the table and click on the file to download it. (Previous versions for older operating systems are available.) |
| − | # Follow the instructions to install | + | # Follow the instructions to install ChimeraX. |
;First tutorial | ;First tutorial | ||
| − | The ''' | + | The '''ChimeraX''' User Guide site has a [https://www.rbvi.ucsf.edu/chimerax/tutorials.html '''set of associated tutorials''']. OPening them in the integrated viewer makes their links executable in the program. |
| − | * Work through | + | * Open ChimeraX and select '''Help''' ▶ '''Quick Start Guide'''. Work through the "Example Atomic-Structure Commands" section. |
| + | |||
| + | * Next, explore the 1BM8 structure: | ||
| + | ** In the ''Models'' pane (open it via the '''Tools''' Menu if it is not open), click on '''2bbv''' to select it, and click the '''Close''' button to remove the model. | ||
| + | ** Type <code>open 1bm8</code> in the commandline to load the 1BM8 structure from the PDB. | ||
| + | ** Ctrl+click on the first beta strand and press up-arrow to expand the selection to the entire strand. Select '''Actions''' ▶ '''Color''' ▶ '''Hot Pink''' to give the strand an interesting contrast. | ||
| + | ** Select '''Actions''' ▶ '''Atoms/Bonds''' ▶ '''Show''' to display side chains. Then Ctrl+click the background to deselect. | ||
| + | ** Ctrl-click either of the Tyrosine sidechains to select an atom, click up-arrow twice to select the entire residue. Then select '''Actions''' ▶ '''Labels''' ▶ '''Residues''' ▶ '''Name Combo'''. This displays the Chain, amino acid, and residue number. Deselect. | ||
| + | ** Type <code>color sequential #2/A palette blue-white-red</code> into the command line. <code>#2/A</code> mains: Model number two, chain A. You can find the model number in the ''Models'' pane, in the '''ID''' column. | ||
| + | ** Select '''Actions''' ▶ '''Atoms/Bonds''' ▶ '''Hide''' and ''Actions''' ▶ '''Labels''' ▶ '''Residues''' ▶ '''Off'''. Now issue a selection that spans the DNA recognition domain and shows the side chains: <code>show #2/a:49-74 target ab</code>. The first part selects, and the <code>target</code> command sends the selection to an action - in this case <c ode>ab</code>, "Atoms/Bonds". Also set the cartoon representation in this region off: <code>hide #2/a:49-74 cartoon</code>. Note that you can '''Edit''' ▶ '''Undo''' and '''Edit''' ▶ '''Redo''' commands. | ||
| + | ** Select '''Select''' ▶ '''Chemistry''' ▶ '''Element''' ▶ '''H''' and '''Actions''' ▶ '''Atoms/Bonds''' ▶ '''Hide''' to hide H-atoms. (Or type <code>hide H</code>.) | ||
| + | ** Select Nitrogen atoms and colour them cornflower blue in this region: <code>color #2/a:49-74@N* cornflower blue</code>. Again, deselect. | ||
| + | |||
| + | That should get you an idea to get started. To note: if you know what you are doing, the command line is always faster than the menu. The menu helps you to explore what you don't know yet. Every menu command is repeated with its command line equivalent in the ''Log'' pane, and every command is linked to its syntax-help. ChimeraX is very powerful, but it undoubtedly has a bit of a learning curve. However, the authors have done a great job to make learning easy. | ||
| + | |||
| + | * Quit ChimeraX, so we can restart it in a sane state. | ||
}} | }} | ||
| Line 168: | Line 175: | ||
{{Smallvspace}} | {{Smallvspace}} | ||
| − | Stereo viewing is easy to learn with a molecular viewer like | + | Stereo viewing is easy to learn with a molecular viewer like ChimeraX. |
| − | Being able to visualize and experience structure in 3D is an essential skill if you are | + | Being able to visualize and experience structure in 3D is an essential skill if you are even somewhat serious about understanding the molecules of molecular biology. This is not sufficiently realized in the field: many molecular biologists have never invested the small effort it takes to learn the skill, and they will tell you that it is not actually necessary, and you can get by regardless, after all they are doing just fine. Of course you are talking to a biased population – unless you have experienced and worked with stereo images, you won't understand how much you are actually missing. Unfortunately, over the years, this attitude has become more and more prevalent as the older generation of structural biologists are retiring, and we are now in a situation where the default viewer on the PDB website, Mol*, aparently does not even support split-screen stereo anymore. Incredible. We are training a whole generation of (structural) molecular biologists who have an inreasingly underdeveloped intuition about the spatial relationships of the 3D-objects they are working with. This means you. You are losing out. I have a strong opinion on this matter: '''Not supporting stereo representation of structures is at its core anti-intellectual. And it is Cargo Cult.''' You should rebel. |
| + | |||
| + | Once you have acquired the skill, you'll regret not having been taught earlier. Speak to people who use stereo vision: seeing molecules in 3D is like the difference between seeing a photograph of a place and actually being there. In 3D you can appreciate size, scale, distance, spatial relations all at a single glance. You can make perfect sense of partially hidden detail of overlapping clouds of atoms and bonds. And you can develop intuitions that are forever inaccessible to those who are only working with the structure's shadows. I insist: '''you can't <u>understand</u> structure unless you experience it in 3D'''. | ||
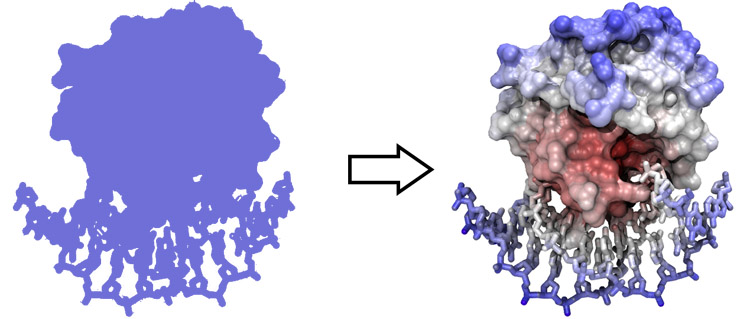
[[Image:2D_3D.jpg|frame|none|From 2D to ''pseudo'' 3D<br> | [[Image:2D_3D.jpg|frame|none|From 2D to ''pseudo'' 3D<br> | ||
| − | Images we see on screen or on paper are two-dimensional projections of three-dimensional objects. | + | Images we see on screen or on paper are two-dimensional projections, "shadows", of three-dimensional objects. |
]] | ]] | ||
| − | Even though hardware devices exist that | + | Even though hardware devices exist that support three-dimensional perception of computer graphics images, there is really no alternative to being able to fuse stereo pair images by just looking at them, without any device. ChimeraX is an excellent tool to practice stereo viewing and develop the skill. Stereo images consist of a left-eye and a right-eye view of the same object, with a slight rotation around the vertical axis (about 5 degrees). Your brain can accurately calculate depth from these two images, if they are presented to the right and left eye separately. This means you need to look at the two images and then fuse them into a single image - this happens when the left eye looks directly at the left image and the right eye at the right image. |
In this tutorial, I teach you a method to learn stereo viewing. The method is pretty foolproof - I have taught this many years in my classes with virtually 100% success rates. But I can only teach you the method – ''learning'' must be done by you. | In this tutorial, I teach you a method to learn stereo viewing. The method is pretty foolproof - I have taught this many years in my classes with virtually 100% success rates. But I can only teach you the method – ''learning'' must be done by you. | ||
| Line 185: | Line 194: | ||
<small>Some people find convergent (cross-eyed) stereo viewing easier to learn. I recommend the divergent (wall-eyed) viewing - not only because it is much more comfortable in my experience, but also because it is the default way in which stereo images in books and manuscripts are presented. The method explained below will '''only''' work for learning to view '''divergent''' stereo pairs.</small> | <small>Some people find convergent (cross-eyed) stereo viewing easier to learn. I recommend the divergent (wall-eyed) viewing - not only because it is much more comfortable in my experience, but also because it is the default way in which stereo images in books and manuscripts are presented. The method explained below will '''only''' work for learning to view '''divergent''' stereo pairs.</small> | ||
| − | + | {{Smallvspace}} | |
| − | |||
====Physiology==== | ====Physiology==== | ||
| Line 201: | Line 209: | ||
{{task|1= | {{task|1= | ||
| − | Here are step by step instructions of how to practice stereo viewing with | + | Here are step by step instructions of how to practice stereo viewing with ChimeraX. |
| + | |||
| + | *Load the Mbp1 APSES domain structure 1BM8 small protein into ChimeraX and display it as a simple backbone model. | ||
| + | **Start ChimeraX. A gallery of thumbnails of recently loaded structures appears. Click on 1bm8. | ||
| + | ** Type <code>color sequential /a palette cornflower:white:teal</code> to set an underlying colour gradient that allows to ditinguish N-terminal from C-terminal structure. | ||
| + | ** Type <code>cartoon hide</code> to hide the cartoon display. | ||
| + | ** Type <code>shape tube /a@CA radius 0.9 bandLength 4</code> to create a tube that smootly traces <tt>CA</tt> atoms, and is coloured according to the atoms it is tracing +- a <tt>bandLength</tt> distance. (Try setting a short <tt>bandLength</tt> for a striped appearance.) | ||
| + | ** Next click on the '''Graphics''' tab, just below the top frame of the window. | ||
| + | |||
| + | ** Select the '''Soft''' option in the '''Lighting''' menu bar at the top, to give good depth contrast and click on ''Shadow''' to produce more depth-cues. All of this will help initially establish the stereo-effect. | ||
| + | |||
| + | This sets up a simple scene that is suitable for practicing stereo viewing. Now we switch stereo on. | ||
| + | |||
| + | * Type <code>camera sbs</code> to switch on side-by-side ("walleye") stereo images. Close the ''Log'' and ''Models'' pane (click the ⓧ ). | ||
| + | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 216: | Line 230: | ||
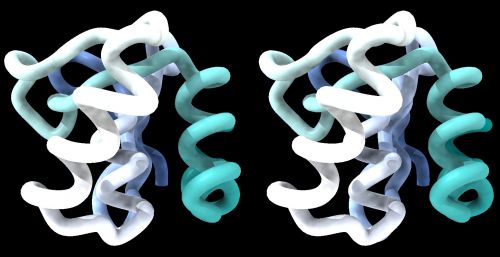
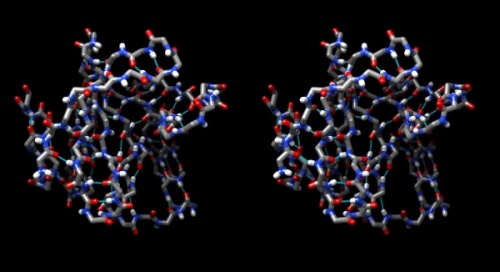
| − | {{stereo|1BM8_basic_stereo.jpg|'''1BM8: Mbp1 transcription factor APSES domain''' rendered as a | + | {{stereo|1BM8_basic_stereo.jpg|'''1BM8: Mbp1 transcription factor APSES domain''' rendered as a tube model, with depth-cueing applied and a colour-ramp emphasizing the fold from N-C terminus. The molecule is shown in '''wall-eye stereo''': the left-hand image is rotated correctly for the left eye. You should resize the window of your molecular viewer (ChimeraX) until ''equivalent'' points are about 15% less than your pupil separation apart.}} |
| − | *For the next step, you need to know how far your eyes are apart. Not just approximately, but pretty accurately. Measure the distance between your pupils in front of a mirror, or have soemone help you. | + | * For the next step, you need to know how far your eyes are apart. Not just approximately, but pretty accurately. Measure the distance between your pupils in front of a mirror, or have soemone help you. |
| − | *Resize the horizontal distance of the viewing window by dragging its lower right-hand corner. This changes the separation of the two views. | + | * Resize the horizontal distance of the viewing window by dragging its lower right-hand corner. This changes the separation of the two views. |
| − | **Resize the window so that two equivalent points on the protein are about 15% closer together on the screen than the pupils of your eyes are apart. Calculate, and measure eg. the separation between a protruding loop on the left and right image. Don't just guess, measure the distance, and adjust your on-screen scene to better than two or three millimetres of the correct separation. | + | ** Resize the window so that two equivalent points on the protein are about 15% closer together on the screen than the pupils of your eyes are apart. Calculate, and measure eg. the separation between a protruding loop on the left and right image. Don't just guess, measure the distance, and adjust your on-screen scene to better than two or three millimetres of the correct separation. |
| − | **Also, the images themselves should be small: about the size of a postage stamp. | + | ** Also, the images themselves should be small: about the size of a postage stamp. |
| + | |||
| + | * Orient the white helix to the front and type <code>rock</code>. (You can stop the rocking motion later by typing <code>stop</code>.) | ||
;Now follow these instructions exactly: | ;Now follow these instructions exactly: | ||
| − | *Touch your nose and forehead to the screen to get your eyes '''directly in front of the two images'''. Make sure you see the right image with your right eye, the left with your left eye. Of course, since you are so close, the images will be blurred. Nevertheless, you should see one solid, three dimensional shape in the centre, plus two peripheral images of the same view on the sides. <small>You see three copies of the same scene, but only the fused, overlapping centre scene appears three-dimensional; the other two become less noticeable as you practice more, your brain simply begins editing them out.</small> | + | * Touch your nose and forehead to the screen to get your eyes '''directly in front of the two images'''. Make sure you see the right image with your right eye, the left with your left eye. Of course, since you are so close, the images will be blurred. Nevertheless, you should see one solid, three dimensional shape in the centre, plus two peripheral images of the same view on the sides. <small>You see three copies of the same scene, but only the fused, overlapping centre scene appears three-dimensional; the other two become less noticeable as you practice more, your brain simply begins editing them out.</small> |
| − | *Without moving your head, resize the window slightly left and right until the centre image overlaps and "fuses". This way you find the ''exact'' distance for the images that works best for you. Slowly rotating the protein with the mouse helps generate the impression of a 3D object floating before you. Gaze at the image in the centre. | + | * Without moving your head, resize the window slightly left and right until the centre image overlaps and "fuses". This way you find the ''exact'' distance for the images that works best for you. Slowly rotating the protein with the mouse helps generate the impression of a 3D object floating before you. Gaze at the image in the centre. |
| − | *Spend some time with this. Don't worry that it is out of focus. Imagine that you are looking at something underwater with your eyes open. But make sure that you see '''one central fused image''' and it appears three-dimensional to you. <small>Don't continue unless you can achieve this. Ask for help if it doesn't work for you.</small> | + | * Spend some time with this. Don't worry that it is out of focus. Imagine that you are looking at something underwater with your eyes open. But make sure that you see '''one central fused image''' and it appears three-dimensional to you. <small>Don't continue unless you can achieve this. Ask for help if it doesn't work for you.</small> |
| − | *Once you see the | + | * Once you see the impression of depth is established, try to move your head backwards slowly, until the structure comes into focus. Do not voluntarily try to focus, since this will induce your eyes to converge and you will lose the 3D effect. You should be relaxed, and passively achieve this effect. Don't force it. After a short distance, you will probably lose the 3D effect. Once you loose the 3D effect, pause, close your eyes, then look somewhere else. Relax, take a deep breath and start over with your forehead on the screen. |
| − | *You will find that you will be able to hold the image for longer, then move your head further back, as you practice this. Give it time, you '''will''' be able to achieve focus on the | + | * You will find that you will be able to hold the image for longer, then move your head further back, as you practice this. Give it time, you '''will''' be able to achieve focus on the 3D object. |
;Now start a practice routine! | ;Now start a practice routine! | ||
| − | * Practice this procedure patiently, three or four times daily for perhaps 3 to 5 minutes. Between lectures is ideal. Keep your scene in | + | * Practice this procedure patiently, three or four times daily for perhaps 3 to 5 minutes. Between lectures is ideal. Keep your scene in ChimeraX open and go to it frequently. But stop, when your head feels funny. Don't force yourself. Better to practice more frequently for shorter periods. |
;Keep it up until you have mastered the skill. | ;Keep it up until you have mastered the skill. | ||
| Line 249: | Line 265: | ||
<div class="mw-collapsible mw-collapsed FAQ-box" data-expandtext="Notes for troubleshooting..." data-collapsetext="Collapse"> | <div class="mw-collapsible mw-collapsed FAQ-box" data-expandtext="Notes for troubleshooting..." data-collapsetext="Collapse"> | ||
| − | What could possibly go wrong? ... <small>Click to expand.</small> | + | What could possibly go wrong? ... <small>Click to expand.</small>▶ |
<div class="mw-collapsible-content" style="padding:10px;"> | <div class="mw-collapsible-content" style="padding:10px;"> | ||
---- | ---- | ||
| − | + | ;What ''should'' it look like? | |
| + | :What are you "supposed" to see? You see two images of an object and you see them with each eye, i.e. in principle there are four images. The two central images (image L as seen with the left eye, image R as seen with the right eye) should overlap in the middle; these two images fuse in your visual system to create '''one 3D image'''. The two peripheral images still remain; since you don't concentrate on them, your visual system will ''edit'' them out of consciousness as you gain experience. | ||
| − | + | ;How do I know I'm seeing the right thing? | |
| + | :A student of the 2020 class suggested to rotate the model so that the white helix is oriented to the side. That way it becomes easier to judge whether the image is properly fused (one white helix <small>or three, if you count the side-images</small>). or still split (two white helices <small>or four, if you count the side-images</small>). If the scene can't be superimposed, resize to reduce the separation. <small>(Thanks Xiang-Ling Li!)</small> | ||
| − | There are two situations which interfere with stereo viewing. One such situation is if the images presented to your eye are of '''unequal size'''. This can happen if you are using glasses with significantly different correction for each eye - the lenses then have different magnifications. The other situation is when equivalent points of the images are vertically '''misaligned''', i.e. one of the images is shifted up or down, or rotated. This can occur when your head is tilted relative to the image. Keep your head straight<ref>That's the same effect as when you are watching a 3D movie in the theatre, with polarized glasses: If you have weak posture or a cute neighbour and slouch to the side, your eyes become misaligned relative to the separated images on the screen, your visual system tries to compensate, but over time you get a headache. This is how stereo-haters are bred.</ref>. | + | ;What if I can't see stero in the first place, for physiological reasons? |
| + | :There are two situations which interfere with stereo viewing. One such situation is if the images presented to your eye are of '''unequal size'''. This can happen if you are using glasses with significantly different correction for each eye - the lenses then have different magnifications. The other situation is when equivalent points of the images are vertically '''misaligned''', i.e. one of the images is shifted up or down, or rotated. This can occur when your head is tilted relative to the image. Keep your head straight<ref>That's the same effect as when you are watching a 3D movie in the theatre, with polarized glasses: If you have weak posture or a cute neighbour and slouch to the side, your eyes become misaligned relative to the separated images on the screen, your visual system tries to compensate, but over time you get a headache. This is how stereo-haters are bred.</ref>. | ||
| − | Images that are difficult to see in 3D are images that are '''rendered differently''' for the left- and right view: non-aligned jagged edges, differing shadows or highlights disturb the stereo effect. To prevent this, try to apply visual effects judiciously. | + | :Images that are difficult to see in 3D are images that are '''rendered differently''' for the left- and right view: non-aligned jagged edges, differing shadows or highlights disturb the stereo effect. To prevent this, try to apply visual effects judiciously. ChimeraX is quite good with it's rendering but e.g. specular highlights in some molecular viewers are quite inconsistent between left and right. (If you are a programmer who writes stereo routines, remember to move the camera location, don't rotate the object because that will alter which parts of the model are shadowed!) |
| − | Of course, if you are practicing '''wall-eyed''' viewing ("side-by-side"), the so-called '''cross-eyed''' stereo images won't work for you. They look like the real thing, but the left-eye view is on the right-hand side and vice versa. You can tell that they are wrong when you achieve the image fusion because the 3-D effect seems to be all wrong. This is because the image becomes inverted in depth: near points appear far away and vice versa. If you are looking at a simple line drawing, you can't tell that this is happening. However if there are any secondary depth cues in the image, such as occlusions, shadows, highlights etc., they are in the wrong places, won't work to enhance the depth effect and just generally look weird. <small>Once you are comfortable with stereo viewing, you can try this deliberately by selecting cross-eyed display in | + | :Of course, if you are practicing '''wall-eyed''' viewing ("side-by-side"), the so-called '''cross-eyed''' stereo images won't work for you. They look like the real thing, but the left-eye view is on the right-hand side and vice versa. You can tell that they are wrong when you achieve the image fusion because the 3-D effect seems to be all wrong. This is because the image becomes inverted in depth: near points appear far away and vice versa. If you are looking at a simple line drawing, you can't tell that this is happening. However if there are any secondary depth cues in the image, such as occlusions, shadows, highlights etc., they are in the wrong places, won't work to enhance the depth effect and just generally look weird. <small>Once you are comfortable with stereo viewing, you can try this deliberately by selecting cross-eyed display in ChimeraX. Inverted space is very strange.</small> |
</div> | </div> | ||
| Line 277: | Line 296: | ||
In this series of tasks we will showcase some of the '''globally''' applied tools that help us study molecular structure. | In this series of tasks we will showcase some of the '''globally''' applied tools that help us study molecular structure. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| Line 303: | Line 304: | ||
{{task|1= | {{task|1= | ||
| − | # | + | # To reset all views and selections, choose '''Tools''' ▶ '''Models'''. Select the 1BM8 model (if it's still loaded from your stereo exercises) and the tube model and click the '''Close''' button to remove them. |
| − | # | + | # In the message bar at the bottom of the viewer window, click on the "lightning bolt" icon at the bottom (this is the "Quick Access Screen"). You should see a button labelled 1BM8 on the right. This is where you will find recent structures. Click <code>1BM8</code> to re-load it<ref>If you ever get to the Quick Access Screen by mistake, just click the lightning bolt icon again to return to the Viewer.</ref>. |
| − | + | # Click on the '''Molecule Display''' tab and the '''b-factor''' icon. The standard colouring scheme is blue - white - red, but you can define your own palette, e.g. from black via red to white, in a {{WP|Black_body_radiation|black-body spectrum}}: <code>color byattribute bfactor palette color byattribute bfactor palette black:black:gray:gray:red:orange:yellow:white</code>. | |
| − | # | + | # Type <code>cartoon hide</code> and <code>show atoms, bonds</code>. You will find that the core of the protein has low temperature factors, and the surface has a number of "glowingly" mobile sidechains and loops. |
| − | |||
| − | # | ||
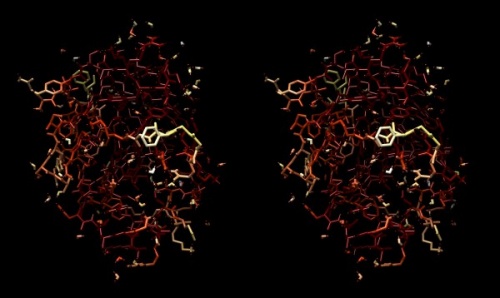
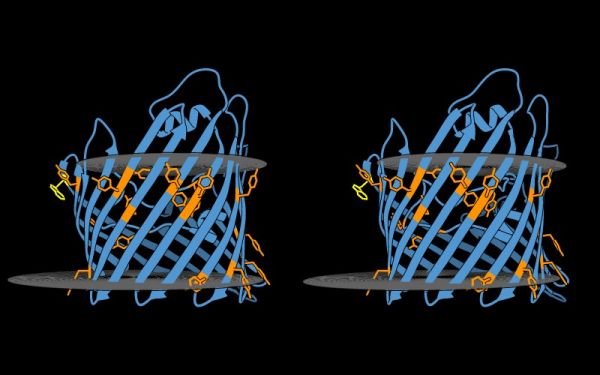
{{stereo|1BM8_thermal_stereo.jpg|'''Structure of the yeast transcription factor Mbp1 DNA binding domain (1BM8)''' coloured by B-factor (thermal factor). The protein bonds are shown in a "stick" model, coloured with a spectrum that emulates black-body radiation. Note that the interior of the protein is less mobile, some of the surface loops are highly mobile (or statically disordered, X-ray structures can't distinguish that) and the discretely bound water molecules that are visible in this high-resolution structure are generally more mobile than the residues they bind to. | {{stereo|1BM8_thermal_stereo.jpg|'''Structure of the yeast transcription factor Mbp1 DNA binding domain (1BM8)''' coloured by B-factor (thermal factor). The protein bonds are shown in a "stick" model, coloured with a spectrum that emulates black-body radiation. Note that the interior of the protein is less mobile, some of the surface loops are highly mobile (or statically disordered, X-ray structures can't distinguish that) and the discretely bound water molecules that are visible in this high-resolution structure are generally more mobile than the residues they bind to. | ||
| Line 319: | Line 318: | ||
====Electrostatics==== | ====Electrostatics==== | ||
| + | {{Smallvspace}} | ||
| + | {{task|1= | ||
| + | * Start by getting ChimeraX and your model in a fresh state. Choose '''File''' ▶ '''Close Session''' and click on the '''1bm8''' thumbnail to reload the structure. | ||
| + | * To visualize the electrostatic potential of the protein, mapped on the surface, first type <code>color cyan</code> for a vividly contrasting color and then <code> coulombic protein range -10,10</code>. | ||
| + | * Use '''Actions''' ▶ '''Surface''' ▶ '''Transparency''' ▶ '''30%''' to make the protein backbone somewhat visible. | ||
| + | * In the ''Graphics'' tab, set the background to '''White''', the lighting to '''Flat''', and the '''Silhouettes''' on. | ||
{{Smallvspace}} | {{Smallvspace}} | ||
| − | + | * Can you identify the putative DNA binding site from the electrostatic potential of the 1BM8 surface? | |
| − | + | ||
| − | + | {{Smallvspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
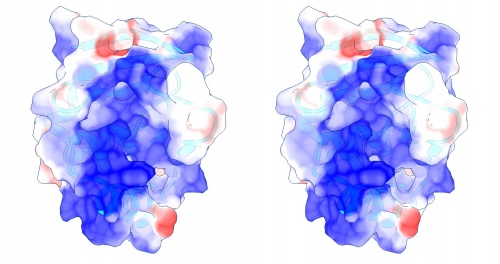
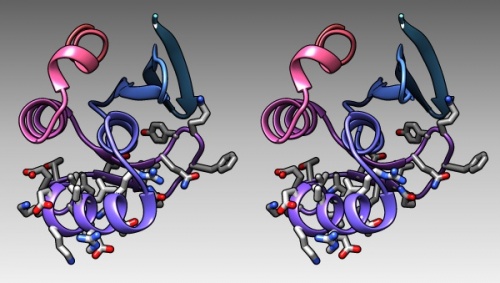
{{stereo|1BM8_coulomb_stereo.jpg|'''Coulomb (electrostatic) potential''' mapped to the solvent accessible surface of the yeast transcription factor Mbp1 DNA binding domain (1BM8). The protein backbone is visible through the transparent surface as a cartoon model, note the helix at the bottom of the structure. This helix has been suggested to play a role in forming the domain's DNA binding site and the positive (blue) electrostatic potential of the region is consistent with binding the negatively charged phosphate backbone of DNA. The other side of the domain has a negative (red) charge excess, which balances the molecule's electric charge overall, but also guides the protein-ligand interaction and supports faster on-rates. | {{stereo|1BM8_coulomb_stereo.jpg|'''Coulomb (electrostatic) potential''' mapped to the solvent accessible surface of the yeast transcription factor Mbp1 DNA binding domain (1BM8). The protein backbone is visible through the transparent surface as a cartoon model, note the helix at the bottom of the structure. This helix has been suggested to play a role in forming the domain's DNA binding site and the positive (blue) electrostatic potential of the region is consistent with binding the negatively charged phosphate backbone of DNA. The other side of the domain has a negative (red) charge excess, which balances the molecule's electric charge overall, but also guides the protein-ligand interaction and supports faster on-rates. | ||
}} | }} | ||
| − | |||
}} | }} | ||
| Line 344: | Line 344: | ||
{{task|1= | {{task|1= | ||
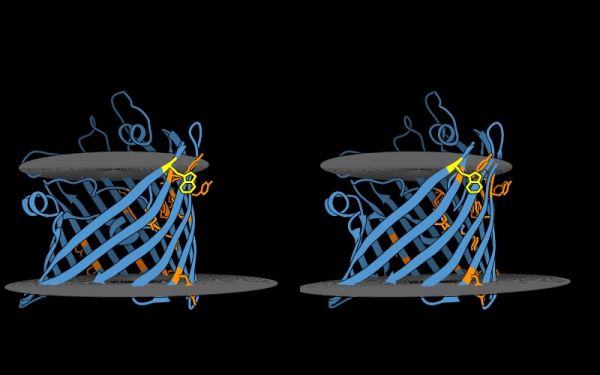
| − | + | * Hydrogen bonds encode the basic folding patterns of the protein. To visualize H-bonds, close and reload the model, hide the cartoon view, select | |
| − | + | '''Select''' ▶ '''Structure''' ▶ '''Backbone''' and '''Actions''' ▶ '''Atoms/Bonds''' ▶ '''Show Only'''. | |
| − | + | * Next, '''Actions''' ▶ '''Color''' ▶ '''Grey'''. | |
| + | * Use '''Tools''' ▶ '''Structure Analysis''' ▶ '''H-Bonds''' to open the H-bond dialogue window. | ||
| + | * Click on the colored square to select a nicer color, a light orange perhaps, for good contrast. | ||
| + | * To emphasize the role of H-bonds in determining the '''basic architecture''' of the protein, change '''Dashes''' to '''0''', increase '''Radius''' to '''0.1Å''', select '''Limit by selection'''and choose '''with both ends selected'''. Click on '''OK''', and deselect. | ||
| + | |||
| + | If the protein were an organism, you would now be looking at its skeleton. | ||
| Line 358: | Line 363: | ||
{{Vspace}} | {{Vspace}} | ||
| − | == | + | ===ChimeraX sequence interface=== |
{{Smallvspace}} | {{Smallvspace}} | ||
| − | In this task we will explore the sequence interface of | + | In this task we will explore the sequence interface of ChimeraX, use it to select specific parts of a molecule, and colour specific regions (or residues) of a molecule separately. |
| | ||
{{task|1= | {{task|1= | ||
| − | + | * Close and reload the 1BM8 model. | |
| − | # | + | |
| − | + | ||
| − | + | ||
| − | + | # Type <code>color sequential /a palette blue:cornflower:gray:white:gray:pink:red</code>. | |
| − | + | * Open the sequence tool: '''Tools''' ▶ '''Sequence''' ▶ '''Show Sequence Viewer''' and click on '''Show''' in the selection diaolgue. The sequence viewer appears as a panel.By default, coloured rectangles overlay the secondary structure elements of the sequence. | |
| − | + | * Hover the mouse over some residues and note that the sequence number and chain is shown as hovertext. | |
| − | + | * Click/drag one residue to select it. <small>(Simply a click wont work, you need to drag a little bit for the selection to catch on.)</small> Note that the residue gets a green overlay in the sequence window, and it also gets selected with a green border in the graphics window. | |
| − | + | * In the bottom of the sequence window, there are instructions how to select (multiple) regions. Clear the selection by Ctrl+click'ing into an empty spot of the viewer. Now select the region that encompasses the residues that have been reported to form the DNA binding subdomain: <code>KRTRILEKEVLKETHEKVQGGFGKYQ</code> (Taylor 2000). Show the side chains of these residues by typing <code>show sel atoms, bonds</code>. | |
| + | * Undisplay the Hydrogen atoms by typing <code>hide H</code>. Choose '''Actions''' ▶ '''Color''' ▶ '''By Element'''. Then add '''silhouettes'''. | ||
| Line 382: | Line 388: | ||
}} | }} | ||
| + | |||
{{Vspace}} | {{Vspace}} | ||
| − | === | + | ==Structure Image Gallery== |
| + | |||
| + | {{Smallvspace}} | ||
| + | |||
| + | <!-- TEMPLATE ======================================================= | ||
| + | ===TITLE=== | ||
| + | |||
| + | {{Smallvspace}} | ||
| + | |||
| + | ;Context | ||
| + | :Description | ||
| + | |||
| + | <div class="image-box"> | ||
| + | |||
| + | [[File:???.jpg|600px]] | ||
| + | |||
| + | Caption ... | ||
| + | |||
| + | </div> | ||
{{Smallvspace}} | {{Smallvspace}} | ||
| − | + | ---- | |
| − | + | <div class="mw-collapsible mw-collapsed" data-expandtext="Expand" data-collapsetext="Collapse" style="width:80%; "> | |
| − | + | ;Instructions for this image ... | |
| − | + | <div class="mw-collapsible-content" style="padding:10px;"> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | ||
| − | |||
| − | |||
| − | |||
| − | + | <div class="note" style=""><small>These instructions were written for "Chimera", the precursor to "ChimeraX", and are no longer accurate. '''Your''' entries need to be written for "ChimeraX"</small> | |
| + | </div> | ||
| + | Instructions ... | ||
| − | + | </div> | |
| + | </div> | ||
| + | <small>({{CC-BY-Small}} NNNN NNNNN <small>(with edits by Boris Steipe)</small>)</small> | ||
{{Vspace}} | {{Vspace}} | ||
| + | ====== END TEMPLATE ================================================= --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | ===1BM8 Worm Plot=== | ||
| + | |||
| + | {{Smallvspace}} | ||
| + | |||
| + | ;Context | ||
| + | :Protein structure representation is just that - a representation. We can add or subtract value in the images we create based on what we make meaningful in our representations. Use of colour is widely embraced, but there is a multitude of properties we may want to denote in our representations: amino acid identity, hydrophobicity, occupancy, b-factor, etc. What can we use in addition to colour to demonstrate the nature of proteins? | ||
| + | |||
| + | :Enter the sausage model. A kind of ribbon representation, the sausage (or "worm") varies in radius according to a specified protein property. | ||
| + | |||
| + | :Perhaps the first sausage model was created by John Kendrew 1957<ref>Chadarevian, Soraya De. Designs for Life: Molecular Biology after World War II. Cambridge University Press, 2011.</ref>, in fact when he submitted his model to the British Science Museum, it was the first formal model of a protein. Kendrew created a wiggly representation of myoglobin out of plasticine<ref>[http://collection.sciencemuseum.org.uk/objects/co13543/kendrews-original-model-of-the-myoglobin-molecule-molecular-models-proteins Kendrew's original model]</ref>, and this revealed a new understanding of how proteins occupy space. | ||
| + | |||
| + | :Today, we can use much more sophisticated methods to generate protein models, and ''in silico'' methods are constantly advancing<ref>[http://www.bonvinlab.org/news/Virtual-reality-meets-proteins/ VERY macro-molecules]</ref>. To demonstrate this, I've coloured 1BM8 (baker's yeast MBP1) according to the secondary structure and rendered a worm plot based on B-factors in yeast MBP1. This helps give an idea of how B-factors are distributed over different structural and functional components of this domain. | ||
| + | |||
| + | <div class="image-box"> | ||
| + | |||
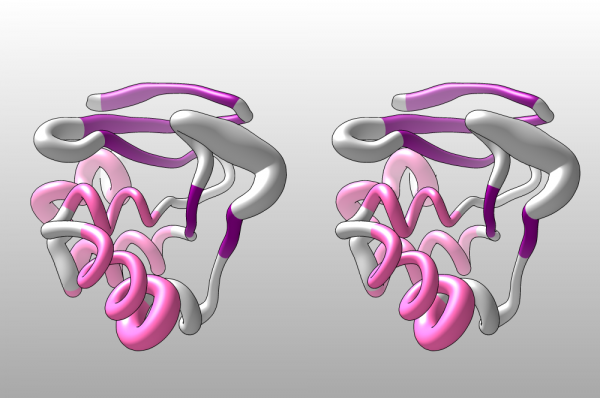
| + | [[File:CH-Worm_1BM8.png|600px]] | ||
| + | |||
| + | The <code>1BM8</code> structure, rendered as a worm plot. A larger radius corresponds to a higher B-factor; this indicates structural disorder. Alpha-helices are coloured pink, beta-strands are maroon. We clearly see that this transcription factor's DNA-binding helix and the adjacent "Wing", of DNA-contacting beta-strands both have unexpectedly high B-factors. This may indicate that these elements are not well integrated with the rest of the domain - which could point to dynamic disorder, i.e. flexibility to facilitate induced fit, or static disorder, arising from relatively high-energy conformations the domain needs to present for detailed DNA binding. Both point to a functional role of these disordered segements. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | {{Smallvspace}} | ||
| + | |||
| + | ---- | ||
| + | |||
| + | <div class="mw-collapsible mw-collapsed" data-expandtext="Expand" data-collapsetext="Collapse" style="width:80%; "> | ||
| + | ;Instructions for this image ... | ||
| + | <div class="mw-collapsible-content" style="padding:10px;"> | ||
| + | |||
| + | <div class="note" style=""><small>These instructions were written for "Chimera", the precursor to "ChimeraX", and are no longer accurate. '''Your''' entries need to be written for "ChimeraX"</small> | ||
| + | </div> | ||
| + | These instructions make use of both Chimera's command line and menu options. Menu items are '''bolded''' and <code>code formatted</code> text should be directly typed or pasted into the Chimera command line | ||
| + | |||
| + | *Open Chimera and choose '''Favorites''' > '''Command Line''' | ||
| + | * In the command line, enter: <code>open 1BM8</code> | ||
| + | |||
| + | You will see a small protein with four helices and 5 beta strands. | ||
| + | |||
| + | * Enter: <code>preset apply pub 1</code> | ||
| + | *Enter: <code>color hot pink,r helix; color dark magenta,r strand; color grey,r coil</code>and colour the protein by secondary structure | ||
| + | * Enter: <code>back grad dark grey,white rgb</code> to change the background | ||
| + | * Choose '''Tools''' > '''Structure Analysis''' > "Render by Attribute" | ||
| + | |||
| + | The "Render/Select by Attribute window will pop up | ||
| + | |||
| + | * From the drop down "Attributes of" menu, select "residues" | ||
| + | * From the drop down "Attribute" menu, select "average" > "b-factor" | ||
| + | |||
| + | How attribute values are mapped to visual representations can be selected in three tabs: Colors, Radii, and Worms. For '''colors''', values will be mapped to a divergent spectrum. This is useful e.g. to visualize B-factors, or conservation scores. '''Radii''' map values to sphere radii of atoms. This is useful e.g. to visualize the uncertainty in atomic positions. Average, per-residue properties can be mapped to the thickness of '''worms''' - a tube representation of the chain-backbone. Here we map B-factors to worms. | ||
| + | |||
| + | * In the '''Worms''' tab, choose Worm-style smooth. Then click '''OK'''. <small>(Or explore the other options, clicking '''Apply''' to view the effect.)</small> | ||
| + | |||
| + | |||
| + | View the complex in wall-eyed stereo and adjust parameters: | ||
| + | * Choose '''Tools''' >'''Viewing Controls''' >"Camera" | ||
| + | * Select: | ||
| + | **camera mode: wall-eye stereo | ||
| + | **projection: perspective | ||
| + | **units: centimetres | ||
| + | **eye separation: 6 (measure distance between your pupils) | ||
| + | **distance to screen: 50 (edit to your normal distance to screen is) | ||
| + | |||
| + | Improve the visualization: | ||
| + | |||
| + | * Choose '''Tools''' > '''Viewing Controls''' > '''Effects''' and turn shadows off | ||
| + | * Turn depth cueing and silhouettes on | ||
| + | * Orient and scale the complex to emphasize what you are trying to demonstrate. Make sure no part of the molecule is accidentally clipped by the viewing window. Make the window quite large, and resize/rescale to minimize unused space at the top and bottom. | ||
| + | |||
| + | Save the image. | ||
| + | |||
| + | ;ChimeraX | ||
| + | :While ChimeraX does not yet support Worm plots natively (as of November 2020), you can export the scene from Chimera as a "{{WP|COLLADA}}" <tt>[.dae]</tt> file, and re-import this to ChimeraX. Open the molecule in Chimera as described above and define your scene '''without''' changing scale or orientation. Use the options under '''File''' > '''Export Scene ...''' to export. Then open your molecule in ChimeraX, again '''without''' changing the orientation, and use '''File''' > '''Open''' to import the <tt>.dae</tt> file. You can further customize the shape by applying ''surface'' properties such as colour, transparency etc. to it. <small>(Thanks to [https://plato.cgl.ucsf.edu/pipermail/chimerax-users/2020-October/001613.html Elaine Meng] for pointing out the option to use COLLADA files in this way.)</small> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | <small>({{CC-BY-Small}} Caitlin Harrigan <small>(with edits by Boris Steipe)</small>)</small> | ||
{{Vspace}} | {{Vspace}} | ||
| + | ===Catalytic triad as convergent evolution=== | ||
| + | |||
| + | {{Smallvspace}} | ||
| + | |||
| + | ;Context | ||
| + | :Subtilisin and Trypsin are proteases, i.e. enzymes that hydrolyze peptide bonds in proteins. X-ray crystallography experiments have found that both Trypsin and Subtilisin have catalytic sites that are characterized by Aspartic acid, Histidine, and Serine in very similar positions relative to each other. <ref>{{#pmid: 10512718}}</ref><ref>{{#pmid: 4508127}}</ref><ref>{{#pmid: 5160720}}</ref> In an example of convergent evolution, despite very different overall structures, both enzymes have independently evolved the same "catalytic triad". A catalytic triad cleaves amide and ester bonds by having the electrophilic oxygen atoms of serine form bonds with the nucleophilic carbons of the carbonyls in amide and esters. Histidine then donates the hydrogen to the nitrogen or oxygen of the amide or ester bonds, respectively, effectively breaking the bond <ref>{{#pmid: 9787641}}</ref>. The structures depicted in the images below clearly display the catalytic triads of both enzymes in very similar positions. However the global architecture of the protein is entirely different. Subtilisin has many alpha-helices (dark green) while trypsin, has more beta strands (light green) and fewer alpha helices and unstructured connecting strands (grey). | ||
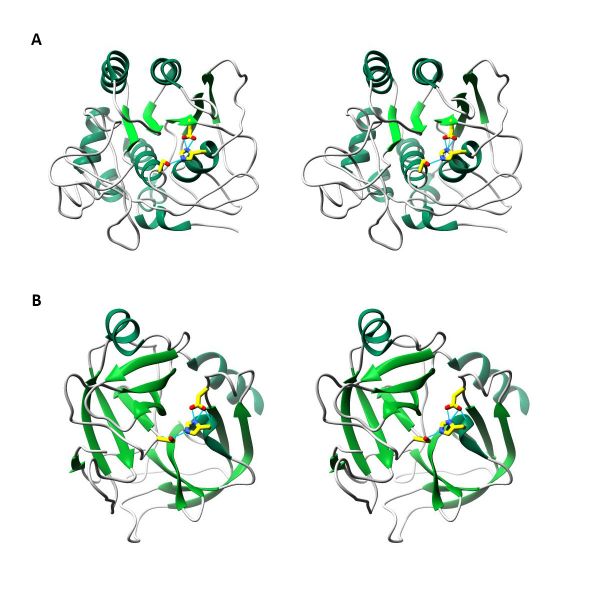
| − | = | + | <div class="image-box"> |
| − | |||
| − | |||
| − | |||
| − | + | [[File:SY-SubtilisinTrypsin.jpg|600px]] | |
| + | Wall-eyed stereo images of structures of Subtilisin ('''A''') and Trypsin ('''B''') with secondary structures coloured in shades of green and side chains of catalytic triad coloured in bright yellow and by atom (oxygen in red, nitrogen in blue). Hydrogen bonds that exist between the catalytic triads are shown in light blue lines. The domains were rotated and scaled to bring the catalytic triad into the same orientation. | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Smallvspace}} | |
| − | + | ---- | |
| − | |||
| − | |||
| − | |||
| + | <div class="mw-collapsible mw-collapsed" data-expandtext="Expand" data-collapsetext="Collapse" style="width:80%; "> | ||
| + | ;Instructions for this image ... | ||
| + | <div class="mw-collapsible-content" style="padding:10px;"> | ||
| + | |||
| + | <div class="note" style=""><small>These instructions were written for "Chimera", the precursor to "ChimeraX", and are no longer accurate. '''Your''' entries need to be written for "ChimeraX"</small> | ||
</div> | </div> | ||
| − | + | Structures of Subtilisin and Trypsin obtained from [http://www.rcsb.org/pdb/home/home.do| Protein Data Bank], PBD ID 1SBT<ref>{{#pmid: 5160720}}</ref> and 1C9P<ref>{{#pmid: 10512718}}</ref> respectively. Only chain A is displayed for Trypsin as chain B is not trypsin. These structures are selected over other subtilisin and trypsin structures available because they exhibit the classic catalytic triad. Images were generated in Chimera and arranged in photoshop. | |
| − | + | ;Image generation (in Chimera and photoshop) | |
| + | : Display in ribbon only, color it gray. | ||
| + | : For trypsin, hide chain b, hide solvents. | ||
| + | : Display side chain for catalytic triads: | ||
| + | <pre> | ||
| + | # Subtilisin catalytic triad: D32, H64, S221 | ||
| + | disp :32, 64, 221 | ||
| − | --> | + | # Trypsin catalytic triad: H57, D102, S195 |
| + | disp :57, 102, 195 | ||
| + | </pre> | ||
| + | : Colour in yellow, then color in heteroatoms: | ||
| + | <pre> | ||
| + | color yellow :32, 64, 221 | ||
| + | color byhet :32, 64, 221 | ||
| + | </pre> | ||
| + | : Favourites > Model Panel > attributes > stick scale, set to 2 | ||
| + | : Display hydrogen bonds that only exist between selected atoms | ||
| + | <pre> | ||
| + | select :32, 64, 221 | ||
| + | hbonds selRestrict both | ||
| + | </pre> | ||
| + | : Tools > Depiction > Colour Secondary Structure. Set the colours to your liking and apply. | ||
| + | : Rotate around so that the triads are in the same configuration for both subtilisin and trypsin. | ||
| + | : Save image as jpeg, use as appears on screen, set width to 1200 pixel. | ||
| + | : In Photoshop: Open both images, resize canvas, display grid, paste both images into the same canvas and use it to align the two different images. | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <small>({{CC-BY-Small}} Suan Chin Yeo <small>(with edits by Boris Steipe)</small>)</small> | ||
{{Vspace}} | {{Vspace}} | ||
| + | ==='''Y''' and '''W''' Delimit Transmembrane Domains=== | ||
| + | {{Smallvspace}} | ||
| − | {{ | + | ;Context |
| + | :Integral membrane proteins generally have a high concentration of tryptophan relative to proteins that are not anchored to the membrane bilayer <ref>{{#pmid:1409540}}</ref>. Tryptophan residues, along with tyrosine, cluster at protein's interface region between the lipophilic bilayer and the aqueous environment of cells <ref>{{#pmid:18550546}}</ref>. Since tryptophan and tyrosine are hydrophobic amino acids, the clustering of such residues into a "belt" may serve to anchor the protein to the bilayer <ref>Sigma-Aldrich. (n.d.) Sigma-Aldrich Amino Acids Reference Chart. Retrieved December 2, 2017 from https://www.sigmaaldrich.com/life-science/metabolomics/learning-center/amino-acid-reference-chart.html</ref>. | ||
| + | |||
| + | <div class="image-box"> | ||
| + | |||
| + | [[File:JS-2POR-1.jpg|600px]] | ||
| + | |||
| + | Tryptophan (yellow) and Tyrosine (orange) are highlighted in atom form within the porin protein structure (PDB: <code>2POR</code>). The entire protein is depicted as embedded in the membrane bilayer, with gray planes representing the membrane boundaries <ref>{{#pmid:11274469}}</ref>. Inclusion of the membrane reveals that tryptophan and tyrosine residues are restricted to the region within the membrane bilayer, thus appearing as a band along the transmembrane region. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | {{Smallvspace}} | ||
| + | |||
| + | <div class="image-box"> | ||
| + | |||
| + | [[File:JS-2POR-2.jpg|600px]] | ||
| + | |||
| + | Rotation of the porin structure reveals that tryptophan and tyrosine do not create a continuous band along the transmembrane region. There is a lack of tryptophan and tyrosine residues on the side of porin in which the protein interacts with other porin proteins to create the homotrimeric membrane channel <ref>{{#pmid:8142898}}</ref>. | ||
| + | </div> | ||
| − | + | {{Smallvspace}} | |
---- | ---- | ||
| + | |||
| + | <div class="mw-collapsible mw-collapsed" data-expandtext="Expand" data-collapsetext="Collapse" style="width:80%; "> | ||
| + | ;Instructions for this image ... | ||
| + | <div class="mw-collapsible-content" style="padding:10px;"> | ||
| + | |||
| + | <div class="note" style=""><small>These instructions were written for "Chimera", the precursor to "ChimeraX", and are no longer accurate. '''Your''' entries need to be written for "ChimeraX"</small> | ||
| + | </div> | ||
| + | I*Open Chimera and on the Rapid Acces screen, click the '''Fetch...''' button to open the '''Fetch Structure by ID ...''' window. | ||
| + | *Enter the PDB ID <code>2POR</code> and click '''Fetch...''' | ||
| + | |||
| + | You will see a beta barrel protein. To display the membrane protein embedded in a bilayer defined by two planes: | ||
| + | *Choose '''Favorites → Side View''' to adjust the viewing window, ensuring the entire protein is visible. | ||
| + | * Choose '''Favorites → Command Line'''. | ||
| + | * In the command line, enter: | ||
| + | <pre> | ||
| + | select :15,148 | ||
| + | </pre> | ||
| + | * Choose '''Tools → Structure Analysis → Axes/Planes/Centroids'''. | ||
| + | * In the '''Structure Measurements''' window, click on '''Define plane...''' button and enter the following parameters: | ||
| + | ** Plane name: Top membrane | ||
| + | ** Replace existing plane: false | ||
| + | ** Color: gray, and reduce the opacity | ||
| + | ** Set disk size to enclose atom projections: true (default) | ||
| + | ** Extra radius (padding): 0.0 (default) | ||
| + | ** Disk thickness 0.1 (default) | ||
| + | * Click '''Apply''' to show the plane. | ||
| + | * In command line, enter line by line: | ||
| + | <pre> | ||
| + | ~select | ||
| + | select :65,195,271,300 | ||
| + | </pre> | ||
| + | * Change the following parameters in the '''Define Plane''' window: | ||
| + | ** Plane name: Bottom membrane. | ||
| + | * Click '''Apply''' to show the plane. | ||
| + | |||
| + | Note that experimental data of membrane spanning B-strands were utilized to define and create the membrane planes in this image <ref>{{#pmid:11274469}}</ref>. | ||
| + | |||
| + | Highlight the tryptophan and tyrosine residues: | ||
| + | * In command line, enter line by line: | ||
| + | <pre> | ||
| + | ~select | ||
| + | color steel blue | ||
| + | select :tyr,trp | ||
| + | show sel | ||
| + | select :tyr | ||
| + | color orange sel | ||
| + | select :trp | ||
| + | color yellow sel | ||
| + | ~sel | ||
| + | </pre> | ||
| + | |||
| + | View the protein in wall-eye stereo: | ||
| + | * Choose '''Tools → Viewing Controls → Camera''' and enter the following parameters: | ||
| + | **Camera mode: wall-eye stereo | ||
| + | **Projection: perspective | ||
| + | **Units: centimetres | ||
| + | **Eye separation: 7.0 (set to personal eye separation measurement) | ||
| + | **Distance to screen: 50 (set to personal distance to screen) | ||
| + | |||
| + | Save the image: | ||
| + | * Choose '''File → Save image'''. | ||
| + | * Select '''JPEG''' file type, '''4X4''' Supersample, and '''wall-eye stereo pair''' image types. | ||
| + | * Set Image width to 800px. | ||
| + | * Uncheck '''Maintain current aspect ratio'''. | ||
| + | * Change '''Image height''' to 500px. | ||
| + | * Click '''Save'''. | ||
| + | |||
| + | Create another image with an alternative view of <code>2por</code>: | ||
| + | * Rotate the protein by dragging the protein on the screen. | ||
| + | * Save the image with the same parameters as outlined above. | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <small>({{CC-BY-Small}} Janeane Santos <small>(with edits by Boris Steipe)</small>)</small> | ||
{{Vspace}} | {{Vspace}} | ||
| − | < | + | == Further reading, links and resources == |
| + | {{#pmid:32881101}} | ||
| + | * [https://www.rbvi.ucsf.edu/chimerax/ '''UCSF ChimeraX home page'''] | ||
| + | <small>This sounds as if one could possible drive Chimera from R scripts via the <code>reticulate::</code> Python interface: | ||
| + | {{#pmid:29340616}} | ||
| + | * [http://www.rcsb.org/pdb/static.do?p=software/software_links/molecular_graphics.html Molecular Graphics Software Links]– a collection of links at the PDB. | ||
| + | |||
| − | + | == Notes == | |
| + | <references /> | ||
{{Vspace}} | {{Vspace}} | ||
| + | |||
<div class="about"> | <div class="about"> | ||
| Line 479: | Line 704: | ||
:2017-08-05 | :2017-08-05 | ||
<b>Modified:</b><br /> | <b>Modified:</b><br /> | ||
| − | : | + | :2020-10-29 |
<b>Version:</b><br /> | <b>Version:</b><br /> | ||
| − | : | + | :2.4 |
<b>Version history:</b><br /> | <b>Version history:</b><br /> | ||
| + | *2.4 Explain the use of COLLADA files for worm plots | ||
| + | *2.3 Added two more Structure Image Examples | ||
| + | *2.2 Clarify: all deliverables with ChimeraX only; add more hints for Stereo Vision troubleshooting; add first entry for Structure Image Gallery | ||
| + | *2.1 Edit policy update | ||
| + | *2.0 Major rewrite: moving from Chimera to ChimeraX | ||
| + | *1.2 Remove assumptions about preferences | ||
| + | *1.1 Charge mapping needs to calculate the surface first | ||
*1.0 First live version | *1.0 First live version | ||
*0.1 First stub - 2016 material combined | *0.1 First stub - 2016 material combined | ||
</div> | </div> | ||
| − | |||
| − | |||
{{CC-BY}} | {{CC-BY}} | ||
| + | [[Category:ABC-units]] | ||
| + | {{EVAL}} | ||
| + | {{LIVE}} | ||
| + | {{EVAL}} | ||
</div> | </div> | ||
<!-- [END] --> | <!-- [END] --> | ||
Latest revision as of 08:22, 29 October 2020
UCSF ChimeraX: Structure Visualization and Analysis
(UCSF ChimeraX; Structure visualization; Structure analysis)
Abstract:
This unit introduces the molecular viewer UCSF ChimeraX, starts you off on a routine to practice stereo viewing of molecular models, and teaches how to use ChimeraX to solve a number of common visualization and analysis tasks.
|
Objectives:
|
Outcomes:
|
Deliverables:
Prerequisites:
This unit builds on material covered in the following prerequisite units:
Contents
Evaluation
This learning unit can be evaluated for a maximum of 5 marks. If you want to submit tasks for this unit for credit you have the following options. If you have any questions about these options, discuss on the Discussion Board.
- Create a new page on the student Wiki as a subpage of your User Page.
- Put all of your writing to submit on this one page.
- When you are done with everything, go to the Quercus Assignments page and open the first Learning Unit that you have not submitted yet. Paste the URL of your Wiki page into the form, and click on Submit Assignment.
Your link can be submitted only once and not edited. But you may change your Wiki page at any time. However only the last version before the due date will be marked. All later edits will be silently ignored.
- Short Report option
- 1. Create a new page on the student Wiki as a subpage of your User Page.
- 2. Alpha helices orient peptide carbonyls parallel to their axis and since the C=O bond is polarized with an excess of electrons near the carboyl oxygen, this creates an induced dipole along the length of the helix, with a positive partial charge towards the beginning of the helix, and a negative partial charge towards its end. To compensate, we often find negativeley charged residues at the beginning of the helix and positively charged residues at the end - to stabilize the helix. However, for DNA binding, electrostatic complementarity would make us assume that alpha helices in DNA binding proteins are generally oriented with the (+)-dipole close to the DNA phosphates, and that there are no compensating charged residues at the termini. Write a short analysis on one of the three following topics - A, B, or C. The goal of this short report is to connect visualization to biology.
- A - Helix dipoles: Nucleosome core
- A.1 Open ChimeraX and load the structure 5XF3.
- A.2 Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings.
- A.3 Write up, illustrate, and interpret your findings in a short report.
- B - Helix dipole: Transcription factor
- B.1 Open ChimeraX and load the structure 4Y60.
- B.2 Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings.
- B.3 Write up, illustrate and interpret your findings in a short report.
- C - Helix dipole: Transcription factor
- C.1 Open ChimeraX and load the structure 4ZSF.
- C.2 Study the complex with respect to this assumption. Produce 2 or more informative stereo-images that illustrate your findings.
- C.3 Write up, illustrate and interpret your findings in a short report.
- 3. When you are done with everything, submit the page via Quercus as described above.
- Option to produce a "protein structure gallery"" entry
- Illustrating a research article well is a crucial skill that can significantly enhance the chances of it being accpted for publication. ChimeraX provides great support for all sorts of visual and quantitative analysis, in this deliverable you showcase some of its options to highlight a biological fact, or illustrate the use of the ChimeraX program. Navigate to the Instructions and Topics page on the Student Wiki to view an example and choose a topic.
- Create a new page on the student Wiki as a subpage of your User Page. Develop your visual analysis there.
- When you are done with everything, submit the page via Quercus as described above.
Contents
Molecular graphics: UCSF ChimeraX
To view molecular structures, we need a tool to visualize the three dimensional relationships of atoms. A molecular viewer is a program that takes 3D structure data and allows you to display and explore it. For a number of reasons, I use the UCSF ChimeraX viewer for this course[1]:
- ChimeraX is open, and free for academia;
- It creates very appealing graphics;
- It is under ongoing development and is well maintained;
- There is a very helpful and professional help-list available;
- It provides an array of useful utilities for structure analysis;
- besides an intuitive, menu driven interface, ChimeraX can be scripted via its command line, or even programmed via its in-built python interpreter;
- and - a remote-control option allows to script ChimeraX display and analysis directly from R.
ChimeraX: First steps
Task:
- Installation
- Access the ChimeraX homepage and navigate to the Download section.
- Find the the newest version for your platform in the table and click on the file to download it. (Previous versions for older operating systems are available.)
- Follow the instructions to install ChimeraX.
- First tutorial
The ChimeraX User Guide site has a set of associated tutorials. OPening them in the integrated viewer makes their links executable in the program.
- Open ChimeraX and select Help ▶ Quick Start Guide. Work through the "Example Atomic-Structure Commands" section.
- Next, explore the 1BM8 structure:
- In the Models pane (open it via the Tools Menu if it is not open), click on 2bbv to select it, and click the Close button to remove the model.
- Type
open 1bm8in the commandline to load the 1BM8 structure from the PDB. - Ctrl+click on the first beta strand and press up-arrow to expand the selection to the entire strand. Select Actions ▶ Color ▶ Hot Pink to give the strand an interesting contrast.
- Select Actions ▶ Atoms/Bonds ▶ Show to display side chains. Then Ctrl+click the background to deselect.
- Ctrl-click either of the Tyrosine sidechains to select an atom, click up-arrow twice to select the entire residue. Then select Actions ▶ Labels ▶ Residues ▶ Name Combo. This displays the Chain, amino acid, and residue number. Deselect.
- Type
color sequential #2/A palette blue-white-redinto the command line.#2/Amains: Model number two, chain A. You can find the model number in the Models pane, in the ID column. - Select Actions' ▶ Atoms/Bonds ▶ Hide and Actions ▶ Labels ▶ Residues ▶ Off. Now issue a selection that spans the DNA recognition domain and shows the side chains:
show #2/a:49-74 target ab. The first part selects, and thetargetcommand sends the selection to an action - in this case <c ode>ab, "Atoms/Bonds". Also set the cartoon representation in this region off:hide #2/a:49-74 cartoon. Note that you can Edit ▶ Undo and Edit ▶ Redo commands. - Select Select ▶ Chemistry ▶ Element ▶ H and Actions ▶ Atoms/Bonds ▶ Hide to hide H-atoms. (Or type
hide H.) - Select Nitrogen atoms and colour them cornflower blue in this region:
color #2/a:49-74@N* cornflower blue. Again, deselect.
That should get you an idea to get started. To note: if you know what you are doing, the command line is always faster than the menu. The menu helps you to explore what you don't know yet. Every menu command is repeated with its command line equivalent in the Log pane, and every command is linked to its syntax-help. ChimeraX is very powerful, but it undoubtedly has a bit of a learning curve. However, the authors have done a great job to make learning easy.
- Quit ChimeraX, so we can restart it in a sane state.
Stereo Viewing
Stereo viewing is easy to learn with a molecular viewer like ChimeraX.
Being able to visualize and experience structure in 3D is an essential skill if you are even somewhat serious about understanding the molecules of molecular biology. This is not sufficiently realized in the field: many molecular biologists have never invested the small effort it takes to learn the skill, and they will tell you that it is not actually necessary, and you can get by regardless, after all they are doing just fine. Of course you are talking to a biased population – unless you have experienced and worked with stereo images, you won't understand how much you are actually missing. Unfortunately, over the years, this attitude has become more and more prevalent as the older generation of structural biologists are retiring, and we are now in a situation where the default viewer on the PDB website, Mol*, aparently does not even support split-screen stereo anymore. Incredible. We are training a whole generation of (structural) molecular biologists who have an inreasingly underdeveloped intuition about the spatial relationships of the 3D-objects they are working with. This means you. You are losing out. I have a strong opinion on this matter: Not supporting stereo representation of structures is at its core anti-intellectual. And it is Cargo Cult. You should rebel.
Once you have acquired the skill, you'll regret not having been taught earlier. Speak to people who use stereo vision: seeing molecules in 3D is like the difference between seeing a photograph of a place and actually being there. In 3D you can appreciate size, scale, distance, spatial relations all at a single glance. You can make perfect sense of partially hidden detail of overlapping clouds of atoms and bonds. And you can develop intuitions that are forever inaccessible to those who are only working with the structure's shadows. I insist: you can't understand structure unless you experience it in 3D.
Even though hardware devices exist that support three-dimensional perception of computer graphics images, there is really no alternative to being able to fuse stereo pair images by just looking at them, without any device. ChimeraX is an excellent tool to practice stereo viewing and develop the skill. Stereo images consist of a left-eye and a right-eye view of the same object, with a slight rotation around the vertical axis (about 5 degrees). Your brain can accurately calculate depth from these two images, if they are presented to the right and left eye separately. This means you need to look at the two images and then fuse them into a single image - this happens when the left eye looks directly at the left image and the right eye at the right image.
In this tutorial, I teach you a method to learn stereo viewing. The method is pretty foolproof - I have taught this many years in my classes with virtually 100% success rates. But I can only teach you the method – learning must be done by you.
Some people find convergent (cross-eyed) stereo viewing easier to learn. I recommend the divergent (wall-eyed) viewing - not only because it is much more comfortable in my experience, but also because it is the default way in which stereo images in books and manuscripts are presented. The method explained below will only work for learning to view divergent stereo pairs.
Physiology
In order to visually fuse stereo image pairs, you need to override a vision reflex that couples divergence and focusing. This needs to be practiced for a while. Usually 5 to 10 minutes of practice twice daily for a week should be quite sufficient. It is not as hard as learning to ride a bicycle, but you need to practice regularly for some time, maybe 10 or 20 sessions of 3 to 5 minute over a period of a week or two. Once you have acquired the skill, it is really very comfortable and can be done effortlessly and for extended periods. You will enter a new world of molecular wonders!
Practice
Task:
Here are step by step instructions of how to practice stereo viewing with ChimeraX.
- Load the Mbp1 APSES domain structure 1BM8 small protein into ChimeraX and display it as a simple backbone model.
- Start ChimeraX. A gallery of thumbnails of recently loaded structures appears. Click on 1bm8.
- Type
color sequential /a palette cornflower:white:tealto set an underlying colour gradient that allows to ditinguish N-terminal from C-terminal structure. - Type
cartoon hideto hide the cartoon display. - Type
shape tube /a@CA radius 0.9 bandLength 4to create a tube that smootly traces CA atoms, and is coloured according to the atoms it is tracing +- a bandLength distance. (Try setting a short bandLength for a striped appearance.) - Next click on the Graphics tab, just below the top frame of the window.
- Select the Soft' option in the Lighting menu bar at the top, to give good depth contrast and click on Shadow to produce more depth-cues. All of this will help initially establish the stereo-effect.
This sets up a simple scene that is suitable for practicing stereo viewing. Now we switch stereo on.
- Type
camera sbsto switch on side-by-side ("walleye") stereo images. Close the Log and Models pane (click the ⓧ ).
The model could look something like this:
1BM8: Mbp1 transcription factor APSES domain rendered as a tube model, with depth-cueing applied and a colour-ramp emphasizing the fold from N-C terminus. The molecule is shown in wall-eye stereo: the left-hand image is rotated correctly for the left eye. You should resize the window of your molecular viewer (ChimeraX) until equivalent points are about 15% less than your pupil separation apart.
- For the next step, you need to know how far your eyes are apart. Not just approximately, but pretty accurately. Measure the distance between your pupils in front of a mirror, or have soemone help you.
- Resize the horizontal distance of the viewing window by dragging its lower right-hand corner. This changes the separation of the two views.
- Resize the window so that two equivalent points on the protein are about 15% closer together on the screen than the pupils of your eyes are apart. Calculate, and measure eg. the separation between a protruding loop on the left and right image. Don't just guess, measure the distance, and adjust your on-screen scene to better than two or three millimetres of the correct separation.
- Also, the images themselves should be small: about the size of a postage stamp.
- Orient the white helix to the front and type
rock. (You can stop the rocking motion later by typingstop.)
- Now follow these instructions exactly
- Touch your nose and forehead to the screen to get your eyes directly in front of the two images. Make sure you see the right image with your right eye, the left with your left eye. Of course, since you are so close, the images will be blurred. Nevertheless, you should see one solid, three dimensional shape in the centre, plus two peripheral images of the same view on the sides. You see three copies of the same scene, but only the fused, overlapping centre scene appears three-dimensional; the other two become less noticeable as you practice more, your brain simply begins editing them out.
- Without moving your head, resize the window slightly left and right until the centre image overlaps and "fuses". This way you find the exact distance for the images that works best for you. Slowly rotating the protein with the mouse helps generate the impression of a 3D object floating before you. Gaze at the image in the centre.
- Spend some time with this. Don't worry that it is out of focus. Imagine that you are looking at something underwater with your eyes open. But make sure that you see one central fused image and it appears three-dimensional to you. Don't continue unless you can achieve this. Ask for help if it doesn't work for you.
- Once you see the impression of depth is established, try to move your head backwards slowly, until the structure comes into focus. Do not voluntarily try to focus, since this will induce your eyes to converge and you will lose the 3D effect. You should be relaxed, and passively achieve this effect. Don't force it. After a short distance, you will probably lose the 3D effect. Once you loose the 3D effect, pause, close your eyes, then look somewhere else. Relax, take a deep breath and start over with your forehead on the screen.
- You will find that you will be able to hold the image for longer, then move your head further back, as you practice this. Give it time, you will be able to achieve focus on the 3D object.
- Now start a practice routine!
- Practice this procedure patiently, three or four times daily for perhaps 3 to 5 minutes. Between lectures is ideal. Keep your scene in ChimeraX open and go to it frequently. But stop, when your head feels funny. Don't force yourself. Better to practice more frequently for shorter periods.
- Keep it up until you have mastered the skill.
After time and with practice, it will become easier and easier to achieve the effect. Also you will become quite independent of the distance of equivalent points, thus you can increase the viewer window size and take advantage of the increased resolution.
It might take you about a week or ten days to master this, with regular training it will become very easy. And, the best thing is, you do not easily forget this skill. It is like riding a bicycle, equalizing pressure in your ears while scuba diving, or circular breathing to play the didgeridoo: once you teach your body what to do, it remembers.
What could possibly go wrong? ... Click to expand.▶
- What should it look like?
- What are you "supposed" to see? You see two images of an object and you see them with each eye, i.e. in principle there are four images. The two central images (image L as seen with the left eye, image R as seen with the right eye) should overlap in the middle; these two images fuse in your visual system to create one 3D image. The two peripheral images still remain; since you don't concentrate on them, your visual system will edit them out of consciousness as you gain experience.
- How do I know I'm seeing the right thing?
- A student of the 2020 class suggested to rotate the model so that the white helix is oriented to the side. That way it becomes easier to judge whether the image is properly fused (one white helix or three, if you count the side-images). or still split (two white helices or four, if you count the side-images). If the scene can't be superimposed, resize to reduce the separation. (Thanks Xiang-Ling Li!)
- What if I can't see stero in the first place, for physiological reasons?
- There are two situations which interfere with stereo viewing. One such situation is if the images presented to your eye are of unequal size. This can happen if you are using glasses with significantly different correction for each eye - the lenses then have different magnifications. The other situation is when equivalent points of the images are vertically misaligned, i.e. one of the images is shifted up or down, or rotated. This can occur when your head is tilted relative to the image. Keep your head straight[2].
- Images that are difficult to see in 3D are images that are rendered differently for the left- and right view: non-aligned jagged edges, differing shadows or highlights disturb the stereo effect. To prevent this, try to apply visual effects judiciously. ChimeraX is quite good with it's rendering but e.g. specular highlights in some molecular viewers are quite inconsistent between left and right. (If you are a programmer who writes stereo routines, remember to move the camera location, don't rotate the object because that will alter which parts of the model are shadowed!)
- Of course, if you are practicing wall-eyed viewing ("side-by-side"), the so-called cross-eyed stereo images won't work for you. They look like the real thing, but the left-eye view is on the right-hand side and vice versa. You can tell that they are wrong when you achieve the image fusion because the 3-D effect seems to be all wrong. This is because the image becomes inverted in depth: near points appear far away and vice versa. If you are looking at a simple line drawing, you can't tell that this is happening. However if there are any secondary depth cues in the image, such as occlusions, shadows, highlights etc., they are in the wrong places, won't work to enhance the depth effect and just generally look weird. Once you are comfortable with stereo viewing, you can try this deliberately by selecting cross-eyed display in ChimeraX. Inverted space is very strange.
Some more examples of stereo scenes can be found on the Stereo vision practice page on this Wiki.
Global properties
In this series of tasks we will showcase some of the globally applied tools that help us study molecular structure.
B-factors
Task:
- To reset all views and selections, choose Tools ▶ Models. Select the 1BM8 model (if it's still loaded from your stereo exercises) and the tube model and click the Close button to remove them.
- In the message bar at the bottom of the viewer window, click on the "lightning bolt" icon at the bottom (this is the "Quick Access Screen"). You should see a button labelled 1BM8 on the right. This is where you will find recent structures. Click
1BM8to re-load it[3]. - Click on the Molecule Display tab and the b-factor icon. The standard colouring scheme is blue - white - red, but you can define your own palette, e.g. from black via red to white, in a black-body spectrum:
color byattribute bfactor palette color byattribute bfactor palette black:black:gray:gray:red:orange:yellow:white. - Type
cartoon hideandshow atoms, bonds. You will find that the core of the protein has low temperature factors, and the surface has a number of "glowingly" mobile sidechains and loops.
Structure of the yeast transcription factor Mbp1 DNA binding domain (1BM8) coloured by B-factor (thermal factor). The protein bonds are shown in a "stick" model, coloured with a spectrum that emulates black-body radiation. Note that the interior of the protein is less mobile, some of the surface loops are highly mobile (or statically disordered, X-ray structures can't distinguish that) and the discretely bound water molecules that are visible in this high-resolution structure are generally more mobile than the residues they bind to.
Electrostatics
Task:
- Start by getting ChimeraX and your model in a fresh state. Choose File ▶ Close Session and click on the 1bm8 thumbnail to reload the structure.
- To visualize the electrostatic potential of the protein, mapped on the surface, first type
color cyanfor a vividly contrasting color and thencoulombic protein range -10,10. - Use Actions ▶ Surface ▶ Transparency ▶ 30% to make the protein backbone somewhat visible.
- In the Graphics tab, set the background to White, the lighting to Flat, and the Silhouettes on.
- Can you identify the putative DNA binding site from the electrostatic potential of the 1BM8 surface?
Coulomb (electrostatic) potential mapped to the solvent accessible surface of the yeast transcription factor Mbp1 DNA binding domain (1BM8). The protein backbone is visible through the transparent surface as a cartoon model, note the helix at the bottom of the structure. This helix has been suggested to play a role in forming the domain's DNA binding site and the positive (blue) electrostatic potential of the region is consistent with binding the negatively charged phosphate backbone of DNA. The other side of the domain has a negative (red) charge excess, which balances the molecule's electric charge overall, but also guides the protein-ligand interaction and supports faster on-rates.
Hydrogen bonds
Task:
- Hydrogen bonds encode the basic folding patterns of the protein. To visualize H-bonds, close and reload the model, hide the cartoon view, select
Select ▶ Structure ▶ Backbone and Actions ▶ Atoms/Bonds ▶ Show Only.
- Next, Actions ▶ Color ▶ Grey.
- Use Tools ▶ Structure Analysis ▶ H-Bonds to open the H-bond dialogue window.
- Click on the colored square to select a nicer color, a light orange perhaps, for good contrast.
- To emphasize the role of H-bonds in determining the basic architecture of the protein, change Dashes to 0, increase Radius to 0.1Å, select Limit by selectionand choose with both ends selected. Click on OK, and deselect.
If the protein were an organism, you would now be looking at its skeleton.
ChimeraX sequence interface
In this task we will explore the sequence interface of ChimeraX, use it to select specific parts of a molecule, and colour specific regions (or residues) of a molecule separately.
Task:
- Close and reload the 1BM8 model.
- Type
color sequential /a palette blue:cornflower:gray:white:gray:pink:red.
- Open the sequence tool: Tools ▶ Sequence ▶ Show Sequence Viewer and click on Show in the selection diaolgue. The sequence viewer appears as a panel.By default, coloured rectangles overlay the secondary structure elements of the sequence.
- Hover the mouse over some residues and note that the sequence number and chain is shown as hovertext.
- Click/drag one residue to select it. (Simply a click wont work, you need to drag a little bit for the selection to catch on.) Note that the residue gets a green overlay in the sequence window, and it also gets selected with a green border in the graphics window.
- In the bottom of the sequence window, there are instructions how to select (multiple) regions. Clear the selection by Ctrl+click'ing into an empty spot of the viewer. Now select the region that encompasses the residues that have been reported to form the DNA binding subdomain:
KRTRILEKEVLKETHEKVQGGFGKYQ(Taylor 2000). Show the side chains of these residues by typingshow sel atoms, bonds. - Undisplay the Hydrogen atoms by typing
hide H. Choose Actions ▶ Color ▶ By Element. Then add silhouettes.
Structure Image Gallery
1BM8 Worm Plot
- Context
- Protein structure representation is just that - a representation. We can add or subtract value in the images we create based on what we make meaningful in our representations. Use of colour is widely embraced, but there is a multitude of properties we may want to denote in our representations: amino acid identity, hydrophobicity, occupancy, b-factor, etc. What can we use in addition to colour to demonstrate the nature of proteins?
- Enter the sausage model. A kind of ribbon representation, the sausage (or "worm") varies in radius according to a specified protein property.
- Perhaps the first sausage model was created by John Kendrew 1957[4], in fact when he submitted his model to the British Science Museum, it was the first formal model of a protein. Kendrew created a wiggly representation of myoglobin out of plasticine[5], and this revealed a new understanding of how proteins occupy space.
- Today, we can use much more sophisticated methods to generate protein models, and in silico methods are constantly advancing[6]. To demonstrate this, I've coloured 1BM8 (baker's yeast MBP1) according to the secondary structure and rendered a worm plot based on B-factors in yeast MBP1. This helps give an idea of how B-factors are distributed over different structural and functional components of this domain.
The 1BM8 structure, rendered as a worm plot. A larger radius corresponds to a higher B-factor; this indicates structural disorder. Alpha-helices are coloured pink, beta-strands are maroon. We clearly see that this transcription factor's DNA-binding helix and the adjacent "Wing", of DNA-contacting beta-strands both have unexpectedly high B-factors. This may indicate that these elements are not well integrated with the rest of the domain - which could point to dynamic disorder, i.e. flexibility to facilitate induced fit, or static disorder, arising from relatively high-energy conformations the domain needs to present for detailed DNA binding. Both point to a functional role of these disordered segements.
- Instructions for this image ...
These instructions make use of both Chimera's command line and menu options. Menu items are bolded and code formatted text should be directly typed or pasted into the Chimera command line
- Open Chimera and choose Favorites > Command Line
- In the command line, enter:
open 1BM8
You will see a small protein with four helices and 5 beta strands.
- Enter:
preset apply pub 1 - Enter:
color hot pink,r helix; color dark magenta,r strand; color grey,r coiland colour the protein by secondary structure - Enter:
back grad dark grey,white rgbto change the background - Choose Tools > Structure Analysis > "Render by Attribute"
The "Render/Select by Attribute window will pop up
- From the drop down "Attributes of" menu, select "residues"
- From the drop down "Attribute" menu, select "average" > "b-factor"
How attribute values are mapped to visual representations can be selected in three tabs: Colors, Radii, and Worms. For colors, values will be mapped to a divergent spectrum. This is useful e.g. to visualize B-factors, or conservation scores. Radii map values to sphere radii of atoms. This is useful e.g. to visualize the uncertainty in atomic positions. Average, per-residue properties can be mapped to the thickness of worms - a tube representation of the chain-backbone. Here we map B-factors to worms.
- In the Worms tab, choose Worm-style smooth. Then click OK. (Or explore the other options, clicking Apply to view the effect.)
View the complex in wall-eyed stereo and adjust parameters:
- Choose Tools >Viewing Controls >"Camera"
- Select:
- camera mode: wall-eye stereo
- projection: perspective
- units: centimetres
- eye separation: 6 (measure distance between your pupils)
- distance to screen: 50 (edit to your normal distance to screen is)
Improve the visualization:
- Choose Tools > Viewing Controls > Effects and turn shadows off
- Turn depth cueing and silhouettes on
- Orient and scale the complex to emphasize what you are trying to demonstrate. Make sure no part of the molecule is accidentally clipped by the viewing window. Make the window quite large, and resize/rescale to minimize unused space at the top and bottom.
Save the image.
- ChimeraX
- While ChimeraX does not yet support Worm plots natively (as of November 2020), you can export the scene from Chimera as a "COLLADA" [.dae] file, and re-import this to ChimeraX. Open the molecule in Chimera as described above and define your scene without changing scale or orientation. Use the options under File > Export Scene ... to export. Then open your molecule in ChimeraX, again without changing the orientation, and use File > Open to import the .dae file. You can further customize the shape by applying surface properties such as colour, transparency etc. to it. (Thanks to Elaine Meng for pointing out the option to use COLLADA files in this way.)
((CC BY) Caitlin Harrigan (with edits by Boris Steipe))
Catalytic triad as convergent evolution
- Context
- Subtilisin and Trypsin are proteases, i.e. enzymes that hydrolyze peptide bonds in proteins. X-ray crystallography experiments have found that both Trypsin and Subtilisin have catalytic sites that are characterized by Aspartic acid, Histidine, and Serine in very similar positions relative to each other. [7][8][9] In an example of convergent evolution, despite very different overall structures, both enzymes have independently evolved the same "catalytic triad". A catalytic triad cleaves amide and ester bonds by having the electrophilic oxygen atoms of serine form bonds with the nucleophilic carbons of the carbonyls in amide and esters. Histidine then donates the hydrogen to the nitrogen or oxygen of the amide or ester bonds, respectively, effectively breaking the bond [10]. The structures depicted in the images below clearly display the catalytic triads of both enzymes in very similar positions. However the global architecture of the protein is entirely different. Subtilisin has many alpha-helices (dark green) while trypsin, has more beta strands (light green) and fewer alpha helices and unstructured connecting strands (grey).
Wall-eyed stereo images of structures of Subtilisin (A) and Trypsin (B) with secondary structures coloured in shades of green and side chains of catalytic triad coloured in bright yellow and by atom (oxygen in red, nitrogen in blue). Hydrogen bonds that exist between the catalytic triads are shown in light blue lines. The domains were rotated and scaled to bring the catalytic triad into the same orientation.
- Instructions for this image ...
Structures of Subtilisin and Trypsin obtained from Protein Data Bank, PBD ID 1SBT[11] and 1C9P[12] respectively. Only chain A is displayed for Trypsin as chain B is not trypsin. These structures are selected over other subtilisin and trypsin structures available because they exhibit the classic catalytic triad. Images were generated in Chimera and arranged in photoshop.
- Image generation (in Chimera and photoshop)
- Display in ribbon only, color it gray.
- For trypsin, hide chain b, hide solvents.
- Display side chain for catalytic triads:
# Subtilisin catalytic triad: D32, H64, S221 disp :32, 64, 221 # Trypsin catalytic triad: H57, D102, S195 disp :57, 102, 195
- Colour in yellow, then color in heteroatoms:
color yellow :32, 64, 221 color byhet :32, 64, 221
- Favourites > Model Panel > attributes > stick scale, set to 2
- Display hydrogen bonds that only exist between selected atoms
select :32, 64, 221 hbonds selRestrict both
- Tools > Depiction > Colour Secondary Structure. Set the colours to your liking and apply.
- Rotate around so that the triads are in the same configuration for both subtilisin and trypsin.
- Save image as jpeg, use as appears on screen, set width to 1200 pixel.
- In Photoshop: Open both images, resize canvas, display grid, paste both images into the same canvas and use it to align the two different images.
((CC BY) Suan Chin Yeo (with edits by Boris Steipe))
Y and W Delimit Transmembrane Domains
- Context
- Integral membrane proteins generally have a high concentration of tryptophan relative to proteins that are not anchored to the membrane bilayer [13]. Tryptophan residues, along with tyrosine, cluster at protein's interface region between the lipophilic bilayer and the aqueous environment of cells [14]. Since tryptophan and tyrosine are hydrophobic amino acids, the clustering of such residues into a "belt" may serve to anchor the protein to the bilayer [15].
Tryptophan (yellow) and Tyrosine (orange) are highlighted in atom form within the porin protein structure (PDB: 2POR). The entire protein is depicted as embedded in the membrane bilayer, with gray planes representing the membrane boundaries [16]. Inclusion of the membrane reveals that tryptophan and tyrosine residues are restricted to the region within the membrane bilayer, thus appearing as a band along the transmembrane region.
Rotation of the porin structure reveals that tryptophan and tyrosine do not create a continuous band along the transmembrane region. There is a lack of tryptophan and tyrosine residues on the side of porin in which the protein interacts with other porin proteins to create the homotrimeric membrane channel [17].
- Instructions for this image ...
I*Open Chimera and on the Rapid Acces screen, click the Fetch... button to open the Fetch Structure by ID ... window.
- Enter the PDB ID
2PORand click Fetch...
You will see a beta barrel protein. To display the membrane protein embedded in a bilayer defined by two planes:
- Choose Favorites → Side View to adjust the viewing window, ensuring the entire protein is visible.
- Choose Favorites → Command Line.
- In the command line, enter:
select :15,148
- Choose Tools → Structure Analysis → Axes/Planes/Centroids.
- In the Structure Measurements window, click on Define plane... button and enter the following parameters:
- Plane name: Top membrane
- Replace existing plane: false
- Color: gray, and reduce the opacity
- Set disk size to enclose atom projections: true (default)
- Extra radius (padding): 0.0 (default)
- Disk thickness 0.1 (default)
- Click Apply to show the plane.
- In command line, enter line by line:
~select select :65,195,271,300
- Change the following parameters in the Define Plane window:
- Plane name: Bottom membrane.
- Click Apply to show the plane.
Note that experimental data of membrane spanning B-strands were utilized to define and create the membrane planes in this image [18].
Highlight the tryptophan and tyrosine residues:
- In command line, enter line by line:
~select color steel blue select :tyr,trp show sel select :tyr color orange sel select :trp color yellow sel ~sel
View the protein in wall-eye stereo:
- Choose Tools → Viewing Controls → Camera and enter the following parameters:
- Camera mode: wall-eye stereo
- Projection: perspective
- Units: centimetres
- Eye separation: 7.0 (set to personal eye separation measurement)
- Distance to screen: 50 (set to personal distance to screen)
Save the image:
- Choose File → Save image.
- Select JPEG file type, 4X4 Supersample, and wall-eye stereo pair image types.
- Set Image width to 800px.
- Uncheck Maintain current aspect ratio.
- Change Image height to 500px.
- Click Save.
Create another image with an alternative view of 2por:
- Rotate the protein by dragging the protein on the screen.
- Save the image with the same parameters as outlined above.
((CC BY) Janeane Santos (with edits by Boris Steipe))
Further reading, links and resources
| Pettersen et al. (2020) UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci . (pmid: 32881101) |
|
[ PubMed ] [ DOI ] UCSF ChimeraX is the next-generation interactive visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX brings (i) significant performance and graphics enhancements; (ii) new implementations of Chimera's most highly used tools, many with further improvements; (iii) several entirely new analysis features; (iv) support for new areas such as virtual reality, light-sheet microscopy, and medical imaging data; (v) major ease-of-use advances, including toolbars with icons to perform actions with a single click, basic "undo" capabilities, and more logical and consistent commands; and (vi) an app store for researchers to contribute new tools. ChimeraX includes full user documentation and is free for noncommercial use, with downloads available for Windows, Linux, and macOS from https://www.rbvi.ucsf.edu/chimerax. This article is protected by copyright. All rights reserved. |
This sounds as if one could possible drive Chimera from R scripts via the reticulate:: Python interface:
| Rodríguez-Guerra Pedregal & Maréchal (2018) PyChimera: use UCSF Chimera modules in any Python 2.7 project. Bioinformatics 34:1784-1785. (pmid: 29340616) |
|
[ PubMed ] [ DOI ] Motivation: UCSF Chimera is a powerful visualization tool remarkably present in the computational chemistry and structural biology communities. Built on a C++ core wrapped under a Python 2.7 environment, one could expect to easily import UCSF Chimera's arsenal of resources in custom scripts or software projects. Nonetheless, this is not readily possible if the script is not executed within UCSF Chimera due to the isolation of the platform. UCSF ChimeraX, successor to the original Chimera, partially solves the problem but yet major upgrades need to be undergone so that this updated version can offer all UCSF Chimera features. Results: PyChimera has been developed to overcome these limitations and provide access to the UCSF Chimera codebase from any Python 2.7 interpreter, including interactive programming with tools like IPython and Jupyter Notebooks, making it easier to use with additional third-party software. Availability and implementation: PyChimera is LGPL-licensed and available at https://github.com/insilichem/pychimera. Contact: jaime.rodriguezguerra@uab.cat or jeandidier.marechal@uab.cat. Supplementary information: Supplementary data are available at Bioinformatics online. |
- Molecular Graphics Software Links– a collection of links at the PDB.
Notes
- ↑ Previous versions of the course used UCSF Chimera and some of the example code and images might still reflect that - don't be confused.
- ↑ That's the same effect as when you are watching a 3D movie in the theatre, with polarized glasses: If you have weak posture or a cute neighbour and slouch to the side, your eyes become misaligned relative to the separated images on the screen, your visual system tries to compensate, but over time you get a headache. This is how stereo-haters are bred.
- ↑ If you ever get to the Quick Access Screen by mistake, just click the lightning bolt icon again to return to the Viewer.
- ↑ Chadarevian, Soraya De. Designs for Life: Molecular Biology after World War II. Cambridge University Press, 2011.
- ↑ Kendrew's original model
- ↑ VERY macro-molecules
- ↑
Rester et al. (1999) Structure of the complex of the antistasin-type inhibitor bdellastasin with trypsin and modelling of the bdellastasin-microplasmin system. Structure Of The Complex Of The Antistasin-type Inhibitor Bdellastasin With Trypsin And Modelling Of The Bdellastasin-microplasmin System 293:93-106. (pmid: 10512718) [ PubMed ] [ DOI ] The serine proteinase plasmin is, together with tissue-type plasminogen activator (tPA) and urokinase-type plasminogen activator (uPA), involved in the dissolution of blood clots in a fibrin-dependent manner. Moreover, plasmin plays a key role in a variety of other activation cascades such as the activation of metalloproteinases, and has also been implicated in wound healing, pathogen invasion, cancer invasion and metastasis. The leech-derived (Hirudo medicinalis) antistasin-type inhibitor bdellastasin represents a specific inhibitor of trypsin and plasmin and thus offers a unique opportunity to evaluate the concept of plasmin inhibition. The complexes formed between bdellastasin and bovine as well as porcine beta-trypsin have been crystallised in a monoclinic and a tetragonal crystal form, containing six molecules and one molecule per asymmetric unit, respectively. Both structures have been solved and refined to 3.3 A and 2.8 A resolution. Bdellastasin turns out to have an antistasin-like fold exhibiting a bis-domainal structure like the tissue kallikrein inhibitor hirustasin. The interaction between bdellastasin and trypsin is restricted to the C-terminal subdomain of bdellastasin, particularly to its primary binding loop, comprising residues Asp30-Glu38. The reactive site of bdellastasin differs from other antistasin-type inhibitors of trypsin-like proteinases, exhibiting a lysine residue instead of an arginine residue at P1. A model of the bdellastasin-microplasmin complex has been created based on the X-ray structures. Our modelling studies indicate that both trypsin and microplasmin recognise bdellastasin by interactions which are characteristic for canonically binding proteinase inhibitors. On the basis of our three-dimensional structures, and in comparison with the tissue-kallikrein-bound and free hirustasin and the antistasin structures, we postulate that the binding of the inhibitors toward trypsin and plasmin is accompanied by a switch of the primary binding loop segment P5-P3. Moreover, in the factor Xa inhibitor antistasin, the core of the molecule would prevent an equivalent rotation of the P3 residue, making exosite interactions of antistasin with factor Xa imperative. Furthermore, Arg32 of antistasin would clash with Arg175 of plasmin, thus impairing a favourable antistasin-plasmin interaction and explaining its specificity.
- ↑
Drenth et al. (1972) A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo. A Comparison Of The Three-dimensional Structures Of Subtilisin BPN' And Subtilisin Novo 36:107-16. (pmid: 4508127) - ↑
Alden et al. (1971) Atomic coordinates for subtilisin BPN' (or Novo). Atomic Coordinates For Subtilisin BPN' (or Novo) 45:337-44. (pmid: 5160720) - ↑
Dodson & Wlodawer (1998) Catalytic triads and their relatives. Catalytic Triads And Their Relatives 23:347-52. (pmid: 9787641) [ PubMed ] [ DOI ] Interactions among the residues in the serine protease Asp-His-Ser catalytic triad, in the special environment of the enzyme-substrate complex, activate the nucleophilic potential of the seryl O gamma. In the subtilisin and trypsin families, the composition and arrangement of the catalytic triad do not vary significantly. However, the mechanisms of action of many other hydrolytic enzymes, which target a wide range of substrates, involve nucleophilic attack by a serine (or threonine) residue. Review of these enzymes shows that the acid-base-ser/thr pattern of catalytic residues is generally conserved, although the individual acids and bases can vary. The variations in sequence and organization illustrate the adaptability shown by proteins in generating catalytic stereochemistry on different main-chain frameworks.
- ↑
Alden et al. (1971) Atomic coordinates for subtilisin BPN' (or Novo). Atomic Coordinates For Subtilisin BPN' (or Novo) 45:337-44. (pmid: 5160720) - ↑
Rester et al. (1999) Structure of the complex of the antistasin-type inhibitor bdellastasin with trypsin and modelling of the bdellastasin-microplasmin system. Structure Of The Complex Of The Antistasin-type Inhibitor Bdellastasin With Trypsin And Modelling Of The Bdellastasin-microplasmin System 293:93-106. (pmid: 10512718) [ PubMed ] [ DOI ] The serine proteinase plasmin is, together with tissue-type plasminogen activator (tPA) and urokinase-type plasminogen activator (uPA), involved in the dissolution of blood clots in a fibrin-dependent manner. Moreover, plasmin plays a key role in a variety of other activation cascades such as the activation of metalloproteinases, and has also been implicated in wound healing, pathogen invasion, cancer invasion and metastasis. The leech-derived (Hirudo medicinalis) antistasin-type inhibitor bdellastasin represents a specific inhibitor of trypsin and plasmin and thus offers a unique opportunity to evaluate the concept of plasmin inhibition. The complexes formed between bdellastasin and bovine as well as porcine beta-trypsin have been crystallised in a monoclinic and a tetragonal crystal form, containing six molecules and one molecule per asymmetric unit, respectively. Both structures have been solved and refined to 3.3 A and 2.8 A resolution. Bdellastasin turns out to have an antistasin-like fold exhibiting a bis-domainal structure like the tissue kallikrein inhibitor hirustasin. The interaction between bdellastasin and trypsin is restricted to the C-terminal subdomain of bdellastasin, particularly to its primary binding loop, comprising residues Asp30-Glu38. The reactive site of bdellastasin differs from other antistasin-type inhibitors of trypsin-like proteinases, exhibiting a lysine residue instead of an arginine residue at P1. A model of the bdellastasin-microplasmin complex has been created based on the X-ray structures. Our modelling studies indicate that both trypsin and microplasmin recognise bdellastasin by interactions which are characteristic for canonically binding proteinase inhibitors. On the basis of our three-dimensional structures, and in comparison with the tissue-kallikrein-bound and free hirustasin and the antistasin structures, we postulate that the binding of the inhibitors toward trypsin and plasmin is accompanied by a switch of the primary binding loop segment P5-P3. Moreover, in the factor Xa inhibitor antistasin, the core of the molecule would prevent an equivalent rotation of the P3 residue, making exosite interactions of antistasin with factor Xa imperative. Furthermore, Arg32 of antistasin would clash with Arg175 of plasmin, thus impairing a favourable antistasin-plasmin interaction and explaining its specificity.
- ↑
Schiffer et al. (1992) The functions of tryptophan residues in membrane proteins. The Functions Of Tryptophan Residues In Membrane Proteins 5:213-4. (pmid: 1409540) [ PubMed ] [ DOI ] Membrane proteins have a significantly higher Trp content than do soluble proteins. This is especially true for the M and L subunits of the photosynthetic reaction center from purple bacteria. The Trp residues are not uniformly distributed through the membrane but are concentrated at the periplasmic side of the complex. In addition, Trp residues are not randomly aligned. Within the protein subunits, many form hydrogen bonds with carbonyl oxygens of the main chain, thereby stabilizing the protein. On the surface of the molecule, they are correctly positioned to form hydrogen bonds with the lipid head groups while their hydrophobic rings are immersed in the lipid part of the bilayer. These observations suggest that Trp residues are involved in the translocation of protein through the membrane and that following translocation, Trp residues serve as anchors on the periplasmic side of the membrane.
- ↑
Sun et al. (2008) The preference of tryptophan for membrane interfaces: insights from N-methylation of tryptophans in gramicidin channels. The Preference Of Tryptophan For Membrane Interfaces: Insights From N-methylation Of Tryptophans In Gramicidin Channels 283:22233-43. (pmid: 18550546) [ PubMed ] [ DOI ] To better understand the structural and functional roles of tryptophan at the membrane/water interface in membrane proteins, we examined the structural and functional consequences of Trp --> 1-methyl-tryptophan substitutions in membrane-spanning gramicidin A channels. Gramicidin A channels are miniproteins that are anchored to the interface by four Trps near the C terminus of each subunit in a membrane-spanning dimer. We masked the hydrogen bonding ability of individual or multiple Trps by 1-methylation of the indole ring and examined the structural and functional changes using circular dichroism spectroscopy, size exclusion chromatography, solid state (2)H NMR spectroscopy, and single channel analysis. N-Methylation causes distinct changes in the subunit conformational preference, channel-forming propensity, single channel conductance and lifetime, and average indole ring orientations within the membrane-spanning channels. The extent of the local ring dynamic wobble does not increase, and may decrease slightly, when the indole NH is replaced by the non-hydrogen-bonding and more bulky and hydrophobic N-CH(3) group. The changes in conformational preference, which are associated with a shift in the distribution of the aromatic residues across the bilayer, are similar to those observed previously with Trp --> Phe substitutions. We conclude that indole N-H hydrogen bonding is of major importance for the folding of gramicidin channels. The changes in ion permeability, however, are quite different for Trp --> Phe and Trp --> 1-methyl-tryptophan substitutions, indicating that the indole dipole moment and perhaps also ring size and are important for ion permeation through these channels.
- ↑ Sigma-Aldrich. (n.d.) Sigma-Aldrich Amino Acids Reference Chart. Retrieved December 2, 2017 from https://www.sigmaaldrich.com/life-science/metabolomics/learning-center/amino-acid-reference-chart.html
- ↑
Jacoboni et al. (2001) Prediction of the transmembrane regions of beta-barrel membrane proteins with a neural network-based predictor. Prediction Of The Transmembrane Regions Of Beta-barrel Membrane Proteins With A Neural Network-based Predictor 10:779-87. (pmid: 11274469) [ PubMed ] [ DOI ] A method based on neural networks is trained and tested on a nonredundant set of beta-barrel membrane proteins known at atomic resolution with a jackknife procedure. The method predicts the topography of transmembrane beta strands with residue accuracy as high as 78% when evolutionary information is used as input to the network. Of the transmembrane beta-strands included in the training set, 93% are correctly assigned. The predictor includes an algorithm of model optimization, based on dynamic programming, that correctly models eight out of the 11 proteins present in the training/testing set. In addition, protein topology is assigned on the basis of the location of the longest loops in the models. We propose this as a general method to fill the gap of the prediction of beta-barrel membrane proteins.
- ↑
Kreusch et al. (1994) Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 A resolution. Structure Of The Membrane Channel Porin From Rhodopseudomonas Blastica At 2.0 A Resolution 3:58-63. (pmid: 8142898) [ PubMed ] [ DOI ] The crystal structure of a membrane channel, homotrimeric porin from Rhodopseudomonas blastica has been determined at 2.0 A resolution by multiple isomorphous replacement and structural refinement. The current model has an R-factor of 16.5% and consists of 289 amino acids, 238 water molecules, and 3 detergent molecules per subunit. The partial protein sequence and subsequently the complete DNA sequence were determined. The general architecture is similar to those of the structurally known porins. As a particular feature there are 3 adjacent binding sites for n-alkyl chains at the molecular 3-fold axis. The side chain arrangement in the channel indicates a transverse electric field across each of the 3 pore eyelets, which may explain the discrimination against nonpolar solutes. Moreover, there are 2 significantly ordered girdles of aromatic residues at the nonpolar/polar borderlines of the interface between protein and membrane. Possibly, these residues shield the polypeptide conformation against adverse membrane fluctuations.
- ↑
Jacoboni et al. (2001) Prediction of the transmembrane regions of beta-barrel membrane proteins with a neural network-based predictor. Prediction Of The Transmembrane Regions Of Beta-barrel Membrane Proteins With A Neural Network-based Predictor 10:779-87. (pmid: 11274469) [ PubMed ] [ DOI ] A method based on neural networks is trained and tested on a nonredundant set of beta-barrel membrane proteins known at atomic resolution with a jackknife procedure. The method predicts the topography of transmembrane beta strands with residue accuracy as high as 78% when evolutionary information is used as input to the network. Of the transmembrane beta-strands included in the training set, 93% are correctly assigned. The predictor includes an algorithm of model optimization, based on dynamic programming, that correctly models eight out of the 11 proteins present in the training/testing set. In addition, protein topology is assigned on the basis of the location of the longest loops in the models. We propose this as a general method to fill the gap of the prediction of beta-barrel membrane proteins.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2020-10-29
Version:
- 2.4
Version history:
- 2.4 Explain the use of COLLADA files for worm plots
- 2.3 Added two more Structure Image Examples
- 2.2 Clarify: all deliverables with ChimeraX only; add more hints for Stereo Vision troubleshooting; add first entry for Structure Image Gallery
- 2.1 Edit policy update
- 2.0 Major rewrite: moving from Chimera to ChimeraX
- 1.2 Remove assumptions about preferences
- 1.1 Charge mapping needs to calculate the surface first
- 1.0 First live version
- 0.1 First stub - 2016 material combined
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.