Difference between revisions of "Perl"
m |
m |
||
| (One intermediate revision by the same user not shown) | |||
| Line 5: | Line 5: | ||
| − | {{ | + | {{fix}} |
| Line 26: | Line 26: | ||
<li><span class="toctext">[[Perl references]]</span></li> | <li><span class="toctext">[[Perl references]]</span></li> | ||
<li><span class="toctext">[[Perl simulation]]</span></li> | <li><span class="toctext">[[Perl simulation]]</span></li> | ||
| − | |||
<li><span class="toctext">[[Perl: Object oriented programming]]</span></li> | <li><span class="toctext">[[Perl: Object oriented programming]]</span></li> | ||
<li><span class="toctext">[[Perl: Ugly programming]]</span></li> | <li><span class="toctext">[[Perl: Ugly programming]]</span></li> | ||
| − | |||
<li><span class="toctext">[[BioPerl]]</span></li> | <li><span class="toctext">[[BioPerl]]</span></li> | ||
</ul> | </ul> | ||
| Line 417: | Line 415: | ||
| | ||
[[Category:Applied_Bioinformatics]] | [[Category:Applied_Bioinformatics]] | ||
| + | [[Category:Perl]] | ||
</div> | </div> | ||
Latest revision as of 14:53, 16 September 2012
Perl
Summary ...
Contents
Introductory reading
Perl
Perl is a programming language.
perl is actually a program that runs commands in the Perl programming language. But from a user's perspective, that really doesn't make a difference.
Perl is:
- free-format (whitespace is optional)
- compiled (everything is looked at before its executed)
- interpreted (works from code, step by step)
... with
- automatic typing and automatic memory management.
What is Perl good for?
- Text processing
- Rapid prototyping
- Easy to learn for easy tasks
- Powerful enough for difficult tasks
- Programming for the Web
- Use of large libraries of useful code modules
- "Magic"
- TIMTOWTDI - "There's more than one way to do it".
What is Perl bad at?
- Need for complex datastructures
- Performance-critical applications
- Complex, long-lived software projects with multiple authors
- "Magic"
- TIMTOWTDI - "There's more than one way to do it".
More information on Perl
... to be found here. No static list would make any sense.
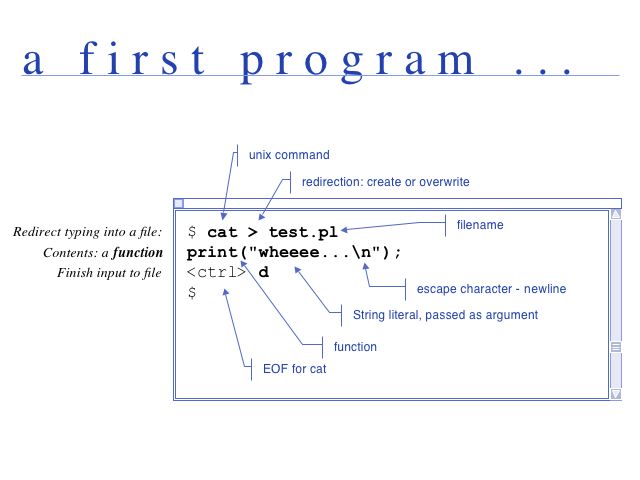
cat in Perl
print()
The print function takes a list as it's argument and writes the list to STDOUT or any other designated "filehandle".
print List;I have installed GeSHi, a syntax highlighter, on this Wiki. Source code is colored so that the different semantics of its elements can be easily seen.
There are three points to make:
(1) Its use is straightforward in principle: list elements are passed to the function, evaluated and written to file.
print("Line $number: ", $line, "\n");(2) Any filehandle can be used as target (STDOUT is default):
open(OUTFILE, ">output.txt");
print(OUTFILE "$value\n";(3) Variables are interpolated but not evaluated:
my $i = 4;
print("value: $i**2\n"); # value: 4**2
print("value: ", $i**2, "\n"); # value: 16The string "\n" denotes a linefeed. It uses the "escape character to either prevent interpolation of special characters or to create special characters from normal ones.
" \n " # newline
" \b " # backspace
" \t " # tab
" \\ " # the backslash itself
" \" " # a double quote
" \$ " # a $ sign (variable is not interpolated)
my $i = 42;
print ("\$i is $i\n"); # $i is 42a cat program
Create a new file by typing
nano cat.plnano or pico are simple text editors, installed on most Unix systems.
Enter the text below exactly as it is written here. Don't mix up the bracket types.
Save by typing <ctrl>o, and exit <ctrl>x.
Make the file executable:
chmod u+x cat.plor
chmod 700 cat.plHere is the code:
#!/usr/bin/perl
use warnings;
use strict;
while (my $line = <STDIN>) {
print($line);
}
exit();Save the file and run the code. Type something and press return. When you are all done, type <ctrl>d to signal the end of input. The program should behave exactly like the cat command. But now we have all single steps of the program at our fingertips, to change and modify them as we wish.
use "strict" and "warnings"
Warnings tell you when Perl thinks you typed things you didn't even mean.
Strict makes Perl complain when variables were not declared - like when they have been mistyped !
Try the error checking functionality:
Replace $line by $lime behind the print statement,
run the program.
"Comment out" use warnings (put a '#' before it), then
change = to == . Run. Abort with <ctrl>c when you are fed up. Remove the # and run this again.
Perl one-liners
With some knowledge of the perl intepreter's run-time flags, one can use (or abuse) perl as a sophisticated command-line utility in a single line. Perl one-liners can be powerful tools, but they can also be quite impenetrable. It is not often that you actually need to work with a one-liner but you may encounter one used elsewhere and it is thus useful to understand how they work.
-e
The flag -e will cause the string that follows it to be executed as a perl script. For example:
perl -e 'for (@INC) {print $_,"\n"}'
will print the path on which perl searches for modules, line by line. The above one-liner is entirely equivalent to running the following Perl program:
#!/usr/bin/perl
for (@INC) {print $_,"\n"}
One would probably not write the program quite as tersely, for readability. But in one-liners, terseness is considered a virtue. Here is a more explicit version of the same:
#!/usr/bin/perl
for (my $i=0; $i < scalar(@INC); $i++) {
print ( $INC[$i], "\n" )
}
- use ' (apostrophes) around the command string and " (quotation marks) within
- actually you could also turn this around, but you can't mix them, otherwise the perl interpreter would not know which is which.
Other flags should not follow the -e - but many other flags can precede it:
- use -M to include modules.
- for example, the follwing will use the standard module Env to access the current path and then modify it to print each directory on the path to a line.
perl -MEnv -e 'for(split(/:/,$PATH)){print $_,"\n"}'
-n
- use -n to loop over input.
- This flag causes to read from the file specified in @ARGV and loop over its contents line by line. The -n flag executes the following bit of code:
while (<>) {
... # your program goes here
}
For example:
perl -ne 'if(/^ATOM .{7}CA..(...)/){print $1," "}' < 2IMM.pdb
prints the three-letter amino acid sequence from PDB file 2IMM.pdb.
-p
- use the flag -p instead of -n,
- ... it automatically prints $_ For example:
perl -pe 'tr/[a-z]/[A-Z]/' < filename
prints the contents of filename in uppercase.
-i
- use the flag -i,
- ... to perform in-place replacements of strings in files. This allows you to substitute all occurrences of a string within one file:
For example:
perl -pi -e 's|../img|../../img|g' < test.html
... substitutes ../../img for each occurrence of ../img in a file, which would be useful if you move a html file into a subdirectory and need to keep the relative path to images intact. For safety (highly recommended!), the -i flag can be used with an argument that is appended to the filename of the original, to create a backup copy; for example in
perl -p -i'.mac' -e 's/\r/\n/g' test.txt
which replaces old-style mac linefeeds with unix linefeeds and saves the original to a backup copy with the extension .mac .
- to remove linefeeds in the data that is to be processed, the flag -l may be used in addition
- This removes the linefeed from input, and adds it back on output ... important if it would otherwise get in the way of processing.
-a and -F
- use the flag -a,
- ... to turn on "autosplit" on whitespace. This causes an implicit @F = split(/\s/,$_) to be executed on every line. This example prints every second word of each line in a file but only if there is a second word.
perl -nae 'print "$F[1]\n" if $F[1]' < inputfile
Adding the -F parameter allows you to specify an arbitrary character as field delimiter for split().
This second one-line script will extract all usernames from /etc/passwd.
perl -na -F: -e 'print "$F[0]\n"' < /etc/passwd
You can use -F/:/ to split on a pattern instead of a string literal. Be careful, because the shell may escape characters preceded by a \ if they are not enclosed in single quotes. Here is an example that will split on tabs and return the first and last field of lines in a tab-delimited file, regardless of how many fields there are.
perl -na -F'/(\t|\n)/' -e 'print "$F[0]\t$F[-2]\n"' < inputfile
Note that we're splitting on either tab or newline (otherwise the newline character will remain stuck to the last field) and we reading the second-to-last element - because the last one is the \n character itself. Why don't we simply use the -l flag instead? Apparently the split is done before the newline is stripped - so -l doesn't have any effect when -a is set.
Creative examples
- Wordcount
Count the number of occurrences of each word in a file and output in order, sorted by frequency.
perl -nle 'split/[^a-z]+/,lc;for(@_){$h{$_}++}}{print"$h{$_}\t$_"for sort{$h{$a}<=>$h{$b}}keys%h' < inputfile
Converts the inputfile to lowercase, splits into words, and then creates a hash where each value is incremented by one when a word is seen. This effectively counts the occurrence of each word. I then iterate over all keys in the hash and sort the entries according to a numerical comparison of the hash values. The resulting pairs are printed.
- Skip lines
Skip a given number of lines at the beginning of a file.
perl -ne 'if($.>3){print}' < inputfile
The Perl special variable '$. holds the current line number of the input.
- Strip tags
Strip tags from a file (this assumes that tags are not broken across lines).
perl -pe 's/\<[^<]*\>//g' < inputfile
- Collapse whitespace
Strips any single whitespace or runs of whitespaces directly at the beginning of a line (^\s+) or at the end of a line (\s+$), collapses all runs of whitespaces into a single blank.
perl -ple 's/(^\s+|\s+$)//g;s/\s+/ /g' < inputfile
Additional reading, examples and resources
- Cultured Perl: one liners 101
- Cultured Perl: one liners 102
- One-liners for bioinformaticians
- The Scriptome - a system of interoperting one-liners for bioinformatics
- Perl One Liners
- Documentation on how to run perl, the Perl interpreter
Installation of perl modules
How to install Perl modules and programs on Unix systems.
- Generic installation
- 1. download the source-archive
- 2. unzip and untar the file
- 3. cd to the newly created directory
- 4. type Makefile.PL
- 5. type make
- 6. type make test
and if the test results appear reasonable:
- 7. type sudo make install
CPAN
CPAN is the Comprehensive Perl Archive Network, a repository of Perl modules.
How do you know you are missing a module?
Type the following Perl one-liner :
- $ perl -e "use <ModuleName>;"
If the Perl interpreter complains about not finding the module it is not installed or not on your Perl path. If you know the module is on the system, you can list your Perl path by typing:
- $ perl -e 'for (@INC){print $_,"\n"}'
... and confirm whether the module is indeed not in one of these directories (e.g. you might have installed it in a local repository because you are modifying or developing it). You can then either move the module, add its directory to @INC in your script, include the full path in your use statement or set the PERl5LIB environment variable to point to your local repository.
How to search CPAN
Use the CPAN search page to search by module, distribution or author.
What is not in CPAN
CPAN usually only contains stable releases; for developer releases search Google for the project home pages. Often open source projects will be on Sourceforge and can also be searched there. CPAN only contains open-source code licensed under GPL or similar, for commercial code, search with Google. If you are looking for more general examples of algorithms and how-to, or have specific coding questions, check out Perl Monks or look into/ask in comp.lang.perl.misc or comp.lang.perl.modules (here via a Web gateway).
- Installation via CPAN
- one module
sudo perl -MCPAN -e "install ''My::Modules''"
or
sudo perl -MCPAN -e "force install ''My::Modules''"- Installation via CPAN
- interactively
Once you have found a module you need, you can use also run the Perl CPAN module to download it interactively. Type
- sudo perl -MCPAN -e shell
... to start an interactive CPAN session. Type ? for help on commands. For example, to install Bundle::Bioperl, I went through the following steps.
$ cd ~/Downloads $ sudo perl -MCPAN -e shell Password: ........ cpan> install Bundle::BioPerl
[...] all modules get fetch'ed, make'd and install'ed. (Well, not quite ... the process paused and asked for the directory where libgd was installed in order to be able to install GD; I opened a second shell and installed libgd from source following instructions [1], then continued ... GD installation will be a separate topic. Also some modules failed subtests and wouldn't install without force...)
cpan> exit $
see "Installing modules" on CPAN for more information.
Links
Further reading and resources