Perl MySQL introduction
Perl and MySQL: Introduction
Summary ...
Contents
Prerequisites
The MySQL server must be running for successful installation, otherwise the test provided with the interfaces will fail.
- DBI needs to be installed
- DBD::mysql needs to be installed. Installation form CPAN is straightforward:
sudo perl -MCPAN -e shell cpan> install Bundle::DBI cpan> install DBD::mysql
... to install the DBI interface and all required supporting files.
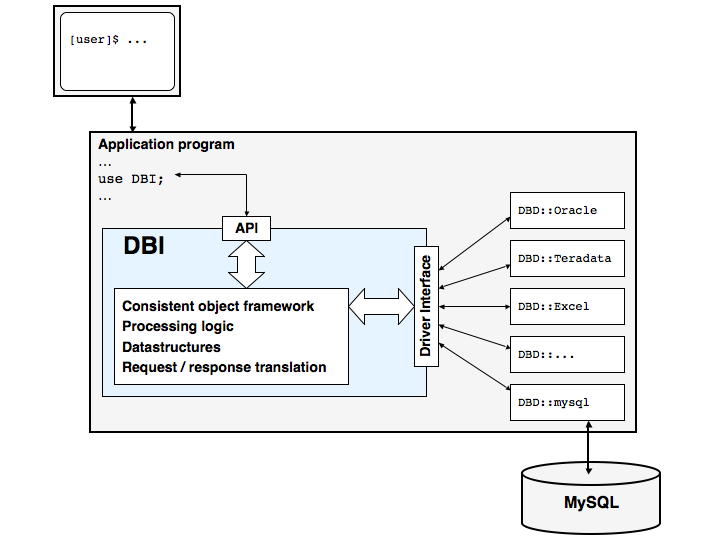
Overview of DBI and DBD architecture
DBI (the DataBase Interface) is a generic interface for relational databases that provides a consistent set of methods and parameters for programs that wish to access a database. DBI communicates with one of a number of DBD::... (DataBase Driver) modules, which handle the database-engine specific details. We are using DBD::mysql. The reason behind this is portability: your program need not depend on any particular database engine, such as MySQL or Oracle, because switching from one to the other is a matter as simple as useing a different DBD module and changing a single connect statement.
The image below shows the architecture of an application that uses DBI for SQL database access:
A first example program
The simplest way to use DBI is by using its do( ) method to execute individual SQL statements. This is most useful for statements that are excuted only once.
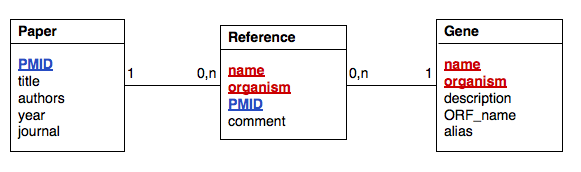
Here is an example datamodel for genes and publications, similar to the one we discussed in the MySQL page.
The following program code connects to the mysql server, creates the database "example" and implements this datamodel. Remember that the user has to exist, and she must have the appropriate privileges to work with a database by that name. Copy and paste the code, then step through it in the debugger.
#!/usr/bin/perl -w use strict; use DBI; # define all the variables my $host = 'localhost'; # MySQL Host my $user = 'developer'; # your username my $password = 'mysecretpassword'; # your password my $database_name = 'example'; # name of database # connect to MySQL on localhost USEing the preexisting database 'test' # The result of the call to 'connect' is a database handle object. my $dbh = DBI->connect("dbi:mysql:database=test;host=$host", $user, $password) or die "Couldn't connect to database: $DBI::errstr\n"; # Use the dbh->do() method to directly execute SQL statements. # The return value is the number of rows affected, or undef on error. $dbh->do("CREATE DATABASE IF NOT EXISTS $database_name;") or die "Couldn't create database: $DBI::errstr\n"; $dbh->do("USE $database_name;"); # If the three tables we are about to create already exist # we DROP them. This way we can run this example code # several times without error and make changes to the table structure if we wish. $dbh->do('DROP TABLE IF EXISTS Gene;'); $dbh->do('DROP TABLE IF EXISTS Paper;'); $dbh->do('DROP TABLE IF EXISTS Reference;'); # Now create the three tables $dbh->do(' CREATE TABLE Gene ( name varchar(100) NOT NULL, organism varchar(255), description text NOT NULL, ORF_name varchar(55), alias varchar(255), PRIMARY KEY (name, organism) ); ') or die "Couldn't create tables: $DBI::errstr\n"; # same for table "Paper" $dbh->do(' CREATE TABLE Paper ( PMID varchar(55) NOT NULL, title varchar(255) NOT NULL, authors varchar(255) NOT NULL, year int(4) NOT NULL, journal varchar(255) NOT NULL, PRIMARY KEY (PMID) ); ') or die "Couldn't create tables: $DBI::errstr\n"; # same for table "Reference" $dbh->do(' CREATE TABLE Reference ( name varchar(100) NOT NULL, organism varchar(255) NOT NULL, PMID varchar(55) NOT NULL, comment text, PRIMARY KEY (name, organism, PMID) ); ') or die "Couldn't create tables: $DBI::errstr\n"; $dbh->disconnect(); exit;
Now verify that the database was created, by conecting to the mysql client. Type the input from the following terminal session:
$ /usr/local/mysql/bin/mysql -u developer -p Enter password: ....... Welcome to the MySQL monitor. Commands end with ; or \g. Your MySQL connection id is 22 to server version: 5.0.18-standard Type 'help;' or '\h' for help. Type '\c' to clear the buffer. mysql> SHOW DATABASES; +--------------------+ | Database | +--------------------+ | information_schema | | example | | test | +--------------------+ 3 rows in set (0.00 sec) mysql> USE example; Reading table information for completion of table and column names You can turn off this feature to get a quicker startup with -A Database changed mysql> SHOW TABLES; +-------------------+ | Tables_in_example | +-------------------+ | Gene | | Paper | | Reference | +-------------------+ 3 rows in set (0.00 sec) mysql> DESCRIBE Paper; +---------+--------------+------+-----+---------+-------+ | Field | Type | Null | Key | Default | Extra | +---------+--------------+------+-----+---------+-------+ | PMID | varchar(55) | NO | PRI | | | | title | varchar(255) | NO | | | | | authors | varchar(255) | NO | | | | | year | int(4) | NO | | | | | journal | varchar(255) | NO | | | | +---------+--------------+------+-----+---------+-------+ 5 rows in set (0.24 sec) mysql> QUIT; Bye $
All is well if your output looks like mine.
This straightforward approach of sending SQL instructions to the database through the database-handle dbh->do(expression); constructs has a limitation: it is not designed to return multivalued data - rather it only returns a single scalar: the number of rows affected by the statement.
In order to capture multivalued data, we can use the dbh->selectrow_arrayref() and dbh->selectall_arrayref() methods. Both methods return references to arrays. This is the common way for Perl subroutines to return more-dimensional data.
A digression: array references in Perl
Consider the following:
#!/usr/bin/perl -w use strict; my @array = make_array(); foreach my $number (@array) { print "$number \n"; } exit; sub make_array { my @a = (1, 1, 3, 5, 8, 13); return @a; }
This works, but is not very efficient. It actually creates the same array twice: once inside the subroutine (@a)and once in the main body of the function (@array). @a is copied to @arrray in the return process. For large arrays this can be a problem and databases can create very large arrays. Subroutines that are written with efficiency in mind thus do not return arrays or hashes directly, but create them once and then return a reference, a single scalar value that identifies the memory position where the first element of that item is located. References in Perl are created by prepending an item with a backslash "\" ...
my $array_ref = \@array;
... and in order to use an item based on its reference, the reference has to be dereferenced, which is simply achieved by prepending the appropriate sigil ...
my @new_array = @$array_ref;
Using array references in the example above thus works as follows:
#!/usr/bin/perl -w use strict; my $a_ref = make_array(); foreach my $number (@$a_ref) { print "$number \n"; } exit; sub make_array { my @a = (1, 1, 3, 5, 8, 13); return \@a; }
Arrayreferences in database handles
As we had mentionend above, the dbh->do() statement returns only a single scalar. But suppose we want to retrieve multivalued results, such as the results of SELECT or SHOW statements. DBI makes these available as references to arrays containing the values of a row, or as references to arrays containing references to arrays containing the values of a row. Its really quite simple.
Consider the return values of the SQL statement SHOW TABLES; We get three rows, each with a single value: the table name. The $dbh->selectrow_arrayref($statement); method returns an array reference to the first row of data from this response. This first row contains the single value "Gene". Append the following code to the database example above (before the dbh->disconnect statement) to see this work.
my $arrayref = $dbh->selectrow_arrayref('SHOW TABLES;'); my @array = @$arrayref; foreach my $element (@array) { print $element,"\n"; }
The result of this code is the single item: "Gene". Let's extend this principle to DESCRIBE the entire Gene table:
my $arrayref = $dbh->selectrow_arrayref('SHOW TABLES;'); my @array = @$arrayref; $arrayref = $dbh->selectrow_arrayref("DESCRIBE $array[0];"); @array = @$arrayref; foreach my $element (@array) { print $element,"\n"; }
This lists the values for "Field", "Type", "Null", "Key", "Default", and "Extra" for the first table in the current database. This example shows how to obtain all values from a single row. But what about multiple rows?
For multiple rows, we use the method $dbh->selectall_arrayref($statement) which returns a reference to an array of references to arrays of values. It's the same thing as before, but because we are returning multiple rows, we need an array that holds multiple references. Then the method returns a reference to that array as the result.
Here is the whole thing in code: first we pick up all three table names. Replace the code above with the following and run this.
my $list_ref = $dbh->selectall_arrayref('SHOW TABLES;'); my @list = @$list_ref; foreach my $row_ref (@list) { my @row = @$row_ref; print $row[0],"\n"; }
Then we extend this by requesting DESCRIBE for each table:
my $list_ref = $dbh->selectall_arrayref('SHOW TABLES;'); my @list = @$list_ref; foreach my $row_ref (@list) { # for each table in database my @row = @$row_ref; print "\n\n", $row[0], ":\n"; # print table name my $a_ref = $dbh->selectall_arrayref("DESCRIBE $row[0];"); foreach my $column_ref (@$a_ref) { # for each column in table foreach my $property (@$column_ref){ # for each property of column if ($property) { # except if undefined print $property, "\t"; } } print "\n"; } }
Run this code and make sure you understand it!
The DBI statement handles
(Note that the usual method of working with statement handles described here can be combined into a single method call on a database handle as shown above, however the description below is the more generic way to proceed.)
The dbh->selectall() methods presented above are actually a shortcut combination the three statement-handle methods sth->prepare(), sth->execute() and sth->fetchall(). Statement handles are the generic approach DBI uses to work with recurring SQL statements.
- sth->prepare()
- prepares an SQL statement object for execution and setup the required datastructures.
- sth->execute()
- execute the statement
- sth->fetchall()
- retrieve the results of the last execution of this object
The key concept here is that statement handles are prepared once, but then can be executed many times, changing only the variable parts of the statement. This is demonstrated in the code example below in which we fill the database we have generated above with data.
#!/usr/bin/perl -w use strict; use DBI; # define all the variables my $host = 'localhost'; # MySQL Host my $user = 'developer'; # your username my $password = 'mysecretpassword'; # your password my $database_name = 'example'; # name of database # connect to MySQL on localhost USEing the preexisting database 'test' # The result of the call to 'connect' is a database handle object. my $dbh = DBI->connect("dbi:mysql:database=$database_name;host=$host", $user, $password) or die "Couldn't connect to database: $DBI::errstr\n"; #initialize some data in a two-dimensional array my @data = ( () ); $data[0][0] = 'RPL10'; $data[0][1] = 'Responsible for joining the 40S and 60S subunits'; $data[0][2] = 'YLR075W'; $data[0][3] = 'Saccharomyces Cerevisiae'; $data[1][0] = 'RPL11A'; $data[1][1] = 'Involved in ribosomal assembly'; $data[1][2] = 'YPR102C'; $data[1][3] = 'Saccharomyces Cerevisiae'; $data[2][0] = 'RPL11B'; $data[2][1] = 'Involved in ribosomal assembly'; $data[2][2] = 'YGR085C'; $data[2][3] = 'Saccharomyces Cerevisiae'; $data[3][0] = 'RPL12A'; $data[3][1] = 'rpl12a rpl12b double mutant exhibits slow growth and slow translation'; $data[3][2] = 'YEL054C'; $data[3][3] = 'Saccharomyces Cerevisiae'; $data[4][0] = 'RPL12B'; $data[4][1] = 'rpl12a rpl12b double mutant exhibits slow growth and slow translation'; $data[4][2] = 'YDR418W'; $data[4][3] = 'Saccharomyces Cerevisiae'; # Use the dbh->prepare() method to create a statement handle # use four placeholders for the four data elements of each row. my $sth = $dbh->prepare('INSERT INTO Gene (name,description,ORF_name,organism) VALUES(?,?,?,?);'); # iterate over the data table and execute the INSERT statements for (my $i=0; $i < scalar(@data); $i++) { $sth->execute( $data[$i][0], $data[$i][1], $data[$i][2], $data[$i][3] ) or die ( "Error on INSERT: $DBI::errstr\n" ); } # a different strategy for the Paper table. # First, create rows with the primary key $sth = $dbh->prepare('INSERT INTO Paper (PMID) VALUES (?);'); $sth->execute('12627396') or die ( "Error on INSERT: $DBI::errstr\n" ); $sth->execute('11893754') or die ( "Error on INSERT: $DBI::errstr\n" ); $sth->execute('11071906') or die ( "Error on INSERT: $DBI::errstr\n" ); $sth->execute('9822666') or die ( "Error on INSERT: $DBI::errstr\n" ); # insert titles via UPDATE ... WHERE statements $sth = $dbh->prepare( 'UPDATE Paper SET title=? WHERE PMID=?;' ); $sth->execute('Translational regulator RpL10p/Grc5p interacts physically...', '12627396') or die ( "Error on UPDATE: $DBI::errstr\n" ); $sth->execute('Normal assembly of 60 S ribosomal subunits is required for...', '11893754') or die ( "Error on UPDATE: $DBI::errstr\n" ); $sth->execute('Factors affecting nuclear export of the 60S ribosomal subunit...', '11071906') or die ( "Error on UPDATE: $DBI::errstr\n" ); $sth->execute('The GTPase center protein L12 is required for correct...', '9822666') or die ( "Error on UPDATE: $DBI::errstr\n" ); # insert remaining information via UPDATE ... WHERE statements $sth = $dbh->prepare( 'UPDATE Paper SET authors=?, year=?, journal=? WHERE PMID=?;' ); # insert titles $sth->execute('Oender Kamil, et al.', '2003', 'Yeast (Chichester, England)', '12627396') or die ( "Error on UPDATE: $DBI::errstr\n" ); $sth->execute('Miyoshi Keita, et al.', '2002', 'The Journal of biological chemistry.', '11893754') or die ( "Error on UPDATE: $DBI::errstr\n" ); $sth->execute('Stage-Zimmermann T, et al.', '2000', 'Molecular biology of the cell.', '11071906') or die ( "Error on UPDATE: $DBI::errstr\n" ); $sth->execute('Briones E, et al.', '1998', 'The Journal of Biological Chemistry.', '9822666') or die ( "Error on UPDATE: $DBI::errstr\n" ); # a third strategy for the Reference table: insert everything directly with dbh->do(); my $org = "Saccharomyces Cerevisiae"; $dbh->do( "INSERT INTO Reference (PMID, name, organism) VALUES('12627396', 'RPL10', '$org');") or die ( "Error on INSERT: $DBI::errstr\n" ); $dbh->do( "INSERT INTO Reference (PMID, name, organism) VALUES('11893754', 'RPL11A', '$org');") or die ( "Error on INSERT: $DBI::errstr\n" ); $dbh->do( "INSERT INTO Reference (PMID, name, organism) VALUES('11893754', 'RPL11B', '$org');") or die ( "Error on INSERT: $DBI::errstr\n" ); $dbh->do( "INSERT INTO Reference (PMID, name, organism) VALUES('11071906', 'RPL11B', '$org');") or die ( "Error on INSERT: $DBI::errstr\n" ); $dbh->do( "INSERT INTO Reference (PMID, name, organism) VALUES('9822666', 'RPL12A', '$org');") or die ( "Error on INSERT: $DBI::errstr\n" ); $dbh->do( "INSERT INTO Reference (PMID, name, organism) VALUES('9822666', 'RPL12B', '$org');") or die ( "Error on INSERT: $DBI::errstr\n" ); # done $dbh->disconnect(); exit;
Combining the two principles we have illustrated in the preceding code fragments, it is now straightforward to code and execute a complicated SELECT statement. In this example, we start from an ORF_name and pick up the gene name, we then search the bibliographic information for all papers that are anotated with the gene name. Note that none of our tables has the entire information for this query, all three need to be combined. Gene establishes the relationship between ORF_name and name, Reference establishes the relationship between name and PMID and finally Paper contains the requested information title, authors, Year for a PMID. Note that we need to explicitly SELECT Gene.name, not just name, since the column name is not unique among the tables FROM which we are selecting in the query. Conversely, e.g. authors is unique and does not need to be qualified as Paper.authors.
#!/usr/bin/perl -w use strict; use DBI; my $host = 'localhost'; # MySQL Host my $user = 'developer'; # your username my $password = 'mysecretpassword'; # your password my $database_name = 'example'; # name of database my $dbh = DBI->connect("dbi:mysql:database=$database_name;host=$host", $user, $password) or die "Couldn't connect to database: $DBI::errstr\n"; # get all the papers that mention gene YGR085C print "\n\n *** Papers that mention YGR085C ***\n\n"; my $query_ORF = 'YGR085C'; my $query = "SELECT Gene.name, authors, year, title "; $query .= " FROM Reference, Paper, Gene "; $query .= " WHERE Gene.ORF_name=\'$query_ORF\' "; $query .= " AND Gene.name=Reference.name "; $query .= " AND Reference.PMID=Paper.PMID;"; my $result_list_ref = $dbh->selectall_arrayref($query) or die("Selection failed: DBI::errstr"); foreach my $row_ref (@$result_list_ref) { # for each row that matches the selection foreach my $column (@$row_ref){ # for each value in the row if ($column) { # except if undefined print $column, "\t"; } } print "\n"; } $dbh->disconnect(); exit;
Whle this (hopefully) works as expected, DBI has a more comfortable way to work with SELECT results: binding result columns directly to Perl variables.
Using bind_columns for return values
We have already used bindings in the code above, when we wrote ? characters as placeholders. This is not a Perl language construct, but syntax that is specific to DBI. We can also use bindings as placeholders for output, as in the following example code. This bind_column method makes working with results, formatted output or further processing quite straightforward.
#!/usr/bin/perl -w use strict; use DBI; my $host = 'localhost'; # MySQL Host my $user = 'developer'; # your username my $password = 'mysecretpassword'; # your password my $database_name = 'example'; # name of database my $dbh = DBI->connect("dbi:mysql:database=$database_name;host=$host", $user, $password) or die "Couldn't connect to database: $DBI::errstr\n"; # get all the papers that mention gene YGR085C print "\n\n *** Papers that mention YGR085C ***\n\n"; my $query_ORF = 'YGR085C'; my $query = "SELECT Gene.name, authors, year, title "; $query .= " FROM Reference, Paper, Gene "; $query .= " WHERE Gene.ORF_name=\'$query_ORF\' "; $query .= " AND Gene.name=Reference.name "; $query .= " AND Reference.PMID=Paper.PMID;"; my $sth = $dbh->prepare($query) or die("Prepare failed: DBI::errstr"); $sth->execute() or die("Execution failed: DBI::errstr"); my ($N, $A, $Y, $T); # each column in the SELECT statement is bound to a reference to a Perl variable $sth->bind_columns(\$N, \$A, \$Y, \$T); # sth->fetch retrieves results row by row while ($sth->fetch) { print "Gene $N\t($A, $Y): \"$T\"\n"; } $dbh->disconnect(); exit;
References
- DBI documentation on CPAN
- DBD::mysql documentation on CPAN
- Perl and databases - an introductory level summary at DC Perl mongers
- A Short Guide to DBI on perl.com
- SQL Database Administration (in Ch 7 of: Perl for System Administration)
Further reading and resources