CSB Web tools
CSB on the Web
This page is a placeholder, or under current development; it is here principally to establish the logical framework of the site. The material on this page is correct, but incomplete.
Important tools and resources for CSB, available on the Web.
Contents
...
Introductory reading
Exercises
References
Further reading and resources
| The NCBI Gene database [ link ] [ page ] Gene is the NCBI's integrated database of gene information in the Entrez system. Records may include Reference Sequences (RefSeqs), maps, pathways, variations, phenotypes, compiled into the database itself, and links to genome-, phenotype-, and locus-specific resources worldwide. The URL links to the record for the human E2F1 transcription factor. For detailed information, see the Gene database information page. | 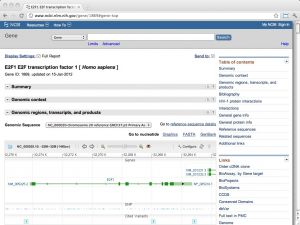 |
| UniProt [ link ] [ page ] UniProt is the protein sequence database of the European Bioinformatics Institute. It is an extraordinarily well constructed, curated, and integrated resource. As a public resource, its results are freely accessible world-wide. The "Knowledge Base" (UniProtKB), which is the database proper, contains two subsections: SwissProt, the manually curated and heavily annotated protein sequence repository; it is approximately equivalent to the NCBI Refseq protein database, albeit with usually higher annotation levels. TrEMBL is much larger and contains sequences that have been computationally translated from the EMBL nucleotide sequence collection. It is approximately equivalent to the NCBI's Entrez protein database. The URL links to the entry for the Saccharomyces cerevisiae cell-cycle regulation transcription factor Mbp1. | 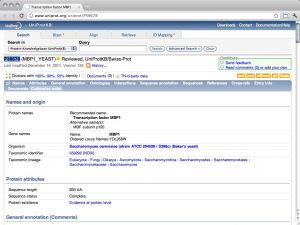 |
| SGD: Saccharomyces Genome Database [ link ] [ page ] The Saccharomyces genome database is a curated database that integrates sequence, structure and function information for yeast molecular biology. It is one of the important model organism databases and can be considered a paradigm for the entire field. The url links to the information page of the cell-cycle regulation transcription factor Mbp1. | 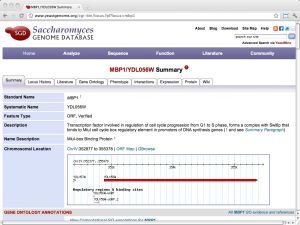 |
| MGI (Mouse Genome Informatics) [ link ] [ page ] The model organism database MGI (Mouse Genome Informatics) is the primary community database resource for the laboratory mouse. It integrates genomics, expression, tumor biology and metabolism information and actively curates GO annotations for mouse genes. The stated goal is to enhance the utility of mouse research for the study of human health and disease. For example, wherever available, human orthologues are cross-referenced with the respective mouse genes. The URL links to the gene details of the mouse orthologue of human E2F1. | 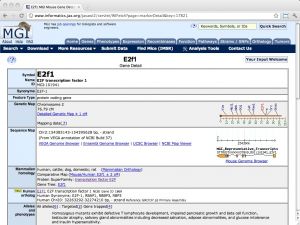 |
|
GO: the Gene Ontology project [ link ] [ page ] Ontologies are important tools to organize and compute with non-standardized information, such as gene annotations. The Gene Ontology project (GO) constructs ontologies for gene and gene product attributes across numerous species. Three major ontologies are being developed: molecular process, biological function and cellular location. Each includes terms, their definition, and their relationships. In addition, genes and gene products are being been annotated with their GO terms and the type of evidence that underlies the annotation. A number of tools such as the AmiGO browser are available to analyse relationships, construct ontologies and curate annotations. Data can be freely downloaded in formats that are convenient for computation. |  |
| The Gene Wiki project [ link ] [ page ] The Gene Wiki project aims to create Wikipedia articles for every human gene whose function has been assigned. This provides pages that are ideally suited for free, community-driven, integrated information resources. Access to the project is through the Gene Wiki Portal, which contains guidelines for contributors. The pages are easy to find since they are linked to the HGNC recognized gene name. For example, the URL links to the human E2F1 transcription factor page. |  |
| Gene/Protein Synonym Database [ link ] [ page ] The ExPASy hosted Gene/Protein Synonym Database collects gene name synonyms from the majority of model organism databases and UniProt, cross-references them and provides a searchable interface. |  |
| HUGO Gene Nomenclature Committee [ link ] [ page ] The HUGO Gene Nomenclature Committee (HGNC) has assigned unique gene symbols and names to more than 32,000 human loci, of which over 19,000 are protein coding. genenames.org is a curated online repository of HGNC-approved gene nomenclature and associated resources including links to genomic, proteomic and phenotypic information, as well as dedicated gene family pages. This site is the definitive resource to resolve gene name ambiguities. The URL links to the search results for Rbp3, which is both a deprecated synonym for the human E2F transcription factor 1, and the official name of retinol binding protein 3. |  |
| Reactome [ link ] [ page ] Reactome is a multi-site collaboration to develop an open source, curated bioinformatics database of human pathways and reactions. It includes annotations, pathways and tools for pathway browsing and analysis, including pathway assignment and overrepresentation analysis of user-supplied data sets. Making use of orthology prediction, Reactome also provides cross-species pathway inference for a large number of model organisms. The URL accesses the E2F mediated regulation of DNA replication. |  |
| GMOD Generic Model Organism Database project [ link ] [ page ] GMOD (the Generic Model Organism Database project), is a collection of open source software tools for creating and managing genome-scale biological databases. GMOD tools are in use at many large and small community databases, especially for Model Organisms. The include the genome browser GBrowse, the CHADO relational database, the GFF annotation databases, and much more The goal is to free developers of community scale biomolecualr databases from reinventing the wheel. A good overview of resources and principles is available on the GMOD wiki. |  |
| NAR database issue [ link ] [ page ] Every year the journal Nucleic Acids Research (NAR) compiles a special issue on important databases in molecular biology (in January), and on important webservers and other resources (in July). The articles are peer-reviewed, and inclusion into the issue is considered a quality endorsement. Both volumes reflect the best practices in the field, as well as its rapidly changing nature. Links to databases and resources are searchable by keyword and topic in the bioinformatics.ca links directory. |  |
| NAR Web Server issue [ link ] [ page ] Every year the journal Nucleic Acids Research (NAR) compiles a special issue on important webservers in molecular biology (in July), and on important databases (in January). The articles are peer-reviewed, and inclusion into the issue is considered a quality endorsement. Both volumes reflect the best practices in the field, as well as its rapidly changing nature. Links to databases and resources are searchable by keyword and topic in the bioinformatics.ca links directory. |  |
| Links directory (bioinformatics.ca) [ link ] [ page ] bioinformatics.ca is the domain of the Canadian Bioinformatics Workshops, currently hosted by the Ontario Institute of Cancer research. The links directory is a curated collection of databases and services that are useful for bioinformatics and computational biology. Links are browsable in several categories, such as Model Organisms, Expression or Sequence Comparison with many subcategories. Importantly, the site contains links to all resources from the NAR database issues and the NAR web server issues in a searchable interface. The URL links to a search for the term "Systems Biology". |  |