Software Development for Research Labs
Boris Steipe
|
Expected Preparations:
|
|||||||
|
|
|||||||
| Keywords: Software development for the research lab | |||||||
|
|
|||||||
|
Objectives:
This unit will …
|
Outcomes:
After working through this unit you …
|
||||||
|
|
|||||||
|
Deliverables: Time management: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit. Journal: Document your progress in your Course Journal. Some tasks may ask you to include specific items in your journal. Don’t overlook these. Insights: If you find something particularly noteworthy about this unit, make a note in your insights! page. |
|||||||
|
|
|||||||
|
Evaluation: NA: This unit is not evaluated for course marks. |
|||||||
Contents
Developing software means much more than writing correct code. It is an engineering process that includes anumber of stakeholders and needs to satisfy a variety of objectives. Here we discuss the process from the perspective of a research lab in molecular or computational biology.
One can make an argument that the creation of software is the most impactful human cultural achievement to date. But writing software well is not easy and much sophisticated methodology has been proposed for software development, primarily addressing the needs of large software companies and enterprise-scale systems. Certainly: once software development becomes the task of teams, and systems become larger than what one person can confidently remember, failure is virtually guaranteed if the task is not organized in a structured way.
But our work often does not fit this paradigm, because in the bioinformatics lab - as in any research lab - the requirements are constantly in flux. The reason is obvious: most of what we produce in science are one-off solutions. Once one analysis runs, we publish the results, and we move on. There is limited value in doing an analysis over and over again. However, this does not mean we can’t profit from applying the basic principles of good development principles. Fortunately that is easy. There actually is only one principle.
Make implicit knowledge explicit.
Everything else follows.
Collaborate
Making project goals explicit and making progress explicit is crucial, everyone has to know what’s going on and what their responsibilities are. Collaboration these days is distributed, and online. Here is a list of options you ought to have tried. :
- Schedule regular face-to-face meetings. If you can’t be in the same room, Google hangout’s may work. Old-time developers often use IRC(W) chat rooms - and newer teams might use discord.
- Zoom, obviously.
- A widely used, integrated tool for chat-type team interactions is slack.
- Software sharing and code collaboration is best managed through a github repository. As a version control system, git is an essential part of all software development.
- Dynamic coauthoring capabilities are offered by Etherpad(W) and its derivatives.
- You have probably collaborated with Google Drive text, spreadsheet, drawings, or presentation documents before. These have superseded many other online offerings.
- A robust system for shared file storage is Dropbox.
- Trello appears to be a nice tool to distribute work-packages and keep up to date with discussions, especially if your “team” is distributed.
There is a lot of turnover with all of these tools and it’s important to backup your important data to alternate locations. Offerings may go out of date, or get absorbed by other services. Are there collaboration tools you like that I haven’t mentioned? Let me know …
Plan
The planning stage involves defining the goals and endpoints of the project. We usually start out with a vague idea of something we would like to achieve. We need to define: * where we are; * where we want to be; * and how we will get there.
For an example of a plan, refer to the 2015 BCB420 Class Project. There, we lay out a plan in three phases: Preparation, Implementation and Results. This is generic, the preparation phase implies an analysis of the problem, which focusses on what will be accomplished, independent of how this will be done. The results of the analysis can be a requirements document (here is a link to a Requirements template - a template to jumpstart such a document) or a less formal collection of goals.
The most important achievement of the plan is to break down the project into manageable parts and define the Milestones that characterize the completion of each part1.
Design
Software Design(W) encompasses a range of activities: conceptualizing the system, defining requirements, structuring components and interfaces, and providing roadmaps for deployment and maintenance. All these are necessary, but none of these matter if you are solving the wrong problem.
In the problems we deal with in a bioinformatics lab, it is a good idea to folllow an architecture(W) centric design process: we explore the requirements with the specific goal of drafting an overall architecture, and we draw up a detailed architecture to make the components and their interfaces explicit. Typically this will involve some modelling and there are different ways to model a system.
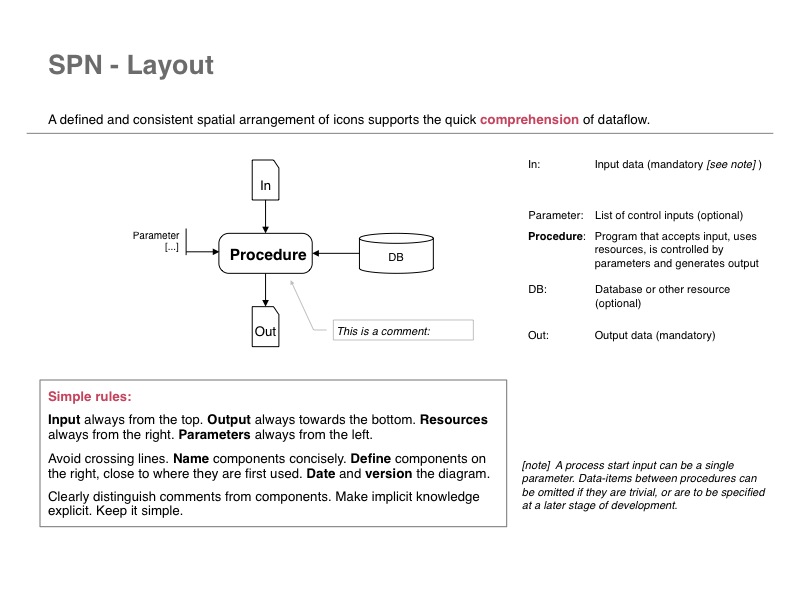
SPN (Structured Process Notation) is one way to define dataflow diagrams for data-driven analysis in bioinformatics. (Introduction_to_SPN-01.jpg)
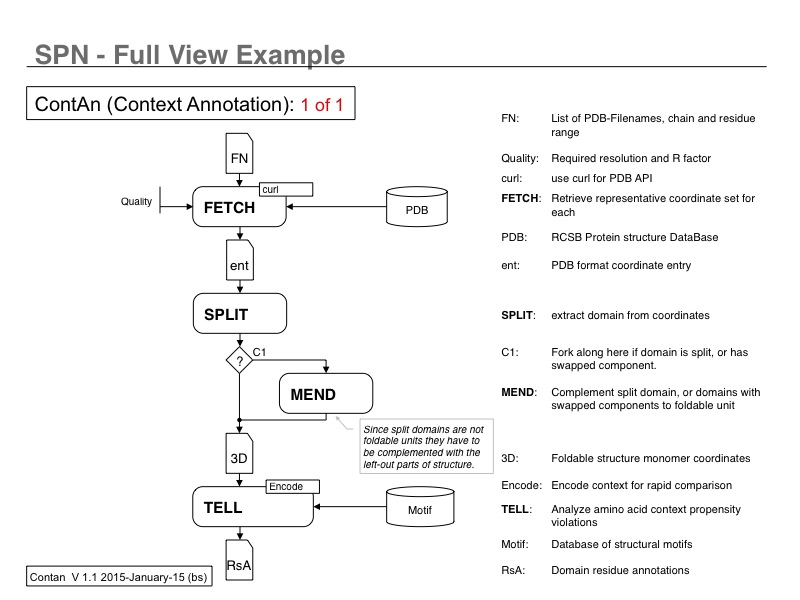
In this example SPN icons are integrated to describe a workflow that annotates protein structure domains at the residue level. (Introduction_to_SPN-02.jpg)
- Structural modelling describes the components and interfaces. The components are typically pieces of software, the interfaces are “contracts” that describe how information passes from one piece to another. Structural models include the Data model that captures how data reflects reality and how reality changes the data in our system;
- Behaviour modelling describes the state changes of our system, how it responds to input and how data flows through the system. In data-driven analysis, a data flow diagram may capture most of what is important about the system.
- (SPN - Structured Process Notation) is a data flow diagram method that I find particularly suited to the kind of code that we often write in bioinformatics: integration of data, transformation, and analytics, all passing through well defined intermediate stages.
Whichever modelling method is adopted, make sure that it is explicit. Many aspects of the widely used UML(W) standard suffer from poor information-design; this can lead to hard-to-understand graphics that obfuscate rather than clarify the issues2.
The design objectives have to be clearly articulated. There are many ways to achieve the same end result, but good design will support the creation of systems that are correct, robust, extensible and maintainable. A very good starting point for the design is a proper separation of concerns(W). The DRY (Do not Repeat Yourself) principle has proven its worth over long time.
More reading
Develop
In the development phase, we actually build our system. It is a misunderstanding if you believe most time will be spent in this phase. Designing a system well is hard. Building it, if it is well designed, is easy. Building it if it is poorly designed is probably impossible.
You may get away with ad hoc methodology if you work alone - at least as long as the complexity of your project is very limited. But even if there is just one co-developer,3 development becomes a totally different story. You need a plan, you need documentation, and you need methodology.
A number of development methodologies and philosophies have been proposed, and they go in and out of fashion. Three successful ideas are Agile development(W), TDD (Test Driven Development)(W) and Literate Programming.
Version Control
Non negotiable. You must develop under version control, and you must understand how to undo changes (including how to understand which point you need to backtrack to).
- Software Carpentry Git Introduction
- Git tutorials
- Introduction to GitHub
- Jenny Bryan’s “Happy Git With R”
You need a mental model of the graph-abstraction that git applies to the development process, the roles that individuals play in the process, and what terms like “forking a repository”, “pull request”, and “merging a commit” mean.
If several people are writing and commiting code, you need some form of continuous integration. On GitHub, CI is part of a larger support framework: CitHub Actions and a wealth of documentation is available.
Fail Safe or Fail Fast?
Testing for correct input is a crucial task for every function, and R especially goes out of its way to coerce input to the type that is needed. This is making functions fail safe. Do consider the opposite philosophy however: “fail fast”, to produce fragile code. You must test anyway whether input is correct, but a good argument can be made that incorrect input should not be fixed, but the function should stop the program and complain loudly and explicitly about what went wrong.4 This - once again - makes implicit knowledge explicit, it helps the caller of the function to understand how to pass correct input, and it prevents code from executing on wrong assumptions. In fact, failing fast may be the real fail safe.
Code
Here is a small list of miscellaneous best-practice items for the phase when actual code is being written:
- Be organized. Keep your files in well-named folders and give your file names some thought.
- Use version control: git is your friend.
- Use an IDE (Integrated Development Environment). Syntax highlighting and code autocompletion are nice, but good support for debugging, especially stepping through code and examining variables, setting breakpoints and conditional breakpoints are essential for development. For Rdevelopment, the R Studio environment is the gold standard, it provides syntax highlighting, a symbolic debugger and it integrates with git and github. If you want AI support (CoPilot) you’ll need to go with Visual Studio until R Studio catches up.
- Design your code to be easily extensible and only loosely coupled. Your requirements will change frequently, make sure your code is modular and nimble to change with your requirements.
- Design reusable code. This may include standardized interface conventions and separating options and operands well.
- DRY (Don’t repeat yourself): create functions or subroutines for tasks that need to be repeated. Or, seen from the other way around: whenever you find yourself repeating code, it’s time to delegate that to a function instead. Incidentally, that also holds for database design: whenever you find yourself intersecting tables with similar patterns, perhaps it’s a good idea to generalize one of the tables and merge the data from the other into it.
- KISS (Keep it simple): resist the temptation for particularly “elegant” language idioms and terse code. “Elegant” idioms are hard to translate into other languages in our multi-language environments, and they are hard to understand by collaborators whose first language may be a different one. Brian Kernighan once wrote: ““Debugging is twice as hard as writing the code in the first place. Therefore, if you write the code as cleverly as possible, you are, by definition, not smart enough to debug it.” Words of wisdom.
- Comment your code. I can’t repeat that often enough. Code is read very much more often than it is written. Unfortunately (for you) the one most likely to have to read and understand your convoluted code is you yourself, half a year later. So do yourself the favour to explain what you are thinking. Don’t comment on what the code does - that ought to be obvious from the code itself - but why you do something the way you do.
- Be consistent.
Keeping Code organized
I have probably mentioned this several time over, but we don’t develop with text-editors, and we don’t code on the command line, even though we could. In order to keep code organized, there are several options (I’m writing this from an R perspective, but you can extrapolate.):
- Command line
- Quick to try out simple ideas and syntax. Commands are stored in “history” and you can navigate through previous commands with the up-arrow and down-arrow key. Major drawback: this is a volatile way to structure your work and you don’t have a proper record of what you are doing.
- Script files
-
Probably the most general approach. Keep everything in one file. This
makes it easy to edit, version, maintain and share code. Most
importantly, you have a record of what you were doing (Reproducible
Research FTW). When you write a paper, write a script file for every
figure and make sure it runs from start to end when you
source()it. You’ll be amazed how often this saves your sanity. - R markdown
- Literate programming. We’ve mentioned it above.
- Shiny apps
- Shiny is a way to build dynamic Web pages that execute embedded R code. See here for more information.
- Jupyter notebooks
- Project Jupyter is a platform to embed code, plots and documentation. It has great advantages and a large following. Be sure to understand what it does. Not everyone agrees with the hype though read here about some of the downsides. The fact that you can’t execute single lines but need to work through code chunk by chunk breaks it for me. This is why I work with “Projects”.
- RStudio projects.
- We use RStudio anyway, since it’s an excellent IDE for R. With projects, we can package code and data into a self contained bundle. Code is available as source and we can edit and execute at will. Most importantly, we can put it all under version control, locally and on github. I think this is the perfect combination. Why don’t we use R notebooks instead? There is a single, all-important reason. Notebooks execute code in chunks, but for teaching/learning purposes it is crucial to be able to step through code line by line, expression by expression.
- R notebooks.
- R notebook authoring is integrated with RStudio capabilities; R notebooks work like Jupyter notebooks.
- https://support.rstudio.com/hc/en-us/articles/200532077-Version-Control-with-Git-and-SVN
- https://support.rstudio.com/hc/en-us/articles/200526207-Using-Projects
Coding style
It should always be your goal to code as clearly and explicitly as possible. Pace yourself, and make sure your reader can follow your flow of thought. More often than not the poor soul who will be confused by a particularly “witty” use of the language will be you, yourself, half a year later.
Coding style has its own learning unit.
Code Reviews
Regularly meet to review other team member’s code. Keep the meetings short, and restrict to no more than 300 lines at a time.
- The code author walks everyone through the code and explains the architecture(W) and the design decisions(W). It should not be necessary to explain what the code does - if such explanation is necessary, that points to opportunities for improvement.
- Briefly consider improvements to coding style as suggestions but don’t spend too much time on them (don’t create a “Bicycle Shed(W)” anti-pattern) - style is not the most important thing about the review. Be constructive and nice - you should encourage your colleagues, not demotivate them.
- Spend most of the time discussing architecture: how does this code fit into the general lay of the land? How would it need to change if its context changes? Is it sufficiently modular to survive? What does it depend on? What depends on it? Does it apply a particular design pattern(W)? Should it? Or has it devolved into an anti-pattern(W)?
- Focus on tests. What is the most dangerous error for the system integrity that the code under review could produce. Are there tests that validate how the code deals with this? Are there tests for the edge cases(W) and corner cases(W)5? This is the best part about the review: bring everyone in the room on board of the real objectives of the project, by considering how one component contributes to it.
- Finally, what’s your gut feeling about the code: is there Code Smell(W)? Are there suboptimal design decisions that perhaps don’t seem very critical at the moment but that could later turn into inappropriate technical debt(W)? Perhaps some refactoring(W) is indicated; solving the same problem again often leads to vastly improved strategies.
However be mindful that code review is a sensitive social issue, and that the primary objective is not to point out errors, but to improve the entire team.
QA
The importance of explicit, structured, proactive QA (Quality Assurance) is all too often not sufficiently appreciated. In my experience this is the single most important reason for projects that ultimately fail to live up to their expectations.
QA needs to keep track of assets that need to be created.
- Is the plan complete or are there gaps?
- Do all components and critical steps have clearly defined milestones?
- Is there a timeline?
- Is the project on track?
QA needs to keep track of assets that exist.
- Has the design been documented to acceptable standards?
- Does the design address the requirements?
- Have the interfaces between modules been specified?
- Is the code written to acceptable standards, is it sufficiently commented, properly modularized?
- Have code reviews been organized?
- Is the code correct? Have test cases been designed (unit tests, integration tests) and has the code been tested? Do runs with true- and false- positives give the expected results? Do comparisons against benchmarks achieve results within acceptable tolerance?
And for both -
- Is the project being documented?
Adding QA ad hoc, as an afterthought is a bad idea. QA makes a project great when coordinated by a capable individual, who catalyzes the whole team to do their best in an uncompromising dedication to excellence.
Deploy and Maintain
In our context, deployment may mean a single run of discovery, and maintenance may be superfluous as the research agenda moves on.
But this does not mean we should ignore best practice in scientific software development: simple, but essential aspects like using version control for our code, using IDEs, writing test cases for all code functions etc. These aspects are very well covered in the open source Software Carpentry project and courses. Free, online, accessible and to the point. Go there and learn:
Further Reading
PracticeUrs Enzler’s “Clean Code Cheatsheet” (at planetgeek.ch) Oriented towards OO developers, but sound principles nevertheless.
Wilson,
Greg et al.. (2014). “Best practices for scientific
computing”. Plos Biology 12(1):e1001745 .
[PMID: 24415924]
[DOI: 10.1371/journal.pbio.1001745]
- Software design(W)
- Software pattern(W)
- Software development process(W)
- Software architecture(W)
- Portal:Software_testing(W)
Ten simple rules for biologists learning to program - Carey and Papin advise novice biologist programmers how to begin. Much of this paper resonates well with our Introduction to R learning units.
Architecture modeling. A quite useful overview of systems modeling, part of the Microsoft Visual Studio documentation.
Kim Waldén and Jean-Marc Nerson: Seamless Object-Oriented Software Architecture: Analysis and Design of Reliable Systems, Prentice Hall, 1995.
The Death of Microservice Madness DWM Kerr’s Blog post covers a recent trend in software engineering - “Microservices” - and discusses some of the pitfalls in adopting this pattern. The post discusses issues that are of general interest beyond the specific pattern, such as granularity, process communication and integration, and dependency management.
Sandve, Geir K et al.. (2013). “Ten simple rules for reproducible computational research”. Plos Computational Biology 9(10):e1003285 .
[PMID: 24204232] [DOI: 10.1371/journal.pcbi.1003285]
Altschul,
Stephen et al.. (2013). “The anatomy of successful
computational biology software”. Nature Biotechnology
31(10):894–7 .
[PMID: 24104757]
[DOI: 10.1038/nbt.2721]
Peng, Roger
D. (2011). “Reproducible research in computational science”.
Science (New York, N.y.) 334(6060):1226–7
.
[PMID: 22144613]
[DOI: 10.1126/science.1213847]
Gaming systems are large and complex. Bill Clark of Riot analyses a taxonomy of technical debt in this blog post (Riot Games Engineering) with examples from League of Legends. Categories include local-, foundational-, data- and naming-debts.
Miscellaneous
Questions, comments
If in doubt, ask! If anything about this contents is not clear to you, do not proceed but ask for clarification. If you have ideas about how to make this material better, let’s hear them. We are aiming to compile a list of FAQs for all learning units, and your contributions will count towards your participation marks.
Improve this page! If you have questions or comments, please post them on the Quercus Discussion board with a subject line that includes the name of the unit.
References
[END]
All projects end; and all ends can be expressed as a milestone. If it doesn’t end, it’s not a project, it’s an activity.↩︎
I would include any kind of overly decorated relationship indicator in this critique, especially in UML association diagrams, or in Crow’s foot notation. Symbols need to be iconic(W), focus on the essence of the message, and resist any decorative fluff.↩︎
And that co-developer may be you, yourself, half a year down the road when you have forgotten what exactly you were thinking when you wrote the first pieces of code.↩︎
I have seen too much code that just stops executing. It is inexcusable for a developer not to take the time to write the very few statements that are needed for the user to understand what was expected, and what happened instead.↩︎
A software engineer walks into a bar and orders a beer. Then he orders 0 beers. Then orders 2147483648 beers. Then he orders a duck. Then orders -1 beers, poured into a bathtub. Then he returns a beer he didn’t order. Then he spills his beer on the floor, shrieks wildly and runs away without paying.↩︎