BIN-SX-Small molecules
"Small Molecule" Structure"
(A small-molecule structure tutorial)
Abstract:
Creating with small molecule structures, finding complexes in the PDB that contain the molecule, and superimposing model and structure.
|
Objectives:
|
Outcomes:
|
Deliverables:
Prerequisites:
You need the following preparation before beginning this unit. If you are not familiar with this material from courses you took previously, you need to prepare yourself from other information sources:
- Biomolecules: The molecules of life; nucleic acids and amino acids; the genetic code; protein folding; post-translational modifications and protein biochemistry; membrane proteins; biological function.
This unit builds on material covered in the following prerequisite units:
Contents
Evaluation
This learning unit can be evaluated for a maximum of 5 marks. To submit material for credit for this unit ...
- Create a new page on the student Wiki as a subpage of your User Page.
- Put all of your writing to submit on this one page.
- When you are done with everything, go to the Quercus Assignments page and open the first Learning Unit that you have not submitted yet. Paste the URL of your Wiki page into the form, and click on Submit Assignment.
Your link can be submitted only once and not edited. But you may change your Wiki page at any time. However only the last version before the due date will be marked. All later edits will be silently ignored.
- Short report option
- 1. Visit the small-molecule selection page on the Student Wiki and choose one small molecule to work with.
- 2. Summarize what this molecule is. Draw the structure of the molecule in one of the molecular editors described in the unit and produce a SMILES string. Find a suitable PDB complex structure and superimpose your model in ChimeraX. Report on the quality of the superposition and create an informative stereo image that illustrates how your model is situated in the ligand binding site relative to the experimentally determined compound.
- Perform all visualization and analysis on the command line (if you use the menu, the "log" will record the command line equivalent and you can copy it from there) and record your commands in a section of your report. The visualization must be fully reproducible from your documentation, i.e. when I copy/paste your commands, I need to get the same image that you are showing (except for rotation/translation/scaling).
- 3. When you are done, submit the link to your page via Quercus as described above.
Contents
Task:
- Read the introductory notes on working with "small molecule" structure.
Modeling small molecules
"Small" molecules are solvent, ligands, substrates, products, prosthetic groups, drugs - in short, essentially everything that is not made by DNA-, RNA-polymerases or the ribosome. Whereas the biopolymers are still front and centre in our quest to understand molecular biology, small molecules are crucial for our quest to interact with the inventory of the cell, create useful products, or advance medicine.
A number of public repositories make small-molecule information available, such as PubChem at the NCBI, the ligand collection at the PDB, the ChEBI database at the European Bioinformatics Institute, the Canadian DrugBank, or the NCI database browser at the US National Cancer Institute. One general way to export topology information from these services is to use SMILES strings—a shorthand notation for the composition and topology of chemical compounds.
Task:
- Caffeine at PubChem
- Access PubChem.
- Enter "caffeine" as a search term in the Compound tab. A number of matches to this keyword search are returned.
- Click on the top hit - 1,3,7-Trimethylxanthine, the Caffeine molecule. Note that the page contains among other items:
- A 2D structural sketch;
- An idealized 3D structural conformer, for which you can download coordinates in several formats;
- The IUPAC name: 1,3,7-trimethylpurine-2,6-dione;
- The CAS identifier
58-08-2which is a unique identifier and can be used as a cross-reference ID; - The SMILES strings
CN1C=NC2=C1C(=O)N(C(=O)N2C)C; - ... and much more.
Task:
- Caffeine at DrugBank
- Access DrugBank.
- Enter "Caffeine" in the search form and. .
- Click on the hit to "Caffeine" itself. Note that the page contains among other items:
- A detailed description
- A 2D structural sketch with a link to 3D options;
- Synonyms, including the IUPAC name: 1,3,7-trimethylpurine-2,6-dione;
- ... and much more.
- Follow the link to the 3D options and note the options for downloading information, including the SMILES string and PDB formatted coordinates.
That's great, but let's sketch our own version of caffeine. Several versions of Peter Ertl's Java Molecular Editor (JME) are offered online, PubChem offers this functionality via its Sketcher tool and the PDB has a similar sketching tool on its ligand search page]
Task:
- Return to the PubChem homepage.
- Click on Draw Structure.
- Sketch the structure of caffeine. I find the editor quite intuitive but clicking on the Help button will give you a quick, structured overview. Make sure you define your double-bonds correctly.
- Export the SMILES string of your compound to your project folder.
Translating SMILES to structure
ChimeraX can translate SMILES strings to coordinates[1].
Task:
- Open ChimeraX.
- Select Tools → Structure Editing → Build Structure.
- In the Build Structure window, select the SMILES string button, paste the string from your file, and click Apply.
- The caffeine molecule will be generated and visualized in the graphics window. This is a "stick" representation.
- You can rotate it with your mouse, pinch to scale, <shift> drag to translate.
- Use the Actions → Atoms/Bonds → ball & stick or sphere menu items to change appearance.
- Use the Actions → Color → by element menu to change colors.
- Change the display back to stick and use Actions → Surface → show to add a solvent accessible surface. Choosing this command triggers the calculation of the surface, which is then available as an individually selectable object. With default pramaters, the surface is a bit rough for this small molecule. Type
surface gridSpacing 0.1to increase the resolution five-fold from its default 0.5A. - By default, the surface inherits the colour of the atoms it envelopes. To change the colour of the surface, use the Actions → Color → all options menu. Click the surfaces button to indicate that the color choice should be applied to the surface object (note what else you can apply color to...), then choose cornflower blue.
- Use the Actions → Surface → transparency → 50% menu to see atoms and bonds that are covered by the surface. I find a soft lighting usually works best:
lighting soft - To begin working with molecules in "true" 3D, type
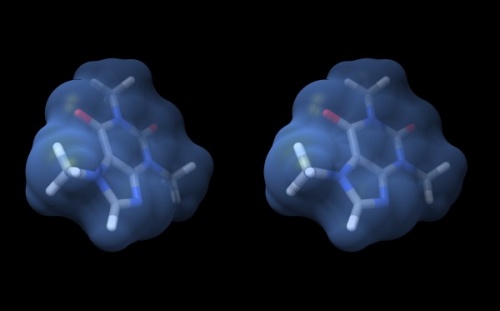
camera sbs. - Your structure should look somthing like what you see below.
Superposition
To investigate a small molecule structure variant in the context of a complex, we need to superimpose it with an existing ligand.
Task:
- Open the structure 3G6M in ChimeraX. This is one of the hits returned from the PDB search for caffeine - a fungal chitinase for which caffeine is a potent inhibitor.
- Choose Select → Residue → CFF (CFF is th PDB three-letter code for this hetero compound), then Select → Invert (selected models), Actions → Atoms/Bonds → hide and Actions → Cartoon → hide to show only the caffeine molecules - there are two. Select the one with residue ID 1, and again Actions → Atoms/Bonds → hide. The remaining CFF molecule has residue ID 427.
To superimpose the structures, we can't use the standard "match" option, because that only works for protein or DNA molecules. Instead, we need to explicitly define matching pairs of atoms through ChimeraX's command line interface. The command line interface is a very powerful way to issue ChimeraX commands, but it has a bit of a learning curve since we need to use a precise model/residue/atom selection syntax.
- Open the User hep page and study the select command options. Note how models are specified with a "#" sigil, residues with a ":" or "::" sigil, and atoms with an "@" sigil.
- The command we need is align, and we need to feed the command atoms in exactly the order of the pairs that the superposition algorithm should superimpose. To identify the atom numbers, we can hover over them with the mouse, or we can select the residue/atom and choose Actions → Label → name. If we superimpose the four nitrogen atoms, the correct command may be:
align #1@N1,N2,N4,N3 to #2:427@N3,N1,N7,N9to superimpose the model we built from the SMILES string onto the structure - but the exact atom names in the model structure depend on how the SMILES string was written. - Note how the two structures are virtually identical - in this case, there are only very small coordinate differences because the conformational degrees of freedom are very much constrained in the xanthin heterocycle. But there are differences nevertheless. One molecule is an idealized structure, the other a structure that has been determined by a high-resolution experiment.
- Turn the protein structure back on, and study how the ligand is bound.
Further reading, links and resources
Notes
- ↑ There are also several online servers that translate SMILES strings to idealized structures, for example the online SMILES translation service at the NCI.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2020-10-07
Version:
- 1.2
Version history:
- 1.2 Edit policy update
- 1.1 2020 Updates - rewrite for changed Websites and using ChimeraX
- 1.0 First live version
- 0.1 First stub
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.