Difference between revisions of "Bioinformatics Main Page"
m |
|||
| Line 8: | Line 8: | ||
</div> | </div> | ||
| − | <small>'''These wiki pages | + | <small>'''These wiki pages provide information and materials, and coordinate activities and projects in the introductory bioinformatics courses and other workshops taught by Boris Steipe at the University of Toronto'''. If you are not one of my students, you can still browse this site, however I can't provide support for your explorations. The material may be useful if you invest some effort in studying it systematically.</small> |
| − | < | + | <div class="alert"> |
| − | |||
| − | |||
| − | |||
| − | + | Welcome to the 2017 version of BCH441, which is being completely revised and reimagined. | |
| − | + | Here is the current draft version of organizational details, activities and contents. The delivery, activities and assessments will comprise an entirely novel format. Updates will be continuously posted here. | |
| − | + | First class-meeting: Tuesday, September 12, 17:00 in LM161. | |
</div> | </div> | ||
| Line 46: | Line 43: | ||
* introduction to systems-level concepts. | * introduction to systems-level concepts. | ||
| − | Practical, hands on tasks and assignments will introduce public data resources and analysis tools. Along with improving general computer literacy, you will learn to use the programming language and statistical workbench '''R''', with a special emphasis on the kind of everyday tasks of data preparation and analysis that have become indispensable for any life-science laboratory. (Yes, you will learn | + | Practical, hands on tasks and assignments will introduce public data resources and analysis tools. Along with improving general computer literacy, you will learn to use the programming language and statistical workbench '''R''', with a special emphasis on the kind of everyday tasks of data preparation and analysis that have become indispensable for any life-science laboratory. (Yes, you will learn to program.) |
| − | The course is complemented by [[Computational_Systems_Biology_Main_Page|'''BCB420 / JTB2020''']] (offered in the Winter Term) which consolidates aspects of | + | The course is complemented by [[Computational_Systems_Biology_Main_Page|'''BCB420 / JTB2020''']] (offered in the Winter Term) which consolidates aspects of computational systems biology in a project context. |
<code>BCH441H1F</code> is the undergraduate course code.<br> | <code>BCH441H1F</code> is the undergraduate course code.<br> | ||
| Line 54: | Line 51: | ||
| − | {{#lst:User:Boris| | + | {{#lst:User:Boris|Instructor}} |
===Dates=== | ===Dates=== | ||
| Line 61: | Line 58: | ||
;First day of class – Tuesday, September 12. You must attend the first class. | ;First day of class – Tuesday, September 12. You must attend the first class. | ||
| − | :We need this time to go over the course delivery and organizational details, and get you an account on the Course Wiki and on the course mailing list. Your personal presence is a requirement of the course. '''Please do not enrol in the course if your travel- or other plans prevent you from attending the first class session. | + | :We need this time to go over the course delivery and organizational details, and get you an account on the Course Wiki and on the course mailing list. Your personal presence is a requirement of the course. '''Please do not enrol in the course if your travel- or other plans prevent you from attending the first class session.''' |
| − | |||
| − | === | + | ==Contents== |
| − | |||
| + | In Fall 2017, the course will be taught for the first time following an entirely new concept: previous material has been decomposed into small "learning units" that are focussed more or less on a single concept. You work through the units independently, in any order that makes sense to you, at your own pace, all with the goal to acquire the knowledge and skills to work on four main "Integrator Units" that bring the contents together. | ||
| − | + | Through this, the course accommodates different levels of preparation more flexibly, probably makes your work more efficient, and implicitly teaches a number of meta-skills such as reporting and time-management. Be mindful though: this format requires a high level of self-motivation and responsibility to do well. In terms of aiming for the highest level of understanding and competence, you will frequently be on your own - just like you are in "real life". On the other hand, you certainly are the best judge of how well prepared you are. Thus there should be no surprises when your deliverables are evaluated. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | === | + | ===Grading and Activities=== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| − | + | This course comprises four key, integrative activities and preparatory "learning units" that lead up to them. Learning units can be completed in any sequence that makes sense to you, at any time until the deadline to submit material. But note that some learning units require evaluation on our "Evaluation Days" (see below), and/or scheduling a test, and that has to be done well in advance. | |
| − | + | There are also a few restrictions on which units you must complete within this course: | |
| − | |||
| − | |||
| − | |||
| + | * you must complete all four <span style="background-color: #e19fa7; border:solid 2px #000000;"> Integrator Units </span> and submit them for marking. These will be worth maximally 40 marks (4 x 10); | ||
| − | = | + | * you can submit a mix of other <span style="background-color: #b3dbce; border:solid 2px #000000;"> learning units </span> worth up to an additional 30 marks for marking. These are typically worth 6 marks each. There are a number of units available, which ones you choose is up to you. |
| − | + | * You must ensure that you have submitted units for evaluation that are worth at least 10% of your final grade by October 31, so they can be marked before the {{dropdate}}. | |
| − | + | * 25% of your mark will be given for your [[FND-Journal|'''Course Journal''']] at the end of class. | |
| − | + | * 5% of your mark will be given for your [[ABC-Insights|'''insights!''' page]] at the end of class. | |
| − | + | Please carefully read the [[ABC-Rubrics|'''evaluation rubrics''']] for each category of deliverables. | |
| + | For graduate students (BCH1441), the marks you receive for learning units and Integrator Units will be scaled by 0.8, and 14 marks are available for your own design of a learning unit covering an aspect of your thesis project. Coordinate this with the instructor well in advance of the {{lastdate}}. | ||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
<table cellpadding="5"> | <table cellpadding="5"> | ||
| Line 140: | Line 107: | ||
<tr class="s2"> | <tr class="s2"> | ||
| − | <td> | + | <td>Integrator Units</td> |
| − | <td> | + | <td>40 marks</td> |
| − | <td> | + | <td>32 marks</td> |
</tr> | </tr> | ||
<tr class="s1"> | <tr class="s1"> | ||
| − | <td> | + | <td>Your selection of other learning units</td> |
| − | <td> | + | <td>30 marks</td> |
| − | <td> | + | <td>24 marks</td> |
</tr> | </tr> | ||
<tr class="s2"> | <tr class="s2"> | ||
| − | <td> | + | <td>Course Journals</td> |
| − | <td> | + | <td>30 marks</td> |
| − | <td> | + | <td>30 marks</td> |
</tr> | </tr> | ||
<tr class="s1"> | <tr class="s1"> | ||
| − | <td> | + | <td>Graduate "Learning Unit Design"</td> |
<td> </td> | <td> </td> | ||
| − | <td> | + | <td>14 marks</td> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
| Line 180: | Line 141: | ||
| + | ;A mix of evaluation methods | ||
| + | :Learning units will be evaluated with a mix of approaches including technical reports, documentation of results in your journal, delivery of R code, quizzes, oral tests and in-class presentations. Details are described in the individual units. | ||
| − | |||
| − | |||
| − | + | ;Test dates | |
| + | :'''Quizzes and in-class presentations''' will be scheduled on the following dates: | ||
| + | :* October 3 | ||
| + | :* October 24 | ||
| + | :* November 14 | ||
| + | :* November 28 | ||
| − | + | :'''Oral tests for Integrator Units''' will be scheduled on November 9 and 10, and November 23 and 24. These are Thursday and Friday dates and we will coordinate your test dates in October. | |
| − | |||
| − | |||
| + | <div class="toccolours mw-collapsible mw-collapsed" style="width:800px"> | ||
| + | '''A final note on marking policy...''' | ||
| + | <div class="mw-collapsible-content"> | ||
| + | I do not adjust marks towards a target mean and variance (i.e. there will be no "belling" of grades), but follow the principles laid out in the marking rubrics. I feel strongly that "normalization" of grade interferes with a collaborative and mutually supportive learning environment. If your classmate gets a great mark because you helped him with a difficult concept, this should never have the effect that it brings down your mark because the class average is "belled-down" by the instructor. Collaborate as much as possible, it is a great way to learn. | ||
| + | <small>But take *utmost* care to follow the instructions on avoiding [[ABC-Plagiarism|'''plagiarism and academic misconduct''']] to the letter, they will be rigorously enforced.</small> | ||
| + | </div> | ||
| + | </div> | ||
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | + | === The learning-units map === | |
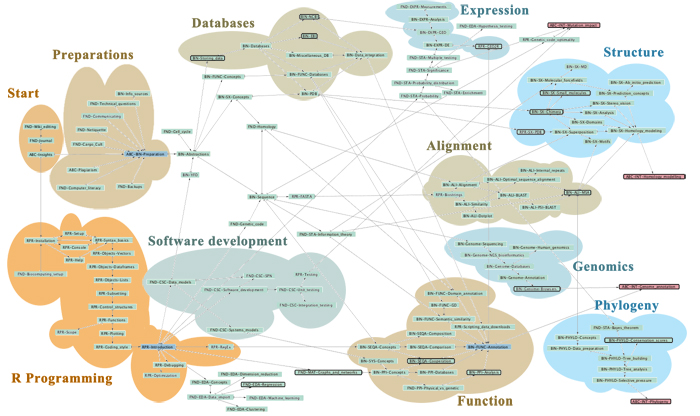
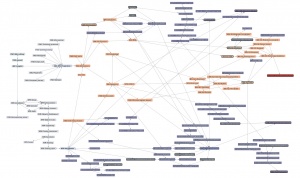
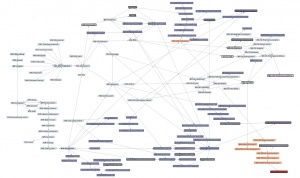
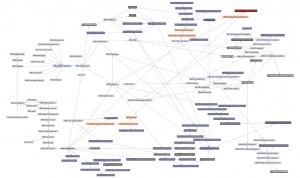
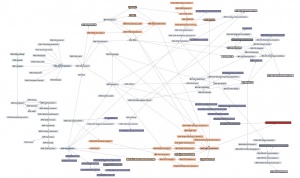
| + | Here is a '''thematic overview''' of the topical areas of this course's learning units: | ||
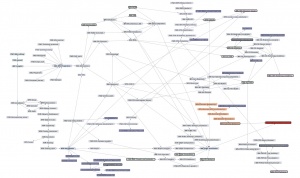
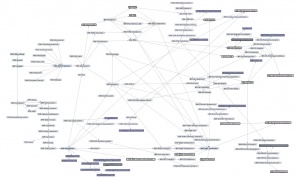
| − | + | [[File:ABC-units_map_themes.jpg]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | And here is the '''detailed map'''. It contains links to all of the units. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ---- | + | * <command>-Click to open the Learning Units Map in a new tab, scale for detail. |
| + | [[File:ABC-units_map.svg|thumb|500px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-units_map.svg|'''A map of the bioinformatics learning units.''']] | ||
| + | * Hover over a learning unit to see its keywords. | ||
| + | * Click on a learning unit to open the associated page. | ||
| + | * The nodes of the learning unit network are colour-coded: | ||
| + | **<span style="background-color: #b3dbce;"> Live units </span> are green | ||
| + | **<span style="background-color: #d9ead5;"> Units under development </span> are light green. These are still in progress. | ||
| + | **<span style="background-color: #f2fafa;"> Stubs </span> (placeholders) are pale. These still need basic contents. | ||
| + | **<span style="background-color: #97bed5;"> Milestone units </span> are blue. These collect a number of prerequisites to simplify the network. | ||
| + | **<span style="background-color: #e19fa7;"> Integrator units </span> are red. These embody the main goals of the course. | ||
| + | **<span style="background-color: #f4d7b7;"> Units that require revision </span> are pale orange. | ||
| + | *Units that have a <span style="background-color: #eeeeee; border:solid 2px #000000;"> black border </span> have deliverables that can be submitted for credit. Choose any you want to submit for credit, up to a maximum worth of 30 marks. | ||
| + | *Arrows point from a prerequisite unit to a unit that requires it. | ||
| − | + | (Unit status will be updated as in-progress units are completed.) | |
| − | |||
| − | |||
| − | |||
| − | |||
| + | {{Vspace}} | ||
| − | + | ====Navigating the course==== | |
| − | + | Everything starts with the following three units: | |
| + | *[[FND-Wiki_editing|Introduction to editing Wiki pages]] | ||
| + | :{{#lst:FND-Wiki_editing|abstract}} | ||
| − | + | *[[FND-Journal|Your Course Journal]] | |
| + | :{{#lst:FND-Journal|abstract}} | ||
| + | *[[ABC-Insights|The "insights!" page]] | ||
| + | :{{#lst:ABC-Insights|abstract}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Everything leads to the ''Integrator Units''. These cover four large areas of bioinformatics that make up the explicit goals of the course: | |
| − | + | (i) algorithms and statistics;<br /> | |
| − | </ | + | (ii) structural modelling and interpretation;<br /> |
| − | </ | + | iii) gene annotation; and;<br /> |
| − | </ | + | (iv) phylogenetic analysis. |
| + | The knowledge and skills you need to work on these ''Integrator Units'' can be obtained from the other learning units that are shown on the [http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-units_map.svg learning units map] as prerequisites. Note that "prerequisites" in this context does not mean you '''must''' do one thing before you can do another, the arrows simply point out which units assume what prior knowledge. You can acquire that knowledge in whatever sequence makes sense to '''you''', and you don't have to learn from the learning units of this course at all. Just make sure that you submit enough general learning units for evaluation along the way. And document what you are doing in your Course Journal. Also, remember that '''all''' the material is cumulative - my evaluation of your work implicitly includes all of the prerequisite material. | ||
| − | + | {{Vspace}} | |
| − | === | + | =====Scenarios===== |
| − | + | Where to begin: possible scenarios for working though the units... | |
| + | <small>(These scenarios are for illustration, you don't have to follow these sequences. There is no implied claim that any of these sequences is better for learning the material than any other. Make your own choices!</small> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | + | <div class="toccolours mw-collapsible mw-collapsed" style="width:800px"> |
| − | + | ;Yvette might have done a project about protein structure in a previous course... | |
| − | + | <div class="mw-collapsible-content"> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | ||
| − | -- | + | <table> |
| − | + | <tr> | |
| − | + | <td>She decides to tackle the [[ABC-INT-Homology_modelling|Homology modelling Integrator Unit]] first, because she is most confident about that material. | |
</td> | </td> | ||
| + | <td>[[File:ABC-Scenario-1-Step-1.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-1.jpg]]</td> | ||
</tr> | </tr> | ||
| − | |||
| − | + | <tr> | |
| − | + | <td>:Obviously, all paths through the learning units begin with the units leading up to the [[ABC-BIN-Preparation|Course Preparation]] milestone, as well as the [[RPR-Introduction|Introduction to R]] milestone. This is where she starts. | |
| − | + | </td> | |
| − | + | <td>[[File:ABC-Scenario-1-Step-2.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-2.jpg]]</td> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <tr | ||
| − | <td | ||
| − | <td | ||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
| − | <tr | + | <tr> |
| − | <td | + | <td>:The homology modelling unit requires the cluster of protein structure units (BIN-SX-...), as well as the sequence alignment units (BIN-ALI-...); |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | :From the database units, she only needs [[BIN-PDB]] for now; | |
</td> | </td> | ||
| + | <td>[[File:ABC-Scenario-1-Step-3.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-3.jpg]]</td> | ||
</tr> | </tr> | ||
| − | |||
| − | + | <tr> | |
| − | + | <td>:For the sequence alignment cluster she needs [[BIN-Sequence]] and [[FND-Homology]] and both of these need [[BIN-Abstractions]]. | |
| − | + | </td> | |
| − | + | <td>[[File:ABC-Scenario-1-Step-4.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-4.jpg]]</td> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <tr | ||
| − | <td | ||
| − | <td | ||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
| − | <tr | + | <tr> |
| − | <td | + | <td>Next she goes for the [[ABC-INT-Phylogeny|Phylogenetic Analysis Integrator Unit]], since it requires only a small number of additional prerequisites and she's a bit busy at that time with midterms in other courses. Some introductory statistics ([[FND-STA-Probability]] and [[FND-STA-Bayes_theorem]] gets her on the way to the BIN-PHYLO-... cluster. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ---- | ||
| − | |||
| − | |||
</td> | </td> | ||
| + | <td>[[File:ABC-Scenario-1-Step-5.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-5.jpg]]</td> | ||
</tr> | </tr> | ||
| − | |||
| − | + | <tr> | |
| − | + | <td>She finds she enjoys working with R, and figuring out algorithms and workflows is like solving puzzles. So she does the [[ABC-INT-Mutation_impact|R programming Integrator Unit]] next, in which she writes code that estimates what mutations in a gene can tell us about that gene's role involvement in a disease phenotype. Adding a small number of software-development focussed units and a bit more statistics gets her on the way. | |
| − | + | </td> | |
| − | + | <td>[[File:ABC-Scenario-1-Step-6.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-6.jpg]]</td> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <tr | ||
| − | <td | ||
| − | <td | ||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
| − | <tr | + | <tr> |
| − | <td | + | <td>Finally, as a kind of capstone, she completes the [[ABC-INT-Genome_annotation|genome annotation Integrator Unit]]. So she fill in the rest of the database units, the units on statistics and analysis of differential expression in genes, the units on networks and protein-protein interactions, and the concepts of protein functions, to complete the [[BIN-FUNC-Annotation]] milestone, ... |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</td> | </td> | ||
| − | <td | + | <td>[[File:ABC-Scenario-1-Step-7.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-7.jpg]]</td> |
| − | + | </tr> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | </td> | + | <tr> |
| + | <td>:... followed by the genomics units (BIN-Genome-...).</td> | ||
| + | <td>[[File:ABC-Scenario-1-Step-8.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-8.jpg]]</td> | ||
</tr> | </tr> | ||
| − | |||
| − | + | <tr> | |
| − | + | <td>And that's it. Units she hasn't covered were optional.</td> | |
| − | + | <td>[[File:ABC-Scenario-1-Step-final.jpg|thumb|300px|none|link=http://steipe.biochemistry.utoronto.ca/abc/assets/ABC-Scenario-1-Step-final.jpg]]</td> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <tr | ||
| − | <td | ||
| − | <td | ||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</table> | </table> | ||
| − | + | </div> | |
| − | + | </div> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | </ | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | {{Smallvspace}} | ||
---- | ---- | ||
| + | {{Smallvspace}} | ||
| − | + | <div class="toccolours mw-collapsible mw-collapsed" style="width:800px"> | |
| − | + | ;Nigel wants to sample different areas of the material broadly before he tackles any of the Integrator Units... | |
| − | + | <div class="mw-collapsible-content"> | |
| − | < | ||
| − | + | * Again: all paths through the learning units begin with the [[ABC-BIN-Preparation|Course Preparation]] milestone, and the [[RPR-Introduction|Introduction to R]] milestone. This is where he starts. | |
| + | * He targets the [[FND-STA-Information_theory|information theory unit]] first, for which he prepares the [[FND-CSC-Data_models|data models unit]] and chooses a [[BIN-YFO|sample organism]], to proceed to the [[BIN-Abstractions|bioinformatics abstractions unit]] and the [[BIN-Sequence|macromolecular sequence unit]]. | ||
| + | * This makes him curious about the relationship between information theory and [[BIN-SX-Molecular_forcefields|molecular forcefields]] so he learns about [[BIN-Databases|database principles]] and [[BIN-SX-Concepts|3D structure concepts]] next, to tackle the [[BIN-PDB|PDB database unit]] and the [[BIN-SX-Chimera|tutorial on the 3D structure viewer "UCSF Chimera"]]. | ||
| + | * Apparently another use of information theory is for quantifying the relatedness of functional annotations. Nigel completes the rest of the database cluster to proceed to learn about the [[BIN-FUNC-GO|Gene Ontology project]] to arrive at the [[BIN-FUNC-Semantic_similarity|"semantic similarity" unit]]. | ||
| + | * It interests Nigel how such fields of mathematics can be employed to study biology: | ||
| + | ** next he works through the statistics cluster (FND-STA-...) to learn about the concepts behind discovering differentially expressed genes, culminating in the [[RPR-GEO2R|GEO2R programming exercise]]; ... | ||
| + | ** ... and follows up with the [[BIN-PPI-Concepts|protein-protein interaction concepts]] via an [[FND-MAT-Graphs_and_networks|introduction to graph theory]]. | ||
| + | * This suggests for him to complete the missing units from the functional annotation cluster (BIN-SEQA-... and BIN-FUNC-...) to complete the [[BIN-FUNC-Annotation]] milestone. | ||
| + | * The greatest importance of functional annotation lies in annotating whole genomes. Nigel learns more about this next with the genomics cluster (BIN-Genome-...). | ||
| + | * He finds it intriguing how so much knowledge of function does not actually rely on the detailed, structural analysis of mechanism. To understand this better, Nigel works through the remainder of the structure units (BIN-SX-...) (for which he incidentally needs the [[BIN-ALI-Alignment]] unit.) | ||
| + | * Next, he realizes that much of what he has worked with so far has implicitly relied on sequence-alignment tools. So he completes the alignment cluster next (Bin-ALI-...). | ||
| + | * And realizing that multiple sequence alignments ([[BIN-ALI-MSA]]) are fundamental to phylogenetic analysis leads him to tackle the phylogenetic analysis cluster next (BIN-PHYLO-...). | ||
| + | * That's it for the learning units. Nigel then completes the four Integrator Units one after another, starting with the [[ABC-INT-Genome_annotation|genome annotation unit]] because that's the biggest one and he wants it safely submitted before the end-of-term rush in his other courses gets to him. | ||
| − | |||
| − | + | </div> | |
| + | </div> | ||
| + | {{Vspace}} | ||
| − | + | ====Timing==== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | You can work through the material entirely at your own pace. There are only two restrictions: a minimum amount of evaluations have to be submitted and marked before the {{dropdate}}, and everything needs to be done by the {{lastdate}}: | |
| − | + | * Work from learning units worth at least 10% of your final grade must have been submitted for evaluation by October 31, so that I can mark it before the {{dropdate}}. If you have not submitted enough work for evaluation by October 31, I will randomly choose an appropriate number of learning units and record a 0 for these. | |
| − | + | * All '''remaining course work''' must have been submitted by the {{lastdate}}. At least one of the Integrator Units will be evaluated in an oral test and there is a limited number of test-dates. There will not be a test-date in December. | |
| − | * | ||
| − | |||
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | ====Submission of items for marking==== | ||
| − | + | Details are listed with each evaluation unit, but in principle you create a separate sub-page of your user page and post your material there. You add an appropriate category tag to the page when it is ready to be evaluated and I can then easily find and mark your page. | |
| − | + | {{Vspace}} | |
| − | + | ====Class time==== | |
| + | Since most of the learning units include hands-on, practical components that you do on your own, we won't need to us class-time for textbook-like delivery of contents.There will be four main activities in class meetings: | ||
| + | * We will always take time for open discussion of topics as they arise. This will be driven by student input and feedback. | ||
| + | * I will discuss marking of some submissions I have received, to make the process more transparent. | ||
| + | * Some evaluations will include quizzes or presentations. These will happen in class on our dedicated "evaluation days". | ||
| + | * We will organize presentations by scientists in the field who will present aspects of their own work that are related to the course contents, e.g. how did this database or that algorithm contribute to solve a real problem in the lab. This will provide you with some sense about how the course material is meaningful in the real world. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Organizational Details== | |
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| + | ===Location=== | ||
| + | :[http://map.utoronto.ca/utsg/building/073 LM 161] (Lash Miller Building) | ||
| − | |||
| − | + | {{#lst:User:Boris|Student_Wiki}} | |
| + | {{#lst:User:Boris|Mailing_list}} | ||
| + | {{#lst:User:Boris|Office_hours}} | ||
| − | |||
| + | ===Prerequisites=== | ||
| + | Introductory courses to biochemistry and molecular biology provide the contents background to the course. Such might be obtained through the listed prerequiste courses: BCH210H1/BCH242Y1; BCH311H1/MGY311Y1/PSL350H1<ref>Please check the [http://www.artsandscience.utoronto.ca/ofr/calendar/crs_bch.htm#BCH441H1 '''official Calendar'''] for the academic year to confirm the "official" prerequisites.</ref>. However I have no way to assess your success in these courses, nor do I know what material actually '''was''' covered. Thus, I generally waive prerequisites. In all cases it is your responsibility to be sufficiently prepared and to make up for material that you have not covered previously. | ||
| − | + | A breakdown of knowledge that I expect you to acquire outside our course, or bring with you from previous courses, is [[FND-prerequisites|'''listed here''']]. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | You must have access to the Internet via your own computer. From time to time it may be necessary to bring your computer to class. If you do not have a laptop computer that is set up to work in the University's wireless network, contact me so we can figure out how to work around any issues. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | + | ===Exclusions & Enrolment controls=== | |
| − | + | :none | |
| − | |||
| − | |||
| + | {{Vspace}} | ||
| − | + | ===Printed material=== | |
| − | + | This is an '''electronic submission only''' course; but if you must print material, you might consider printing double-sided. Learn how, at the [http://printdoublesided.sa.utoronto.ca/ Print-Double-Sided Student Initiative]. Printing of course material is expressly discouraged since the material is updated frequently. | |
| − | === | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| − | |||
| − | |||
| + | == Resources == | ||
| + | ;Course framework | ||
| + | * [[ABC-Rubrics]]. | ||
| + | * [[FND-prerequisites]] | ||
| + | *The [http://steipe.biochemistry.utoronto.ca/abc/students Student Wiki]. | ||
| − | |||
| − | |||
;Course related | ;Course related | ||
| − | *The [http://groups.google.com/group/ | + | *The [http://groups.google.com/group/bch441_2017 2017 Course Google Group]. |
*The [http://steipe.biochemistry.utoronto.ca/abc/students Student Wiki]. | *The [http://steipe.biochemistry.utoronto.ca/abc/students Student Wiki]. | ||
| − | |||
| − | |||
*[http://www.writing.utoronto.ca/advice Writing advice] from the UofT Writing Centre <small>(including: how to avoid plagiarism)</small> | *[http://www.writing.utoronto.ca/advice Writing advice] from the UofT Writing Centre <small>(including: how to avoid plagiarism)</small> | ||
| − | |||
| − | + | <!-- | |
| − | |||
;Contents related | ;Contents related | ||
| + | *<small>Previous [[BCH441_Final_Exam|Final Exams]] (maybe less relevant since the format has changed in 2013)</small> | ||
*The [[UCSF Chimera]] tutorial | *The [[UCSF Chimera]] tutorial | ||
*The [[Stereo Vision]] tutorial | *The [[Stereo Vision]] tutorial | ||
| − | + | *The [[Aminoacid tutorial]] | |
| − | *The [[Database Identifiers]] tutorial | + | *The [[Database Identifiers]] tutorial |
*The [[R tutorial|Introduction to '''R''']] tutorial | *The [[R tutorial|Introduction to '''R''']] tutorial | ||
| Line 672: | Line 411: | ||
*[http://nar.oxfordjournals.org/content/44/W1.toc NAR July-2016 '''Web server''' issue] | *[http://nar.oxfordjournals.org/content/44/W1.toc NAR July-2016 '''Web server''' issue] | ||
<br> | <br> | ||
| + | --> | ||
{{Vspace}} | {{Vspace}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 13:24, 11 September 2017
BCH441 - Bioinformatics
Welcome to the BCH441 Course Wiki.
These wiki pages provide information and materials, and coordinate activities and projects in the introductory bioinformatics courses and other workshops taught by Boris Steipe at the University of Toronto. If you are not one of my students, you can still browse this site, however I can't provide support for your explorations. The material may be useful if you invest some effort in studying it systematically.
Welcome to the 2017 version of BCH441, which is being completely revised and reimagined.
Here is the current draft version of organizational details, activities and contents. The delivery, activities and assessments will comprise an entirely novel format. Updates will be continuously posted here.
First class-meeting: Tuesday, September 12, 17:00 in LM161.
The Course
BCH441 (BCH1441) is an introduction to current bioinformatics for life science students and the specialists in the BCB Program. The course provides an overview of the sources of biomolecular data, data annotation and integration, and the interpretation of results through evidence-based reasoning. This includes the components – sequence, structure, and function, the relationships in phylogeny and in the networks of interactions and regulation, and the “systems” through which we conceptually organize our knowledge.
Specific contents include:
- large, public biomolecular data resources,
- DNA and protein sequences and sequence analysis,
- pairwise and multiple sequence alignment,
- fast database searches to discover homologues,
- protein structure interpretation and homology modeling,
- phylogenetic analysis - tree building and interpretation,
- work with genome-scale data,
- functional annotation with Gene Ontology and other resources,
- relationships discovered through co-expression and protein-protein interactions, and
- introduction to systems-level concepts.
Practical, hands on tasks and assignments will introduce public data resources and analysis tools. Along with improving general computer literacy, you will learn to use the programming language and statistical workbench R, with a special emphasis on the kind of everyday tasks of data preparation and analysis that have become indispensable for any life-science laboratory. (Yes, you will learn to program.)
The course is complemented by BCB420 / JTB2020 (offered in the Winter Term) which consolidates aspects of computational systems biology in a project context.
BCH441H1F is the undergraduate course code.
BCH1441H1F is the cross-listed course code for graduate students.
Instructor
Dates
BCH441/BCH1441 is a Fall Term course; contact times are Tuesdays, 17:00 to 20:00. These are listed nominally as Tutorials T5 and Lectures T6-8, but we will use the time in variable configurations.
- First day of class – Tuesday, September 12. You must attend the first class.
- We need this time to go over the course delivery and organizational details, and get you an account on the Course Wiki and on the course mailing list. Your personal presence is a requirement of the course. Please do not enrol in the course if your travel- or other plans prevent you from attending the first class session.
Contents
In Fall 2017, the course will be taught for the first time following an entirely new concept: previous material has been decomposed into small "learning units" that are focussed more or less on a single concept. You work through the units independently, in any order that makes sense to you, at your own pace, all with the goal to acquire the knowledge and skills to work on four main "Integrator Units" that bring the contents together.
Through this, the course accommodates different levels of preparation more flexibly, probably makes your work more efficient, and implicitly teaches a number of meta-skills such as reporting and time-management. Be mindful though: this format requires a high level of self-motivation and responsibility to do well. In terms of aiming for the highest level of understanding and competence, you will frequently be on your own - just like you are in "real life". On the other hand, you certainly are the best judge of how well prepared you are. Thus there should be no surprises when your deliverables are evaluated.
Grading and Activities
This course comprises four key, integrative activities and preparatory "learning units" that lead up to them. Learning units can be completed in any sequence that makes sense to you, at any time until the deadline to submit material. But note that some learning units require evaluation on our "Evaluation Days" (see below), and/or scheduling a test, and that has to be done well in advance.
There are also a few restrictions on which units you must complete within this course:
- you must complete all four Integrator Units and submit them for marking. These will be worth maximally 40 marks (4 x 10);
- you can submit a mix of other learning units worth up to an additional 30 marks for marking. These are typically worth 6 marks each. There are a number of units available, which ones you choose is up to you.
- You must ensure that you have submitted units for evaluation that are worth at least 10% of your final grade by October 31, so they can be marked before the Template:Dropdate.
- 25% of your mark will be given for your Course Journal at the end of class.
- 5% of your mark will be given for your insights! page at the end of class.
Please carefully read the evaluation rubrics for each category of deliverables.
For graduate students (BCH1441), the marks you receive for learning units and Integrator Units will be scaled by 0.8, and 14 marks are available for your own design of a learning unit covering an aspect of your thesis project. Coordinate this with the instructor well in advance of the Template:Lastdate.
| Activity | Weight BCH441 - (Undergraduates) |
Weight BCH1441 - (Graduates) |
| Integrator Units | 40 marks | 32 marks |
| Your selection of other learning units | 30 marks | 24 marks |
| Course Journals | 30 marks | 30 marks |
| Graduate "Learning Unit Design" | 14 marks | |
| Total | 100 marks | 100 marks |
- A mix of evaluation methods
- Learning units will be evaluated with a mix of approaches including technical reports, documentation of results in your journal, delivery of R code, quizzes, oral tests and in-class presentations. Details are described in the individual units.
- Test dates
- Quizzes and in-class presentations will be scheduled on the following dates:
- October 3
- October 24
- November 14
- November 28
- Oral tests for Integrator Units will be scheduled on November 9 and 10, and November 23 and 24. These are Thursday and Friday dates and we will coordinate your test dates in October.
A final note on marking policy...
I do not adjust marks towards a target mean and variance (i.e. there will be no "belling" of grades), but follow the principles laid out in the marking rubrics. I feel strongly that "normalization" of grade interferes with a collaborative and mutually supportive learning environment. If your classmate gets a great mark because you helped him with a difficult concept, this should never have the effect that it brings down your mark because the class average is "belled-down" by the instructor. Collaborate as much as possible, it is a great way to learn.
But take *utmost* care to follow the instructions on avoiding plagiarism and academic misconduct to the letter, they will be rigorously enforced.
The learning-units map
Here is a thematic overview of the topical areas of this course's learning units:
And here is the detailed map. It contains links to all of the units.
- <command>-Click to open the Learning Units Map in a new tab, scale for detail.
- Hover over a learning unit to see its keywords.
- Click on a learning unit to open the associated page.
- The nodes of the learning unit network are colour-coded:
- Live units are green
- Units under development are light green. These are still in progress.
- Stubs (placeholders) are pale. These still need basic contents.
- Milestone units are blue. These collect a number of prerequisites to simplify the network.
- Integrator units are red. These embody the main goals of the course.
- Units that require revision are pale orange.
- Units that have a black border have deliverables that can be submitted for credit. Choose any you want to submit for credit, up to a maximum worth of 30 marks.
- Arrows point from a prerequisite unit to a unit that requires it.
(Unit status will be updated as in-progress units are completed.)
Everything starts with the following three units:
This should be the first learning unit you work with, since your Course Journal will be kept on a Wiki, as well as all other deliverables. This unit includes an introduction to authoring Wikitext and the structure of Wikis, in particular how different pages live in separate "Namespaces". The unit also covers the standard markup conventions - "Wikitext markup" - the same conventions that are used on Wikipedia - as well as some extensions that are specific to our Course- and Student Wiki. We also discuss page categories that help keep a Wiki organized, licensing under a Creative Commons Attribution license, and how to add licenses and other page components through template codes.
Keeping a journal is an essential task in a laboratory. To practice keeping a technical journal, you will document your activities as you are working through the material of the course. A significant part of your term grade will be given for this Course Journal. This unit introduces components and best practice for lab- and course journals and includes a wiki-source template to begin your own journal on the Student Wiki.
In paralell with your other work, you will maintain an insights! page on which you collect valuable insights and learning experiences of the course. Through this you ask yourself: what does this material mean - for the field, and for myself.
Everything leads to the Integrator Units. These cover four large areas of bioinformatics that make up the explicit goals of the course:
(i) algorithms and statistics;
(ii) structural modelling and interpretation;
iii) gene annotation; and;
(iv) phylogenetic analysis.
The knowledge and skills you need to work on these Integrator Units can be obtained from the other learning units that are shown on the learning units map as prerequisites. Note that "prerequisites" in this context does not mean you must do one thing before you can do another, the arrows simply point out which units assume what prior knowledge. You can acquire that knowledge in whatever sequence makes sense to you, and you don't have to learn from the learning units of this course at all. Just make sure that you submit enough general learning units for evaluation along the way. And document what you are doing in your Course Journal. Also, remember that all the material is cumulative - my evaluation of your work implicitly includes all of the prerequisite material.
Scenarios
Where to begin: possible scenarios for working though the units...
(These scenarios are for illustration, you don't have to follow these sequences. There is no implied claim that any of these sequences is better for learning the material than any other. Make your own choices!
- Yvette might have done a project about protein structure in a previous course...
| She decides to tackle the Homology modelling Integrator Unit first, because she is most confident about that material. | |
| :Obviously, all paths through the learning units begin with the units leading up to the Course Preparation milestone, as well as the Introduction to R milestone. This is where she starts. | |
:The homology modelling unit requires the cluster of protein structure units (BIN-SX-...), as well as the sequence alignment units (BIN-ALI-...);
|
|
| :For the sequence alignment cluster she needs BIN-Sequence and FND-Homology and both of these need BIN-Abstractions. | |
| Next she goes for the Phylogenetic Analysis Integrator Unit, since it requires only a small number of additional prerequisites and she's a bit busy at that time with midterms in other courses. Some introductory statistics (FND-STA-Probability and FND-STA-Bayes_theorem gets her on the way to the BIN-PHYLO-... cluster. | |
| She finds she enjoys working with R, and figuring out algorithms and workflows is like solving puzzles. So she does the R programming Integrator Unit next, in which she writes code that estimates what mutations in a gene can tell us about that gene's role involvement in a disease phenotype. Adding a small number of software-development focussed units and a bit more statistics gets her on the way. | |
| Finally, as a kind of capstone, she completes the genome annotation Integrator Unit. So she fill in the rest of the database units, the units on statistics and analysis of differential expression in genes, the units on networks and protein-protein interactions, and the concepts of protein functions, to complete the BIN-FUNC-Annotation milestone, ... | |
| :... followed by the genomics units (BIN-Genome-...). | |
| And that's it. Units she hasn't covered were optional. |
- Nigel wants to sample different areas of the material broadly before he tackles any of the Integrator Units...
- Again: all paths through the learning units begin with the Course Preparation milestone, and the Introduction to R milestone. This is where he starts.
- He targets the information theory unit first, for which he prepares the data models unit and chooses a sample organism, to proceed to the bioinformatics abstractions unit and the macromolecular sequence unit.
- This makes him curious about the relationship between information theory and molecular forcefields so he learns about database principles and 3D structure concepts next, to tackle the PDB database unit and the tutorial on the 3D structure viewer "UCSF Chimera".
- Apparently another use of information theory is for quantifying the relatedness of functional annotations. Nigel completes the rest of the database cluster to proceed to learn about the Gene Ontology project to arrive at the "semantic similarity" unit.
- It interests Nigel how such fields of mathematics can be employed to study biology:
- next he works through the statistics cluster (FND-STA-...) to learn about the concepts behind discovering differentially expressed genes, culminating in the GEO2R programming exercise; ...
- ... and follows up with the protein-protein interaction concepts via an introduction to graph theory.
- This suggests for him to complete the missing units from the functional annotation cluster (BIN-SEQA-... and BIN-FUNC-...) to complete the BIN-FUNC-Annotation milestone.
- The greatest importance of functional annotation lies in annotating whole genomes. Nigel learns more about this next with the genomics cluster (BIN-Genome-...).
- He finds it intriguing how so much knowledge of function does not actually rely on the detailed, structural analysis of mechanism. To understand this better, Nigel works through the remainder of the structure units (BIN-SX-...) (for which he incidentally needs the BIN-ALI-Alignment unit.)
- Next, he realizes that much of what he has worked with so far has implicitly relied on sequence-alignment tools. So he completes the alignment cluster next (Bin-ALI-...).
- And realizing that multiple sequence alignments (BIN-ALI-MSA) are fundamental to phylogenetic analysis leads him to tackle the phylogenetic analysis cluster next (BIN-PHYLO-...).
- That's it for the learning units. Nigel then completes the four Integrator Units one after another, starting with the genome annotation unit because that's the biggest one and he wants it safely submitted before the end-of-term rush in his other courses gets to him.
Timing
You can work through the material entirely at your own pace. There are only two restrictions: a minimum amount of evaluations have to be submitted and marked before the Template:Dropdate, and everything needs to be done by the Template:Lastdate:
- Work from learning units worth at least 10% of your final grade must have been submitted for evaluation by October 31, so that I can mark it before the Template:Dropdate. If you have not submitted enough work for evaluation by October 31, I will randomly choose an appropriate number of learning units and record a 0 for these.
- All remaining course work must have been submitted by the Template:Lastdate. At least one of the Integrator Units will be evaluated in an oral test and there is a limited number of test-dates. There will not be a test-date in December.
Submission of items for marking
Details are listed with each evaluation unit, but in principle you create a separate sub-page of your user page and post your material there. You add an appropriate category tag to the page when it is ready to be evaluated and I can then easily find and mark your page.
Class time
Since most of the learning units include hands-on, practical components that you do on your own, we won't need to us class-time for textbook-like delivery of contents.There will be four main activities in class meetings:
- We will always take time for open discussion of topics as they arise. This will be driven by student input and feedback.
- I will discuss marking of some submissions I have received, to make the process more transparent.
- Some evaluations will include quizzes or presentations. These will happen in class on our dedicated "evaluation days".
- We will organize presentations by scientists in the field who will present aspects of their own work that are related to the course contents, e.g. how did this database or that algorithm contribute to solve a real problem in the lab. This will provide you with some sense about how the course material is meaningful in the real world.
Organizational Details
Location
- LM 161 (Lash Miller Building)
Student Wiki
Many of the class activities will take place interactively on a separate Wiki site (the "Student Wiki"). You will create a personalized user page there, and use it to submit materials as required.
This Wiki is not accessible to the general public, you need an account that we will be registered after the first class-session.
Contact
Course communication will take place on the Quercus discussion section. We'll see how this goes. If it's not suitable for our needs we'll find an alternative.
Office hours
(Virtual) face to face meetings are by appointment, if required. However, we will be able to resolve almost all issues by e-mail. You will find that discussions by e-mail are both more efficient and effective than meetings. Moreover e-mail discussions leave you with a document trail of what was discussed, can contain links to information sources, and we can share points of general interest more easily with the class.
Prerequisites
Introductory courses to biochemistry and molecular biology provide the contents background to the course. Such might be obtained through the listed prerequiste courses: BCH210H1/BCH242Y1; BCH311H1/MGY311Y1/PSL350H1[1]. However I have no way to assess your success in these courses, nor do I know what material actually was covered. Thus, I generally waive prerequisites. In all cases it is your responsibility to be sufficiently prepared and to make up for material that you have not covered previously.
A breakdown of knowledge that I expect you to acquire outside our course, or bring with you from previous courses, is listed here.
You must have access to the Internet via your own computer. From time to time it may be necessary to bring your computer to class. If you do not have a laptop computer that is set up to work in the University's wireless network, contact me so we can figure out how to work around any issues.
Exclusions & Enrolment controls
- none
Printed material
This is an electronic submission only course; but if you must print material, you might consider printing double-sided. Learn how, at the Print-Double-Sided Student Initiative. Printing of course material is expressly discouraged since the material is updated frequently.
Resources
- Course framework
- Course related
- The 2017 Course Google Group.
- The Student Wiki.
- Writing advice from the UofT Writing Centre (including: how to avoid plagiarism)
Notes
- ↑ Please check the official Calendar for the academic year to confirm the "official" prerequisites.