Bioinformatics Introduction Sequence
Sequence
| Data | Sequence | Structure | Phylogeny | Function |
Warning – this page is currently under construction (2016-12-26).
Do not use before this warning has been removed.
Contents
- 1 The Sequence Unit
- 2 Introduction
- 3 An S. punctatus homologue to Mbp1
- 4 Analyze
- 5 R Sequence Analysis Tools
- 6 Alignment
- 7 Pairwise Alignments: Optimal
- 8 Heuristic pairwise alignments: BLAST
- 9 Heuristic profile-based alignment: PSI BLAST
- 10 Model Based Alignments: PSSMs and HMMs
- 11 SMART domain annotation
- 12 Multiple Sequence Alignment
- 13 Links and resources
- 14 Sequence annotation
- 15 Downloading Protein Data From the Web
- 16 R code: load alignment and compute information scores
- 17 Calculating conservation scores
- 18 Genomics
- 19 The UCSC genome browser
- 20 Links and resources
- 21 Footnotes and references
- 22 Ask, if things don't work for you!
The Sequence Unit
This Unit is part of a brief introduction to bioinformatics. The material is more or less interleaved with the Sequence.R Project File which is part of the RStudio project associated with this material. Refer to the course/workshop page for installation instructions.
This Unit introduces a target organism which has recently been genome-sequenced in which we will search for novel information about APSES domain transcription factors, it discusses pairwise- and multiple sequence alignment, and BLAST for fast database searches, it explores some methods for sequence analysis and introduces work with entire genomes.
Introduction
Sequence alignment is a very large, and important topic.
One of the foundations of bioinformatics is the empirical observation that related sequences conserve structure, and often function. Much of what we know about a protein's physiological function is based on the conservation of that function as the species evolves. Indeed, conservation is a defining aspect of what can rightly be said to be a protein's "function" in the first place. Conservation - or its opposite: variation - is a consequence of selection under constraints: protein sequences change as a consequence of DNA mutations, this changes the protein's structure, this in turn changes functions and that has multiple effects on a species' reproductive fitness. Detrimental variants may be removed. Variation that is tolerated is largely neutral and therefore found only in positions that are neither structurally nor functionally critical. Conservation patterns can thus provide evidence for many different questions: structural conservation among proteins with similar 3D-structures, functional conservation among homologues with comparable roles, or amino acid propensities as predictors for protein engineering and design tasks.
We assess conservation by comparing sequences between related proteins. This is the basis on which we can make inferences from well-studied model organisms for species that have not been studied as deeply. The foundation is to measure protein sequence similarity. If two sequences are much more similar than we could expect from chance, we hypothesize that their similarity comes from shared ancestry plus conservation. The measurement of sequence similarity however requires sequence alignment[1].
A carefully done sequence alignment is a cornerstone for the annotation of the essential properties a gene or protein. It can already tell us a lot about which proteins we expect to have similar functions in different species.
Multiple sequence alignments (MSAs) are further useful to resolve ambiguities in the precise placement of "indels"[2] and to ensure that columns in alignments actually contain amino acids that evolve in a similar context. MSAs serve as input for
- functional annotation;
- protein homology modelling;
- phylogenetic analyses, and
- sensitive homology searches in databases.
Below we will explore the essentials of
- optimal global and local pairwise alignment;
- Fast BLAST searches to determine best matches in large databases, and reciprocal best matches;
- PSI BLAST searches for exhaustive matches;
- Domain annotation by sequence alignment to statistical models;
- Multiple sequence alignments; and
- Genomes.
This is the scenario: the genome sequence of early-diverging fungus Spizellomyces punctatus has been recently published[3]. Does it contain proteins that are related to yeast Mbp1? If there are more, which one is the most closely related protein? Is its DNA binding domain conserved? How can we identify all related genes in S. punctatus? And, what can we learn from that collection of sequences?
An S. punctatus homologue to Mbp1
BLAST is a fast algorithm for searching related sequences in large databases. We will explore it in more detail below, for now, we just briefly use it to find the sequence in S. punctatus that is most closely related to MBP1_SACCE.
Task:
- Return to the Mbp1 protein page at the NCBI and follow the link to Run BLAST under "Analyze this sequence" in the right-hand column.
- This allows you to perform a sequence similarity search. You need to set two parameters:
- As Database, select Reference proteins (refseq_protein) from the drop down menu;
- In the Organism field, type
spizellomyces punctatus. The field will autocomplete to two choices, choose the DAOM BR117 strain (taxid:645134), as that is the strain which has been sequenced.
- Click on BLAST to start the search. This should find a handful of genes, all of them in S. punctatus. If you find none, or hundreds, or they are not all in the same species, you did something wrong. Ask on the mailing list and make sure to fix the problem.
- Look at the top "hit" in the Alignments section. It should be the hypothetical protein SPPG_00022, and it should have two matching Ranges reported.
- In the header information for each hit is a link to its database entry, right next to Sequence ID. It says
XP_016612325.1... follow that link. - Note the RefSeq ID, and the search results %ID, E-value, whether one or more similar regions were found etc. in your Journal. We will refer to this sequence as "S. punctatus Mbp1" or
MBP1_SPIPUor similar in the future. - Finally access the UniProt ID mapping service to retrieve the UniProt ID for the protein. Paste the RefSeq ID and choose RefSeq Protein as the From: option and UniProtKB as the To: option.
- If the mapping works, the UniProt ID will be in the Entry: column of the table that is being returned. Click the link and have a look at the UniProt entry page while you're there.
Add MBP1_SPIPU to the database
Task:
- (Re)-open the RStudio project and open the
Sequence.Rfile. - Work through Part One: Adding Data.
Analyze
Let us perform a few simple sequence analyses using the online EMBOSS tools. EMBOSS (the European Molecular Biology laboratory Open Software Suite) combines a large number of simple but fundamental sequence analysis tools. The tools can be installed locally on your own machine, or run via a public Web interface. Google for EMBOSS explorer, public access points include http://emboss.bioinformatics.nl/ .
Access an EMBOSS Explorer service and familiarize yourself with the offerings in the EMBOSS package. I find some of the nucleic acid tools indispensable in the lab, such as restriction-site mapping tools, and I frequently use the alignment tools Needle and Water, but by and large the utility of many of the components–while fast, efficient and straightforward to use– suffers from lack of reference and comparison and from terse output. The routines show their conceptual origin in the 1970s and 1980s. We will encounter alternatives in later assignments.
Task:
- Local composition
- Find
pepinfounder the PROTEIN COMPOSITION heading. - Retrieve the YFO Mbp1 related sequence from your R database, e.g. with something like
cat(db$protein[db$protein$name == "UMAG_1122"), "sequence"] - Copy and paste the sequence into the input field.
- Run with default parameters.
- Scroll to the figures all the way at the bottom.
- Do the same in a separate window for yeast Mbp1.
- Try to compare ... (kind of hard without reference, overlay and expectation, isn't it?)
Task:
- Global composition
- Find
pepstatsunder the PROTEIN COMPOSITION heading. - Paste the YFO Mbp1 sequence into the input field.
- Run with default parameters.
- Do the same in a separate window for yeast Mbp1.
- Try to compare ... are there significant and unexpected differences?
Task:
- Transmembrane sequences
- Find
tmap. Also findshuffleseq. - Use your YFO sequence to annotate transmembrane helices for your protein and for a few shuffled sequences. The YFO is not expected to have TM helices, nor are the shuffled sequences expected to have any. If you do find some, these are most likely "false positives".
- Also compare the following positive control: Gef1 - a yeast chloride channel with 10 trans-membrane helices and outward localized N-terminus:
>gi|6322500|ref|NP_012574.1| Gef1p [Saccharomyces cerevisiae S288c]
MPTTYVPINQPIGDGEDVIDTNRFTNIPETQNFDQFVTIDKIAEENRPLSVDSDREFLNSKYRHYREVIW
DRAKTFITLSSTAIVIGCIAGFLQVFTETLVNWKTGHCQRNWLLNKSFCCNGVVNEVTSTSNLLLKRQEF
ECEAQGLWIAWKGHVSPFIIFMLLSVLFALISTLLVKYVAPMATGSGISEIKVWVSGFEYNKEFLGFLTL
VIKSVALPLAISSGLSVGKEGPSVHYATCCGYLLTKWLLRDTLTYSSQYEYITAASGAGVAVAFGAPIGG
VLFGLEEIASANRFNSSTLWKSYYVALVAITTLKYIDPFRNGRVILFNVTYDRDWKVQEIPIFIALGIFG
GLYGKYISKWNINFIHFRKMYLSSWPVQEVLFLATLTALISYFNEFLKLDMTESMGILFHECVKNDNTST
FSHRLCQLDENTHAFEFLKIFTSLCFATVIRALLVVVSYGARVPAGIFVPSMAVGATFGRAVSLLVERFI
SGPSVITPGAYAFLGAAATLSGITNLTLTVVVIMFELTGAFMYIIPLMIVVAITRIILSTSGISGGIADQ
MIMVNGFPYLEDEQDEEEEETLEKYTAEQLMSSKLITINETIYLSELESLLYDSASEYSVHGFPITKDED
KFEKEKRCIGYVLKRHLASKIMMQSVNSTKAQTTLVYFNKSNEELGHRENCIGFKDIMNESPISVKKAVP
VTLLFRMFKELGCKTIIVEESGILKGLVTAKDILRFKRIKYREVHGAKFTYNEALDRRCWSVIHFIIKRF
TTNRNGNVI- Evaluate the output: does the algorithm (wrongly) predict TM-helices in your protein? In the shuffled sequences? Does it find all ten TM-helices in Gef1?
Task:
- Motifs
- Find
pepcoil, an algorithm to detect coiled coil motifs. - Run this with the YFO Mbp1 sequence and yeast Mbp1.
- Try to compare ... do both sequences have coiled-coil motif predictions? Are they annotated in approximately comparable regions of the respective sequence?
Coiled coils
A Brief First Encounter of Regular Expressions
- Regular expressions are a concise description language to define patterns for pattern-matching in strings.
Truth be told, many programmers have a love-hate relationship with regular expressions. The syntax of regular expressions is very powerful and expressive, but also terse, not always intuitive, and sometimes hard to understand. I'll introduce you to a few principles here that are quite straightforward and they will probably cover 99% of the cases you will encounter.
Here is our test-case: the sequence of Mbp1, copied from the NCBI Protein database page for yeast Mbp1.
1 msnqiysary sgvdvyefih stgsimkrkk ddwvnathil kaanfakakr trilekevlk
61 ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf dftqtdgsas pppapkhhha
121 skvdrkkair sastsaimet krnnkkaeen qfqsskilgn ptaaprkrgr pvgstrgsrr
181 klgvnlqrsq sdmgfprpai pnssisttql psirstmgpq sptlgileee rhdsrqqqpq
241 qnnsaqfkei dledglssdv epsqqlqqvf nqntgfvpqq qssliqtqqt esmatsvsss
301 pslptspgdf adsnpfeerf pgggtspiis miprypvtsr pqtsdindkv nkylsklvdy
361 fisnemksnk slpqvllhpp phsapyidap idpelhtafh wacsmgnlpi aealyeagts
421 irstnsqgqt plmrsslfhn sytrrtfpri fqllhetvfd idsqsqtvih hivkrksttp
481 savyyldvvl skikdfspqy rielllntqd kngdtalhia skngdvvffn tlvkmgaltt
541 isnkegltan eimnqqyeqm miqngtnqhv nssntdlnih vntnnietkn dvnsmvimsp
601 vspsdyityp sqiatnisrn ipnvvnsmkq masiyndlhe qhdneikslq ktlksisktk
661 iqvslktlev lkesskdeng eaqtnddfei lsrlqeqntk klrkrliryk rlikqkleyr
721 qtvllnklie detqattnnt vekdnntler lelaqeltml qlqrknklss lvkkfednak
781 ihkyrriire gtemnieevd ssldvilqtl iannnknkga eqiitisnan sha
//
Task:
Navigate to http://regexpal.com and paste the sequence into the lower box. This site is one of a number of online regular expression testers; their immediate, visual feedback is invaluable when you are developing regular expression patterns.
Lets try some expressions:
- Most characters are matched literally.
- Type "
a" in to the upper box and you will see all "a" characters matched. Then replaceawithq. - Now type "
aa" instead. Thenkrnnkk. Sequences of characters are also matched literally.
- The pipe character | that symbolizes logical OR can be used to define that more than one character should match
i(s|m|q)nmatchesisnORimnORiqn. Note how we can group with parentheses, and try what would happen without them.
- We can more conveniently specify more than one character to match if we place it in square brackets.
[lq]matcheslORq.[familyvw]matches hydrophobic amino acids.
- Within square brackets, we can specify "ranges".
[1-5]matches digits from 1 to 5.
- Within square brackets, we can specify characters that should NOT be matched, with the caret,
^. [^0-9]matches everything EXCEPT digits.[^a-z]matches everything that is not a lower-case letter. That's what we need (try it).
One of the R functions that uses regular expressions is the function gsub(). It replaces characters that match a "regex" with other characters. That is useful for our purpose: we can
- match all characters that are NOT a letter, and
- replace them by - nothing: the empty string
"".
This deletes them.
Task:
- study the code in the
An excursion into regular expressionssection of the R script
R Sequence Analysis Tools
It's interesting to see this collection of tools that were carefully designed some twenty years ago, as an open source replacement for a set of software tools - the GCG package - that was indispensable for molecular biology labs in the 80s and 90s, but whose cost had become prohibitive. Fundamentally this is a building block approach, and the field has turned to programming solutions instead.
As for functionality, much more sophisticated functions are available on the Web: do take a few minutes and browse the curated Web services directory of bioinformatics.ca.
As for versatility, R certainly has the edge. Let's explore some of the functions available in the seqinr package that you already encountered in the introductory R tutorial. They are comparatively basic - but it is easy to prepare our own analysis.
Task:
- Study the code in the
Sequence Analysissection of the R script
- That is all.
Alignment
Preparation: Updated Database Functions
The database contents and tables will change over time in this course. This means we need a mechanism to update the database, without throwing away previous work.
Task:
- Open the BCH441 project scripts in RStudio by selecting File → Recent Projects → BCH441_216
- Load the newest versions of scripts and data by pulling from the master file on GitHub.
- Study the code in the
Database maintenancesection of theBCH441_A04.Rscript
DotPlots and the Mutation Data Matrix
Before we start calculating alignments, we should get a better sense of the underlying sequence similarity. A Dotplot is a perfect tool for that, because it displays alignment-free similarity information. Let's make a dotplot that uses the BLOSUM62 Mutation Data Matrix to measure pairwise amino acid similarity. The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the BLOSUM62 matrix[4].
The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the BLOSUM62 matrix[5].
Scoring matrices are also available in the Bioconductor Biostrings package.
BLOSUM62
A R N D C Q E G H I L K M F P S T W Y V B J Z X *
A 4 -1 -2 -2 0 -1 -1 0 -2 -1 -1 -1 -1 -2 -1 1 0 -3 -2 0 -2 -1 -1 -1 -4
R -1 5 0 -2 -3 1 0 -2 0 -3 -2 2 -1 -3 -2 -1 -1 -3 -2 -3 -1 -2 0 -1 -4
N -2 0 6 1 -3 0 0 0 1 -3 -3 0 -2 -3 -2 1 0 -4 -2 -3 4 -3 0 -1 -4
D -2 -2 1 6 -3 0 2 -1 -1 -3 -4 -1 -3 -3 -1 0 -1 -4 -3 -3 4 -3 1 -1 -4
C 0 -3 -3 -3 9 -3 -4 -3 -3 -1 -1 -3 -1 -2 -3 -1 -1 -2 -2 -1 -3 -1 -3 -1 -4
Q -1 1 0 0 -3 5 2 -2 0 -3 -2 1 0 -3 -1 0 -1 -2 -1 -2 0 -2 4 -1 -4
E -1 0 0 2 -4 2 5 -2 0 -3 -3 1 -2 -3 -1 0 -1 -3 -2 -2 1 -3 4 -1 -4
G 0 -2 0 -1 -3 -2 -2 6 -2 -4 -4 -2 -3 -3 -2 0 -2 -2 -3 -3 -1 -4 -2 -1 -4
H -2 0 1 -1 -3 0 0 -2 8 -3 -3 -1 -2 -1 -2 -1 -2 -2 2 -3 0 -3 0 -1 -4
I -1 -3 -3 -3 -1 -3 -3 -4 -3 4 2 -3 1 0 -3 -2 -1 -3 -1 3 -3 3 -3 -1 -4
L -1 -2 -3 -4 -1 -2 -3 -4 -3 2 4 -2 2 0 -3 -2 -1 -2 -1 1 -4 3 -3 -1 -4
K -1 2 0 -1 -3 1 1 -2 -1 -3 -2 5 -1 -3 -1 0 -1 -3 -2 -2 0 -3 1 -1 -4
M -1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 0 -2 -1 -1 -1 -1 1 -3 2 -1 -1 -4
F -2 -3 -3 -3 -2 -3 -3 -3 -1 0 0 -3 0 6 -4 -2 -2 1 3 -1 -3 0 -3 -1 -4
P -1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7 -1 -1 -4 -3 -2 -2 -3 -1 -1 -4
S 1 -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4 1 -3 -2 -2 0 -2 0 -1 -4
T 0 -1 0 -1 -1 -1 -1 -2 -2 -1 -1 -1 -1 -2 -1 1 5 -2 -2 0 -1 -1 -1 -1 -4
W -3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 -4 -3 -2 11 2 -3 -4 -2 -2 -1 -4
Y -2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7 -1 -3 -1 -2 -1 -4
V 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4 -3 2 -2 -1 -4
B -2 -1 4 4 -3 0 1 -1 0 -3 -4 0 -3 -3 -2 0 -1 -4 -3 -3 4 -3 0 -1 -4
J -1 -2 -3 -3 -1 -2 -3 -4 -3 3 3 -3 2 0 -3 -2 -1 -2 -1 2 -3 3 -3 -1 -4
Z -1 0 0 1 -3 4 4 -2 0 -3 -3 1 -1 -3 -1 0 -1 -2 -2 -2 0 -3 4 -1 -4
X -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -4
* -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 1
Task:
- Study this and make sure you understand what this table is, how it can be used, and what a reasonable range of values for identities and pairscores for non-identical, similar and dissimilar residues is. Ask on the mailing list in case you have questions. This piece of data is the foundation of any sequence alignment. without it, no sensible alignment could be produced!
- Figure out the following values:
- Compare an identical match of histidine with an identical match of serine. What does this mean?
- How similar are lysine and leucine, as compared to leucine and isoleucine? Is this what you expect?
- PAM matrices are sensitive to an interesting artefact. Since W and R can be interchanged with a single point mutation, the probability of observing W→R and R→W exchanges in closely related sequences is much higher than one would expect from the two amino acid's biophysical properties. (Why?) PAM matrices were compiled from hypothetical point exchanges and then extrapolated. Therefore these matrices assign a relatively high degree of similarity to (W, R), that is not warranted considering what actually happens in nature. Do you see this problem in the BLOSUM matrix? If BLOSUM does not have this issue, why not?
Next, let's apply the scoring matrix for actual comparison:
Task:
- Return to your RStudio session.
- If you've been away from it for a while, it's probably a good idea to update to the newest versions of scripts and data by pulling from the master file on GitHub.
- Study and work through the code in the
Dotplot and MDMsection of theBCH441_A04.Rscript
Pairwise Alignments: Optimal
Optimal pairwise sequence alignment is the mainstay of sequence comparison. To consider such alignments in practice, we'll align the same sequences that we have just mapped in the dotplot exercise: Mbp1 and its YFO relative. For simplicity, I will call the two proteins MBP1_SACCE and MBP1_YFO through the remainder of the assignment. Your dotplots should have shown you two regions of similarity: a highly similar region focussed somewhere around the N-terminal 100 amino acids, and a more extended, but somewhat less similar region in the middle of the sequences. You can think of the sequence alignment algorithm as building the similarity matrix, and then discovering the best path along high-scoring diagonals.
Optimal Sequence Alignment: EMBOSS online tools
Online programs for optimal sequence alignment are part of the EMBOSS tools. The programs take FASTA files or raw text files as input.
Local optimal sequence alignment using "water"
Task:
- Fetch the sequences for
MBP1_SACCEandMBP1_YFOfrom your database. You can simply select them by name (if you have given your sequence the suggested name when you eneterd it into your database): paste the following into the console:
- to print the
MBP1_SACCEprotein sequence to the console
myDB$protein$sequence[myDB$protein$name == "MBP1_SACCE"]- to print the
MBP1_YFOprotein sequence to the console:
YFOseq <- paste("MBP1_", biCode(YFO), sep="")
myDB$protein$sequence[myDB$protein$name == YFOseq](If this didn't work, fix it. Did you give your sequence the right name?)
- Access the EMBOSS Explorer site (if you haven't done so yet, you might want to bookmark it.)
- Look for ALIGNMENT LOCAL, click on water, paste your sequences and run the program with default parameters.
- Study the results. You will probably find that the alignment extends over most of the protein, but does not include the termini.
- Considering the sequence identity cutoff we discussed in class (25% over the length of a domain), do you believe that the N-terminal domains (the APSES domains) are homologous?
- Change the Gap opening and Gap extension parameters to high values (e.g. 30 and 5). Then run the alignment again.
- Note what is different.
Global optimal sequence alignment using "needle"
Task:
- Look for ALIGNMENT GLOBAL, click on needle, paste the
MBP1_SACCEandMBP1_YFOsequences again and run the program with default parameters. - Study the results. You will find that the alignment extends over the entire protein, likely with long indels at the termini.
Optimal Sequence Alignment with R: Biostrings
Biostrings has extensive functions for sequence alignments. They are generally well written and tightly integrated with the rest of Bioconductor's functions. There are a few quirks however: for example alignments won't work with lower-case sequences[6].
Task:
- Return to your RStudio session.
- Once again, if you've been away from it for a while, it's always a good idea to update to pull updtaes from the master file on GitHub.
- Study and work through the code in the
Biostrings Pairwise Alignmentsection of theBCH441_A04.Rscript
Heuristic pairwise alignments: BLAST
BLAST is by a margin the most important computational tool of molecular biology. It is so important, that we have already used BLAST in Assignment 3 even before properly introducing the algorithm and the principles, to find the most similar sequence to MBP1_SACCE in YFO.
In this part of the assignment we will use BLAST to perform Reciprocal Best Matches.
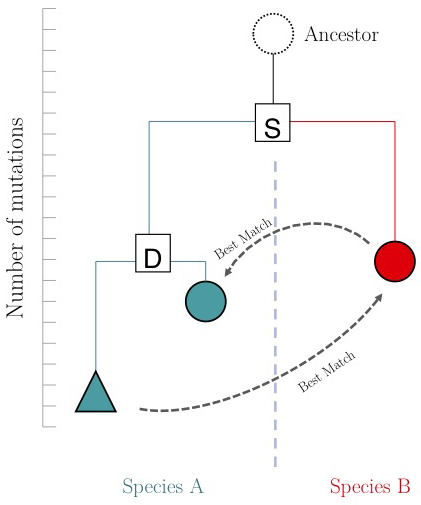
One of the important questions of model-organism based inference is: which genes perform the same function in two different organisms. In the absence of other information, our best guess is that these are the two genes that are mutually most similar. The keyword here is mutually. If MBP1_SACCE from S. cerevisiae is the best match to RES2_SCHPO in S. pombe, the two proteins are only mutually most similar if RES2_SCHPO is more similar to MBP1_SACCE than to any other S. cerevisiae protein. We call this a Reciprocal Best Match, or "RBM"[7].
The argument is summarized in the figure on the right: genes that evolve under continuos selective pressure on their function have relatively lower mutation rates and are thus more similar to each other, than genes that undergo neo- or sub-functionalization after duplication.
However, there is a catch: proteins are often composed of multiple domains that implement distinct roles of their function. Under the assumptions above we could hypothesize:
- a gene in YFO that has the "same" function as the Mbp1 cell-cycle checkpoint switch in yeast should be an RBM to Mbp1;
- a gene that binds to the same DNA sites as Mbp1 should have a DNA-binding domain that is an RBM to the DNA binding domain of Mbp1.
Thus we'll compare RBMs in YFO for full-length Mbp1_SACCE and its DNA-binding domain, and see if the results are the same.
Full-length RBM
You have already performed the first half of the experiment: matching from S. cerevisiae to YFO. The backward match is simple.
Task:
- Access BLAST and follow the link to the protein blast program.
- Enter the RefSeq ID for
MBP1_YFOin the Query sequence field. - Select
refseq_proteinas the database to search in, and enterSaccharomyces cerevisiae (taxid:4932)to restrict the organism for which hits are reported. - Run BLAST. Examine the results.
If your top-hit is NP_010227, you have confirmed the RBM between Mbp1_SACCE and Mbp1_YFO. If it is not, let me know. I expect this to be the same and would like to verify your results if it is not[8].
RBM for the DNA binding domain
The DNA-binding domain of Mbp1_SACCE is called an APSES domain. If the RBM between Saccharomyces cerevisiae Mbp1 and YFO is truly an orthologue, we expect all of the protein's respective domains to have the RBM property as well. But let's not simply assume what we can easily test. We'll define the sequence of the APSES domain in MBP1_SACCE and YFO and see how these definitions reflect in a BLAST search.
Defining the range of the APSES domain annotation
The APSES domain is a well-defined type of DNA-binding domain that is ubiquitous in fungi and unique in that kingdom. Structurally it is a member of the Winged Helix-Turn-Helix family. Recently it was found that it is homologous to the somewhat shorter, prokaryotic KilA-N domain; thus the APSES domain was retired from pFam and instances were merged into the KilA-N family. However InterPro has a KilA-N entry but still recognizes the APSES domain.
KilA-N domain boundaries in Mbp1 can be derived from the results of a CDD search with the ID 1BM8_A (the Mbp1 DNA binding domain crystal structure). The KilA-N superfamily domain alignment is returned.
- (pfam 04383): KilA-N domain; The amino-terminal module of the D6R/N1R proteins defines a novel, conserved DNA-binding domain (the KilA-N domain) that is found in a wide range of proteins of large bacterial and eukaryotic DNA viruses. The KilA-N domain family also includes the previously defined APSES domain. The KilA-N and APSES domains may also share a common fold with the nucleic acid-binding modules of the LAGLIDADG nucleases and the amino-terminal domains of the tRNA endonuclease.
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
1BM8A 16 IHSTGSIMKRKKDDWVNATHILKAANFAKaKRTRILEKEVLKETHEKVQ---------------GGFGKYQGTWVPLNIA 80
Cdd:pfam04383 3 YNDFEIIIRRDKDGYINATKLCKAAGETK-RFRNWLRLESTKELIEELSeennvdkseiiigrkGKNGRLQGTYVHPDLA 81
90
....*....|....
1BM8A 81 KQLA----EKFSVY 90
Cdd:pfam04383 82 LAIAswisPEFALK 95
Note that CDD and SMART are not consistent in how they apply pFam 04383 to the Mbp1 sequence. See annotation below.
The CDD KilA-N domain definition begins at position 16 of the 1BM8 sequence. But virtually all fungal APSES domains have a longer, structurally defined, conserved N-terminus. Blindly applying the KilA-N domain definition to these proteins would lose important information. For most purposes we will prefer the sequence spanned by the 1BM8_A structure. The sequence is given below, the KilA-N domain is coloured dark green. By this definition the APSES domain is 99 amino acids long and comprises residues 4 to 102 of the NP_010227 sequence.
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
1BM8A 1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKETHEKVQGGFGKYQGTWVPLNIA 80
90
....*....|....*....
1BM8A 81 KQLAEKFSVYDQLKPLFDF 99
- Yeast APSES domain sequence in FASTA format
>APSES_MBP1 Residues 4-102 of S. cerevisiae Mbp1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF
- Synopsis of ranges
| Domain | Link | Length | Boundary | Range (Mbp1) | Range (1BM8) |
| KilA-N: pfam04383 (CDD) | CDD alignment | 72 | STGSI ... KFSVY | 21 - 93 | 18 - 90 |
| KilA-N: pfam04383 (SMART) | Smart main page | 79 | IHSTG ... YDQLK | 19 - 97 | 16 - 94 |
| KilA-N: SM01252 (SMART) | Smart main page | 84 | TGSIM ... DFTQT | 22 - 105 | 19 - 99... |
| APSES: Interpro IPR003163 | (Interpro) | 130 | QIYSA ... IRSAS | 3 - 133 | 1 - 99... |
| APSES (1BM8) | – | 99 | QIYSA ... PLFDF | 4 - 102 | 1 - 99 |
Executing the forward search
Task:
- Access BLAST and follow the link to the protein blast program.
- Forward search:
- Paste only the APSES domain sequence for
MBP1_SACCEin the Query sequence field (copy the sequence from above). - Select
refseq_proteinas the database to search in, and enter the correct taxonomy ID for YFO in the Organism field. - Run BLAST. Examine the results.
- If the top hit is the same protein you have already seen, oK. If it's not add it to your protein database in RStudio.
- Paste only the APSES domain sequence for
With this we have confirmed the sequence with the most highly conserved APSES domain in YFO. Can we take the sequence for the reverse search from the alignment that BLAST returns? Actually, that is not a good idea. The BLAST alignment is not guaranteed to be optimal. We should do an optimal sequnece alignment instead. That is: we use two different tools here for two different purposes: we use BLAST to identify one protein as the most similar among many alternatives and we use optimal sequence alignment to determine the best alignment between two sequences. That best alignment is what we will annotate as the YFO APSES domain.
Alignment to define the YFO APSES domain for the reverse search
Task:
- Return to your RStudio session.
- Study and work through the code in the
APSES Domain annotation by alignmentsection of theBCH441_A04.Rscript
Executing the reverse search
Task:
- Paste the the APSES domain sequence for the YFO best-match and enter it into Query sequence field of the BLAST form.
- Select
refseq_proteinas the database to search in, and enterSaccharomyces cerevisiae (taxid:4932)to restrict the organism for which hits are reported. - Run BLAST. Examine the results.
- Select
If your top-hit is again NP_010227, you have confirmed the RBM between the APSES domain of Mbp1_SACCE and Mbp1_<YFO>. If it is not, let me know. There may be some organisms for which the full-length and APSES RBMs are different and I would like to discuss these cases.
Heuristic profile-based alignment: PSI BLAST
It is (deceptively) easy to perform BLAST searches via the Web interface, but to use such powerful computational tools to their greatest advantage takes a considerable amount of care, caution and consideration.
PSI-BLAST allows to perform very sensitive searches for homologues that have diverged so far that their pairwise sequence similarity has become insignificant. It achieves this by establishing a profile of sequences to align with the database, rather than searching with individual sequences. This deemphasizes parts of the sequence that are variable and inconsequential, and focusses on the parts of greater structural and functional importance. As a consequence, the signal to noise ratio is greatly enhanced.
In this part of the assignment, we will set ourselves the task to use PSI-BLAST and find all orthologs and paralogs of the APSES domain containing transcription factors in YFO. We will use these sequences for multiple alignments, calculation of conservation etc.
The first methodical problem we have to address is what sequence to search with. The full-length Mbp1 sequence from Saccharomyces cerevisiae or its RBM from YFO are not suitable: They contain multiple domains (in particular the ubiquitous Ankyrin domains) and would create broad, non-specific profiles. The APSES domain sequence by contrast is structurally well defined. The KilA-N domain, being shorter, is less likely to make a sensitive profile. Indeed one of the results of our analysis will be to find whether APSES domains in fungi all have the same length as the Mbp1 domain, or whether some are indeed much shorter, like the KILA-N domain, as suggested by the Pfam alignment.
The second methodical problem we must address is how to perform a sensitive PSI-BLAST search in one organism. We need to balance two conflicting objectives:
- If we restrict the PSI-BLAST search to YFO, PSI-BLAST has little chance of building a meaningful profile - the number of homologues that actually are in YFO is too small. Thus the search will not become very sensitive.
- If we don't restrict our search, but search in all species, the number of hits may become unwieldily large. It becomes increasingly difficult to closely check all hits as to whether they have good coverage. Also we need to evaluate the fringe cases of marginal E-value: should a new sequence be added to the profile, or should we hold off on it for one or two iterations, to see whether its E-value drops significantly. By all means, we need to avoid profile corruption.
Perhaps this is still be manageable when we are searching in fungi, but imagine you are working with a bacterial protein, or a protein that is conserved across the entire tree of life: your search may find tens of thousands of sequences. And by next year, thousands more will have been added.
Therefore we have to find a middle ground: add enough organisms (sequences) to compile a sensitive profile, but not so many that we can no longer individually assess the sequences that contribute to the profile. We need to define a broadly representative but manageable set of species - to exploit the transitivity of homology - even if we are interested only in matches in one species: YFO. Please reflect on this and make sure you understand why we include sequences in a PSI-BLAST search that we are not actually interested in.
We need a subset of species
- that represent as large a range as possible on the evolutionary tree;
- that are as well distributed as possible on the tree; and
- whose genomes are fully sequenced.
Selecting species for a PSI-BLAST search
To select species, we will use an approach that is conceptually simple: select a set of species according to their shared taxonomic rank in the tree of life. Biological classification provides a hierarchical system that describes evolutionary relatedness for all living entities. The levels of this hierarchy are so called taxonomic ranks. These ranks are defined in Codes of Nomenclature that are curated by the self-governed international associations of scientists working in the field. The number of ranks is not specified: there is a general consensus on seven principal ranks (see below, in bold) but many subcategories exist and may be newly introduced. It is desired–but not mandated–that ranks represent clades (a group of related species, or a "branch" of a phylogeny), and it is desired–but not madated–that the rank is sharply defined. The system is based on subjective dissimilarity. Needless to say that it is in flux.
If we follow a link to an entry in the NCBI's Taxonomy database, eg. Saccharomyces cerevisiae S228c, the strain from which the original "yeast genome" was sequenced in the late 1990s, we see the following specification of its taxonomic lineage:
cellular organisms; Eukaryota; Opisthokonta;
Fungi; Dikarya; Ascomycota; Saccharomyceta;
Saccharomycotina; Saccharomycetes;
Saccharomycetales; Saccharomycetaceae;
Saccharomyces; Saccharomyces cerevisiae
These names can be mapped into taxonomic ranks, since the suffixes of these names e.g. -mycotina, -mycetaceae are specific to defined ranks. (NCBI does not provide this mapping, but Wikipedia is helpful here.)
| Rank | Suffix | Example |
| Domain | Eukaryota (Eukarya) | |
| Subdomain | Opisthokonta | |
| Kingdom | Fungi | |
| Subkingdom | Dikarya | |
| Phylum | Ascomycota | |
| rankless taxon[9] | -myceta | Saccharomyceta |
| Subphylum | -mycotina | Saccharomycotina |
| Class | -mycetes | Saccharomycetes |
| Subclass | -mycetidae | |
| Order | -ales | Saccharomycetales |
| Family | -aceae | Saccharomycetaceae |
| Subfamily | -oideae | |
| Tribe | -eae | |
| Subtribe | -ineae | |
| Genus | Saccharomyces | |
| Species | Saccharomyces cerevisiae |
You can see that there is no common mapping between the yeast lineage listed at the NCBI and the commonly recognized categories - not all ranks are represented. Nor is this consistent across species in the taxonomic database: some have subfamily ranks and some don't. And the tree is in no way normalized - some of the ranks have thousands of members, and for some, only a single extant member may be known, or it may be a rank that only relates to the fossil record.
But the ranks do provide some guidance to evolutionary divergence. Say you want to choose four species across the tree of life for a study, you should choose one from each of the major domains of life: Eubacteria, Euryarchaeota, Crenarchaeota-Eocytes, and Eukaryotes. Or you want to study a gene that is specific to mammals. Then you could choose from the clades listed in the NCBI taxonomy database under Mammalia (a class rank, and depending how many species you would want to include, use the subclass-, order-, or family rank (hover over the names to see their taxonomic rank.)
I have chosen the 10 species below to define a well-distributed search-space for PSI-BLAST. Of course you must also include YFO in the selection (if YFO is not in this list already).
To enter these 10 species as an Entrez restriction, they need to be formatted as below. (One could also enter species one by one, by pressing the (+) button after the organism list)
"Wallemia mellicola"[organism] OR
"Puccinia Graminis"[organism] OR
"Ustilago maydis"[organism] OR
"Cryptococcus neoformans"[organism] OR
"Coprinopsis cinerea"[organism] OR
"Schizosaccharomyces pombe"[organism] OR
"Aspergillus nidulans"[organism] OR
"Neurospora crassa"[organism] OR
"Bipolaris oryzae"[organism] OR
"Saccharomyces cerevisiae"[organism]
Executing the PSI-BLAST search
We have a list of species. Good. Next up: how do we use it.
Task:
- Navigate to the BLAST homepage.
- Select protein BLAST.
- Paste the MBP1_SACCE APSES domain sequence into the search field:
>APSES_MBP1 Residues 4-102 of S. cerevisiae Mbp1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF
- Select refseq as the database.
- Copy the Entrez restrictions from above and add the correct name for YFO to the list if it is not there already. (Obviously, you can't find sequences in YFO if YFO is not included among the genomes you are searching in.) Paste the list into the Entrez Query field.
- In the Algorithm section, select PSI-BLAST.
- Click on BLAST.
Evaluate the results carefully. Since we did not change the algorithm parameters, the threshold for inclusion was set at an E-value of 0.005 by default, and that may be a bit too lenient, i.e. it might include sequences that are not homologous. If you look at the table of your hits– in the Sequences producing significant alignments... section– there may also be a few sequences that have a low query coverage of less than 80%. Let's exclude these from the profile initially: not to worry, if they are true positives, the will come back with improved E-values and greater coverage in subsequent iterations. But if they were false positives, their E-values will rise and they will drop out of the profile and not contaminate it.
Task:
- In the header section, click on Formatting options and in the line "Format for..." set the with inclusion threshold to
0.001(This means E-values can't be above 10-03 for the sequence to be included.) - Click on the Reformat button (top right).
- In the table of sequence descriptions (not alignments!), click on Query cover to sort the table by coverage, not by score.
- Deselect the check mark next to these sequences in the second-to-rightmost column Select for PSI blast.
- Then scroll to Run PSI-BLAST iteration 2 ... and click on
Go.
This is now the "real" PSI-BLAST at work: it constructs a profile from all the full-length sequences and searches with the profile, not with any individual sequence. Note that we are controlling what goes into the profile in two ways:
- we are explicitly removing sequences with poor coverage; and
- we are requiring a more stringent minimum E-value for each sequence.
Task:
- Again, study the table of hits. Sequences highlighted in yellow have met the search criteria in the second iteration and are proposed for inclusion in the next iteration. Note that the coverage of (some) of the previously excluded sequences is now above 80%. These are the ones you need to check carefully: do you agree that they should be included? If there is any doubt, perhaps because of a really marginal E-value, poor coverage or a function annotation that is not compatible with your query, it is safer to exclude a sequence than to risk profile corruption. If the sequence is a true positive, it will return to the list in later iterations, usually with a better E-value as the profile improves. It's a good idea to note such sequences in your journal so you can keep track of how their E-values change.
- Let's exclude partial matches one more time. Again, deselect all sequences with less than 80% coverage. Then run the third iteration.
- Iterate the search in this way, successively relaxing the coverage threshold, until no more "New" sequences are added to the profile. The search has converged. Obviously the result depends on your data, but it would be unusual if the search had not converged after 6 iterations or so, and there is probably a mistake if there are more than 70 hits or so.
- Now look at the list of excluded hits (if any), the hits that are reasonable but didn't quite make the cut. Are there any from YFO that seem like they should actually be included? Perhaps their E-value is only marginally above the threshold? If that's the case, try returning the E-value threshold to the default 0.005 and see what happens...
Once no "new" sequences have been added, we would always get the same result on additional iterations because there are no more changes to the profile. We say that the search has converged. Time to harvest.
Task:
- In the header section of the BLAST report, click on Taxonomy reports and find YFO in the Organism Report section. These are your APSES domain homologs. All of them. There is a link to the alignment, the BLAST score, the E-value, and a link to the entry in RefSeq.
- From the report copy the sequence identifiers from YFO, with E-values above your defined threshold to your notebook.
For example, the list of Saccharomyces genes contains the following information:
Saccharomyces cerevisiae S288c [ascomycetes] taxid 559292
4e-37 ref|NP_010227.1| Mbp1p [Saccharomyces cerevisiae S288c]
2e-30 ref|NP_011036.1| Swi4p [Saccharomyces cerevisiae S288c]
4e-27 ref|NP_012881.1| Phd1p [Saccharomyces cerevisiae S288c]
4e-27 ref|NP_013729.1| Sok2p [Saccharomyces cerevisiae S288c]
7e-06 ref|NP_012165.1| Xbp1p [Saccharomyces cerevisiae S288c]
Xbp1 is a special case. It has only very low coverage, but that is because it has a long domain insertion and the N-terminal match often is not recognized by alignment because the gap scores for long indels are unrealistically large. For now, I keep that sequence with the others.
Task:
- To add the sequences to your database, open each of the links to the RefSeq record for YFO organism into a separate tab.
- Find the UniProt IDs
- Go through the (short) section
add PSI BLAST resultsin the Assignment 04 R-script.
So much for using PSI-BLAST. The last step seems a bit tedious, adding all this information by hand. There's got to be a better way, right?
But for now, we'll have a look at what the sequences tell us.
Model Based Alignments: PSSMs and HMMs
- Position Specific Scoring Matrices (PSSMs)
The sensitivity of PSI-BLAST is based on the alignment of profiles of related sequences. The profiles are represented as position specific scoring matrices compiled from the alignment of hits, first to the original sequence and then to the profile. Incidentally, this process can also be turned around, and a collection of pre-compiled PSSMs can be used to annotate protein sequence: this is the principle employed by RPS-BLAST, the tool that identifies conserved domains at the beginning of every BLAST search, and has been used to build the CDD database of conserved domains (for a very informative help-page on CDD see here.
- Hidden Markov Models (HMMs)
An approach to represent such profile information that is more general than PSSMs is a Hidden Markov model (HMM) and the standard tool to use HMMs in Bioinformatics is HMMER, written by Sean Eddy. HMMER has allowed to represent the entirety of protein sequences as a collection of profiles, stored in databases such as Pfam, Interpro, and SMART. While the details are slightly different, all of these services allow to scan sequences for the presence of domains. Importantly thus, the alignment results are not collections of full-length protein families, but annotate to domain families, i.e. full length proteins are decomposed into their homologous domains. This is a very powerful approach towards the functional annotation of unknown sequences.
In this section, we will annotate the YFO sequence with the domains it contains, using the database of domain HMMs curated by SMART in Heidelberg and Pfam at the EMBL. We will then compare these annotations with those determined for the orthologues in the reference species. In this way we can enhance the information about one protein by determining how its features are conserved.
SMART domain annotation
The SMART database at the EMBL in Heidelberg integrates a number of feature detection tools including Pfam domain annotation and its own, HMM based SMART domain database. You can search by sequence, or by accession number and retrieve domain annotations and more.
SMART search
Task:
- Access the SMART database at http://smart.embl-heidelberg.de/
- Click the lick to access SMART in the normal mode.
- Paste the YFO Mbp1 UniProtKB Accession number into the Sequence ID or ACC field. If you were not able to find a UniProt ID, paste the sequence instead.
- Check all the boxes for:
- outlier homologues (also including homologues in the PDB structure database)
- PFAM domains (domains defined by sequence similarity in the PFAM database)
- signal peptides (using the Gunnar von Heijne's SignalP 4.0 server at the Technical University in Lyngby, Denmark)
- internal repeats (using the programs ariadne and prospero at the Wellcome Trust Centre for Human Genetics at Oxford University, England)
- Click on Sequence SMART to run the search and annotation. (In case you get an error like: "Sorry, your entry seems to have no SMART domain ...", try again with the actual sequence instead of the accession number.)
Study the results.
- Note down the following information so you can enter the annotation in the protein database for YFO:
- From the section on "Confidently predicted domains ..."
- The start and end coordinates of the KilA-N domain (...according to SMART, not Pfam, in case the two differ).
- All start and end coordinates of low complexity segments
- All start and end coordinates of ANK (Ankyrin) domains
- Start and end coordinates of coiled coil domain(s) I expect only one.
- Start and end coordinates of AT hook domain(s) I expect at most one - not all Mbp1 orthologues have one.
- From the section on "Features NOT shown ..."
- All start and end coordinates of low complexity segments for which the Reason is "overlap".
- Any start and end coordinates of overlapping coiled coil segments.
- I expect all other annotations - besides the overlapping KilA-N domain defined by Pfam - to arise from the succession of ankyrin domains that the proteins have, both Pfam_ANK.. domains, as well as internal repeats. However, if there are other features I have not mentioned here, feel encouraged to let me know.
- From the section on "Outlier homologues ..."
- Start and end coordinates of a PDB:1SW6|B annotation (if you have one): this is a region of sequence similarity to a protein for which the 3D structural coordinate are known.
- Of course there should also be annotations to the structure of 1BM8 / 1MB1 and/or 1L3G - all of which are structures of the Mbp1 APSES domain that we have already annotated as an"APSES fold" feature previously. And there will be BLAST annotations to Ankyrin domains. We will not annotate these separately either.
- From the section on "Confidently predicted domains ..."
- Follow the links to the database entries for the information so you know what these domains and features are.
Next we'll enter the features into our database, so we can compare them with the annotations that I have prepared from SMART annotations of Mbp1 orthologues from the ten reference fungi.
Visual comparison of domain annotations in R
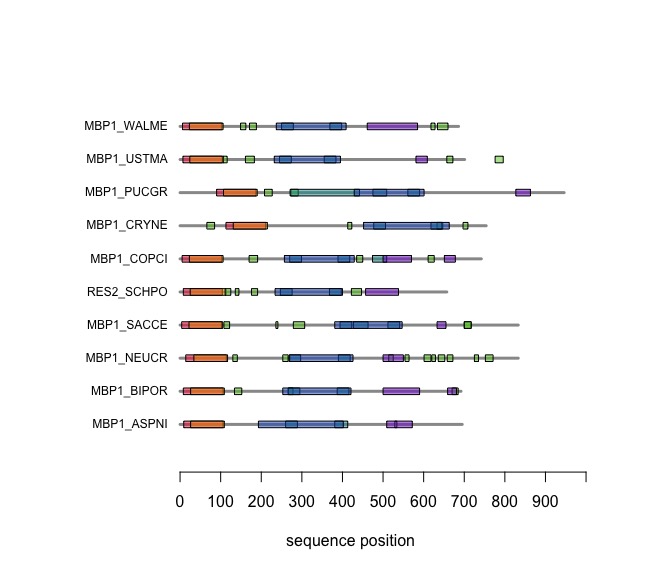
The versatile plotting functions of R allow us to compare domain annotations. The distribution of segments that are annotated as "low-complexity, presumably disordered, is particularly interesting: these are functional features that are often not associated with sequence similarity but may have arisen from convergent evolution. Those would not be detectable through sequence alignment - which is after all based on amino acid pair scores and therefore context independent.
In the following code tutorial, we create a plot similar to the CDD and SMART displays. It is based on the SMART domain annotations of the six-fungal reference species for the course.
Task:
- Return to your RStudio session.
- Make sure you have saved
myDBas instructed previously. Then quit the program, restart, and re-open the project via the File → Recent projects ... menu. This is to clear out-of-date assignments and functions from the workspace. - Do not type
init()yet, but pull the most recent version of files from github. Then typeinit(). - Study and work through the code in the
SMART domain annotationssection of theBCH441_A04.Rscript. This includes entering your domain and other feature annotations into the database. - At the end of the script, print out your plot of the domain annotations for MB1_YFO and the reference proteins. Bring this plot with you for the next quiz.
- Can this plot be improved? What would you do differently to maximize its utility from an information-design point of view?
When you execute the code, your plot should look similar to this one:
A note on the R code up to this point: You will find that we have been writing a lot of nested expressions for selections that join data from multiple tables of our data model. When I teach R workshops for graduate students, postdocs and research fellows, I find that the single greatest barrier in their actual research work is the preparation of data for analysis: filtering, selecting, cross-referencing, and integrating data from different sources. By now, I hope you will have acquired a somewhat robust sense for achieving this. You can imagine that there are ways to simplify those tasks with functions you write, or special resources from a variety of different packages you cab install. But the "pedestrian" approach we have been taking in our scripts has the advantage of working from a very small number of principles, with very few syntactic elements.
Multiple Sequence Alignment
In order to perform a multiple sequence alignment, we obviously need a set of homologous sequences. This is not trivial. All interpretation of MSA results depends absolutely on how the input sequences were chosen. Should we include only orthologues, or paralogues as well? Should we include only species with fully sequenced genomes, or can we tolerate that some orthologous genes are possibly missing for a species? Should we include all sequences we can lay our hands on, or should we restrict the selection to a manageable number of representative sequences? All of these choices influence our interpretation:
- orthologues are expected to be functionally and structurally conserved;
- paralogues may have divergent function but have similar structure;
- missing genes may make paralogs look like orthologs; and
- selection bias may weight our results toward sequences that are over-represented and do not provide a fair representation of evolutionary divergence.
Computing an MSA in R
Let's use the Bioconductor msa package to align the sequences we have. Study and run the following code
Task:
- Return to your RStudio session.
- Make sure you have saved
myDBas instructed previously. - Bring code and data resources up to date:
- pull the most recent version of the project from GitHub
- type
init()to lod the most recent files and functions - re-merge your current
myDB
- Study and work through the code in the
Multiple sequence alignmentssection of theBCH441_A04.Rscript. - Note that the final task asks you to print out some results and bring them to class for the next quiz.
Sequence alignment editors
Really excellent software tools have been written that help you visualize and manually curate multiple sequence alignments. If anything, I think they tend to do too much. Past versions of the course have used Jalview, but I have heard good things of AliView (and if you are on a Mac seqotron might interest you, but I only cover software that is free and runs on all three major platforms).
Right now, I am just mentioning the two alignment editors. If you have experience with comparing them, let us know.
- [Jalview] an integrated MSA editor and sequence annotation workbench from the Barton lab in Dundee. Lots of functions.
- [AliView] from Uppsala: fast, lean, looks to be very practical.
Computing alignments
try two MSA's algorithms and load them in Jalview. Locally: which one do you prefer? Modify the consensus. Annotate domains.
The EBI has a very convenient page to access a number of MSA algorithms. This is especially convenient when you want to compare, e.g. T-Coffee and Muscle and MAFFT results to see which regions of your alignment are robust. You could use any of these tools, just paste your sequences into a Webform, download the results and load into Jalview. Easy.
But even easier is to calculate the alignments directly from Jalview. available. (Not today. Bummer.)
No. Claculate an external alignment and import.
- Calculate a MAFFT alignment using the Jalview Web service option
Task:
- In Jalview, select Web Service → Alignment → MAFFT with defaults.... The alignment is calculated in a few minutes and displayed in a new window.
- Calculate a MAFFT alignment when the Jalview Web service is NOT available
Task:
- In Jalview, select File → Output to Textbox → FASTA
- Copy the sequences.
- Navigate to the MAFFT Input form at the EBI.
- Paste your sequences into the form.
- Click on Submit.
- Close the Jalview sequence window and either save your MAFFT alignment to file and load in Jalview, or simply 'File → Input Alignment → from Textbox, paste and click New Window.
In any case, you should now have an alignment.
Task:
- Choose Colour → Hydrophobicity and → by Conservation. Then adjust the slider left or right to see which columns are highly conserved. You will notice that the Swi6 sequence that was supposed to align only to the ankyrin domains was in fact aligned to other parts of the sequence as well. This is one part of the MSA that we will have to correct manually and a common problem when aligning sequences of different lengths.
Links and resources
Eddy (2004) Where did the BLOSUM62 alignment score matrix come from?. Nat Biotechnol 22:1035-6. (pmid: 15286655) [ PubMed ] [ DOI ] Many sequence alignment programs use the BLOSUM62 score matrix to score pairs of aligned residues. Where did BLOSUM62 come from?
- Biostrings Quick Overview ( summary of Biostrings functions (PDF))
Sequence annotation
In this assignment we will first download a number of APSES domain containing sequences into our database - and we will automate the process. Then we will annotate them with domain data. First manually, and then again, we will automate this. Next we will extract the APSES domains from our database according to the annotations. And finally we will align them, and visualize domain conservation in the 3D model to study parts of the protein that are conserved.
Downloading Protein Data From the Web
In Assignment 3 we created a schema for a local protein sequence collection, and implemented it as an R list. We added sequences to this database by hand, but since the information should be cross-referenced and available based on a protein's RefSeq ID, we should really have a function that automates this process. It is far too easy to make mistakes and enter erroneous information otherwise.
Task:
Work through the following code examples.# To begin, we load some libraries with functions # we need... # httr sends and receives information via the http # protocol, just like a Web browser. if (!require(httr, quietly=TRUE)) { install.packages("httr") library(httr) } # NCBI's eUtils send information in XML format; we # need to be able to parse XML. if (!require(XML, quietly=TRUE)) { install.packages("XML") library(XML) } # stringr has a number of useful utility functions # to work with strings. E.g. a function that # strips leading and trailing whitespace from # strings. if (!require(stringr, quietly=TRUE)) { install.packages("stringr") library(stringr) } # We will walk through the process with the refSeqID # of yeast Mbp1 refSeqID <- "NP_010227" # UniProt. # The UniProt ID mapping service supports a "RESTful # API": responses can be obtained simply via a Web- # browsers request. Such requests are commonly sent # via the GET or POST verbs that a Webserver responds # to, when a client asks for data. GET requests are # visible in the URL of the request; POST requests # are not directly visible, they are commonly used # to send the contents of forms, or when transmitting # larger, complex data items. The UniProt ID mapping # sevice can accept long lists of IDs, thus using the # POST mechanism makes sense. # R has a POST() function as part of the httr package. # It's very straightforward to use: just define the URL # of the server and send a list of items as the # body of the request. # uniProt ID mapping service URL <- "http://www.uniprot.org/mapping/" response <- POST(URL, body = list(from = "P_REFSEQ_AC", to = "ACC", format = "tab", query = refSeqID)) response # If the query is successful, tabbed text is returned. # and we capture the fourth element as the requested # mapped ID. unlist(strsplit(content(response), "\\s+")) # If the query can't be fulfilled because of a problem # with the server, a WebPage is rturned. But the server status # is also returned and we can check the status code. I have # lately gotten many "503" status codes: Server Not Available... if (response$status_code == 200) { # 200: oK uniProtID <- unlist(strsplit(content(response), "\\s+"))[4] if (is.na(uniProtID)) { warning(paste("UniProt ID mapping service returned NA.", "Check your RefSeqID.")) } } else { uniProtID <- NA warning(paste("No uniProt ID mapping available:", "server returned status", response$status_code)) } uniProtID # Let's see what we got... # This should be "P39678" # (or NA if the query failed)
Next, we'll retrieve data from the various NCBI databases.It is has become unreasonably difficult to screenscrape the NCBI site since the actual page contents are dynamically loaded via AJAX. This may be intentional, or just overengineering. While NCBI offers a subset of their data via the eutils API and that works well enough, some of the data that is available to the Web browser's eyes is not served to a program.
The eutils API returns data in XML format. Have a look at the following URL in your browser to see what that looks like:
http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=protein&term=NP_010227
# In order to parse such data, we need tools from the # XML package. # First we build a query URL... eUtilsBase <- "http://eutils.ncbi.nlm.nih.gov/entrez/eutils/" # Then we assemble an URL that will search for get the # unique, NCBI internal identifier, the GI number, # for our refSeqID... URL <- paste(eUtilsBase, "esearch.fcgi?", # ...using the esearch program # that finds an entry in an # NCBI database "db=protein", "&term=", refSeqID, sep="") # Copy the URL and paste it into your browser to see # what the response should look like. URL # To fetch a response in R, we use the function htmlParse() # with our URL as its argument. response <- htmlParse(URL) response # This is XML. We can take the response apart into # its indvidual components with the xmlToList function. xmlToList(response) # Note how the XML "tree" is represented as a list of # lists of lists ... # If we know exactly what elelement we are looking for, # we can extract it from this structure: xmlToList(response)[["body"]][["esearchresult"]][["idlist"]][["id"]] # But this is not very robus, it would break with the # slightest change that the NCBI makes to their response # and the NCBI changes things A LOT! # Somewhat more robust is to specify the type of element # we want - its the text contained in an <id>...</id> # elelement, and use the XPath XML parsing language to # retrieve it. # getNodeSet() lets us fetch tagged contents by # applying toString.XMLNode() to it... node <- getNodeSet(response, "//id/text()") unlist(lapply(node, toString.XMLNode)) # "6320147 " # We will be doing this a lot, so we write a function # for it... node2string <- function(doc, tag) { # an extractor function for the contents of elements # between given tags in an XML response. # Contents of all matching elements is returned in # a vector of strings. path <- paste("//", tag, "/text()", sep="") nodes <- getNodeSet(doc, path) return(unlist(lapply(nodes, toString.XMLNode))) } # using node2string() ... GID <- node2string(response, "id") GID # The GI is the pivot for all our data requests at the # NCBI. # Let's first get the associated data for this GI URL <- paste(eUtilsBase, "esummary.fcgi?", "db=protein", "&id=", GID, "&version=2.0", sep="") response <- htmlParse(URL) URL response taxID <- node2string(response, "taxid") organism <- node2string(response, "organism") taxID organism # Next, fetch the actual sequence URL <- paste(eUtilsBase, "efetch.fcgi?", "db=protein", "&id=", GID, "&retmode=text&rettype=fasta", sep="") response <- htmlParse(URL) URL response fasta <- node2string(response, "p") fasta seq <- unlist(strsplit(fasta, "\\n"))[-1] # Drop the first elelment, # it is the FASTA header. seq # Next, fetch the crossreference to the NCBI Gene # database URL <- paste(eUtilsBase, "elink.fcgi?", "dbfrom=protein", "&db=gene", "&id=", GID, sep="") response <- htmlParse(URL) URL response geneID <- node2string(response, "linksetdb/id") geneID # ... and the actual Gene record: URL <- paste(eUtilsBase, "esummary.fcgi?", "&db=gene", "&id=", geneID, sep="") response <- htmlParse(URL) URL response name <- node2string(response, "name") genome_xref <- node2string(response, "chraccver") genome_from <- node2string(response, "chrstart")[1] genome_to <- node2string(response, "chrstop")[1] name genome_xref genome_from genome_to # So far so good. But since we need to do this a lot # we need to roll all of this into a function. # I have added the function to the dbUtilities code # so you can update it easily. # Run: updateDbUtilities("55ca561e2944af6e9ce5cf2a558d0a3c588ea9af") # If that is successful, try these three testcases myNewDB <- createDB() tmp <- fetchProteinData("NP_010227") # Mbp1p tmp myNewDB <- addToDB(myNewDB, tmp) myNewDB tmp <- fetchProteinData("NP_011036") # Swi4p tmp myNewDB <- addToDB(myNewDB, tmp) myNewDB tmp <- fetchProteinData("NP_012881") # Phd1p tmp myNewDB <- addToDB(myNewDB, tmp) myNewDB
This new
fetchProteinData()function seems to be quite convenient. I have compiled a set of APSES domain proteins for ten fungi species and loaded the 48 protein's data into an R database in a few minutes. This "reference database" will be automatically loaded for you with the next dbUtilities update. Note that it will be recreated every time you start up R. This means two things: (i) if you break something in the reference database, it's not a problem. (ii) if you store your own data in it, it will be lost. In order to add your own genes, you need to make a working copy for yourself.
Computer literacy
- Digression - some musings on computer literacy and code engineering.
It's really useful to get into a consistent habit of giving your files a meaningful name. The name should include something that tells you what the file contains, and something that tells you the date or version. I give versions major and minor numbers, and - knowing how much things always change - I write major version numbers with a leading zero eg.
04so that they will be correctly sorted by name in a directory listing. The same goes for dates: always writeYYYY-MM-DDto ensure proper sorting.On the topic of versions: creating the database with its data structures and the functions that operate on them is an ongoing process, and changes in one part of the code may have important consequences for another part. Imagine I made a poor choice of a column name early on: changing that would need to be done in every single function of the code that reads or writes or analyzes data. Once the code reaches a certain level of complexity, organizing it well is just as important as writing it in the first place. In the new update of
dbUtilities.R, a database has a$versionelement, and every function checks that the database version matches the version for which the function was written. Obviously, this also means the developer must provide tools to migrate contents from an older version to a newer version. And since migrating can run into trouble and leave all data in an inconsistent and unfixable state, it's a good time to remind you to back up important data frequently. Of course you will want to save your database once you've done any significant work with it. And you will especially want to save the databases you create for your Term Project. But you should also (and perhaps more importantly) save the script that you use to create the database in the first place. And on that note: when was the last time you made a full backup of your computer's hard-drive? Too long ago? I thought so.- Backup your hard-drive now.
If your last backup at the time of next week's quiz was less than two days ago, you will receive a 0.5 mark bonus.
New Database
Here is some sample code to work with the new database, enter new protein data for YFO, save it and load it again when needed.
# You don't need to load the reference database refDB. If # everything is set up correctly, it gets loaded at startup. # (Just so you know: you can turn off that behaviour if you # ever should want to...) # First you need to load the newest version of dbUtilities.R updateDButilities("7bb32ab3d0861ad81bdcb9294f0f6a737b820bf9") # If you get an error: # Error: could not find function "updateDButilities" # ... then it seems you didn't do the previous update. # Try getting the update with the new key but the previous function: # updateDbUtilities() # # If that function is not found either, confirm that your ~/.Rprofile # actually loads dbUtilites.R from your project directory. # As a desperate last resort, you could uncomment # the following piece of code and run the update # without verification... # # URL <- "http://steipe.biochemistry.utoronto.ca/abc/images/f/f9/DbUtilities.R" # download.file(URL, paste(PROJECTDIR, "dbUtilities.R", sep="")), method="auto") # source(paste(PROJECTDIR, "dbUtilities.R", sep="")) # # But be cautious: there is no verification. You yourself need # to satisfy yourself that this "file from the internet" is what # it should be, before source()'ing it... # After the file has been source()'d, refDB exists. ls(refDB) # check the contents of refDB: refDB$protein$name refDB$taxonomy # list refSeqIDs for saccharomyces cerevisiae genes. refDB$protein[refDB$protein$taxID == 559292, "refSeqID"] # To add some genes from YFO, I proceed as follows. # Obviously, you need to adapt this to your YFO # and the sequences in YFO that you have found # with your PSI-BLAST search. # Let's assume my YFO is the fly agaric (amanita muscaria) # and its APSES domain proteins have the following IDs # (these are not refSeq btw. and thus unlikely # to be found in UniProt) ... # KIL68212 # KIL69256 # KIL65817 # # First, I create a copy of the database with a name that # I will recognize to be associated with my YFO. amamuDB <- refDB # Then I fetch my protein data ... tmp1 <- fetchProteinData("KIL68212") tmp2 <- fetchProteinData("KIL69256") tmp3 <- fetchProteinData("KIL65817") # ... and if I am satisfied that it contains what I # want, I add it to the database. amamuDB <- addToDB(amamuDB, tmp1) amamuDB <- addToDB(amamuDB, tmp2) amamuDB <- addToDB(amamuDB, tmp3) # Then I make a local backup copy. Note the filename and # version number :-) save(amamuDB, file="amamuDB.01.RData") # Now I can explore my new database ... amamuDB$protein[amamuDB$protein$taxID == 946122, "refSeqID"] # ... but if anything goes wrong, for example # if I make a mistake in checking which # rows contain taxID 946122 ... amamuDB$protein$taxID = 946122 # Ooops ... what did I just do wrong? # ... wnat happened instead? amamuDB$protein$taxID # ... I can simply recover from my backup copy: load("amamuDB.01.RData") amamuDB$protein$taxID
Task:
- Create your own version of the protein database by adding all the genes from YFO that you have discovered with the PSI-BLAST search for the APSES domain. Save it.
R code: load alignment and compute information scores
As discussed in the lecture, Shannon information is calculated as the difference between expected and observed entropy, where entropy is the negative sum over probabilities times the log of those probabilities:
*a review of regex range characters +?*{min,max}, and greedy. *build an AT-hook motif matcher https://en.wikipedia.org/wiki/AT-hook
Here we compute Shannon information scores for aligned positions of the APSES domain, and plot the values in R. You can try this with any part of your alignment, but I have used only the aligned residues for the APSES domain for my example. This is a good choice for a first try, since there are (almost) no gaps.Task:
- Export only the sequences of the aligned APSES domains to a file on your computer, in FASTA format as explained below. You could call this:
Mbp1_All_APSES.fa.- Use your mouse and clik and drag to select the aligned APSES domains in the alignment window.
- Copy your selection to the clipboard.
- Use the main menu (not the menu of your alignment window) and select File → Input alignment → from Textbox; paste the selection into the textbox and click New Window.
- Use File → save as to save the aligned siequences in multi-FASTA format under the filename you want in your R project directory.
- Explore the R-code below. Be sure that you understand it correctly. Note that this code does not implement any sampling bias correction, so positions with large numbers of gaps will receive artificially high scores (the alignment looks like the gap charecter were a conserved character).
# CalculateInformation.R # Calculate Shannon information for positions in a multiple sequence alignment. # Requires: an MSA in multi FASTA format # It is good practice to set variables you might want to change # in a header block so you don't need to hunt all over the code # for strings you need to update. # setwd("/your/R/working/directory") mfa <- "MBP1_All_APSES.fa" # ================================================ # Read sequence alignment fasta file # ================================================ # read MFA datafile using seqinr function read.fasta() library(seqinr) tmp <- read.alignment(mfa, format="fasta") MSA <- as.matrix(tmp) # convert the list into a characterwise matrix # with appropriate row and column names using # the seqinr function as.matrix.alignment() # You could have a look under the hood of this # function to understand beter how to convert a # list into something else ... simply type # "as.matrix.alignment" - without the parentheses # to retrieve the function source code (as for any # function btw). ### Explore contents of and access to the matrix of sequences MSA MSA[1,] MSA[,1] length(MSA[,1]) # ================================================ # define function to calculate entropy # ================================================ entropy <- function(v) { # calculate shannon entropy for the aa vector v # Note: we are not correcting for small sample sizes # here. Thus if there are a large number of gaps in # the alignment, this will look like small entropy # since only a few amino acids are present. In the # extreme case: if a position is only present in # one sequence, that one amino acid will be treated # as 100% conserved - zero entropy. Sampling error # corrections are discussed eg. in Schneider et al. # (1986) JMB 188:414 l <- length(v) a <- rep(0, 21) # initialize a vector with 21 elements (20 aa plus gap) # the set the name of each row to the one letter # code. Through this, we can access a row by its # one letter code. names(a) <- unlist(strsplit("acdefghiklmnpqrstvwy-", "")) for (i in 1:l) { # for the whole vector of amino acids c <- v[i] # retrieve the character a[c] <- a[c] + 1 # increment its count by one } # note: we could also have used the table() function for this tot <- sum(a) - a["-"] # calculate number of observed amino acids # i.e. subtract gaps a <- a/tot # frequency is observations of one amino acid # divided by all observations. We assume that # frequency equals probability. a["-"] <- 0 for (i in 1:length(a)) { if (a[i] != 0) { # if a[i] is not zero, otherwise leave as is. # By definition, 0*log(0) = 0 but R calculates # this in parts and returns NaN for log(0). a[i] <- a[i] * (log(a[i])/log(2)) # replace a[i] with # p(i) log_2(p(i)) } } return(-sum(a)) # return Shannon entropy } # ================================================ # calculate entropy for reference distribution # (from UniProt, c.f. Assignment 2) # ================================================ refData <- c( "A"=8.26, "Q"=3.93, "L"=9.66, "S"=6.56, "R"=5.53, "E"=6.75, "K"=5.84, "T"=5.34, "N"=4.06, "G"=7.08, "M"=2.42, "W"=1.08, "D"=5.45, "H"=2.27, "F"=3.86, "Y"=2.92, "C"=1.37, "I"=5.96, "P"=4.70, "V"=6.87 ) ### Calculate the entropy of this distribution H.ref <- 0 for (i in 1:length(refData)) { p <- refData[i]/sum(refData) # convert % to probabilities H.ref <- H.ref - (p * (log(p)/log(2))) } # ================================================ # calculate information for each position of # multiple sequence alignment # ================================================ lAli <- dim(MSA)[2] # length of row in matrix is second element of dim(<matrix>). I <- rep(0, lAli) # initialize result vector for (i in 1:lAli) { I[i] = H.ref - entropy(MSA[,i]) # I = H_ref - H_obs } ### evaluate I I quantile(I) hist(I) plot(I) # you can see that we have quite a large number of columns with the same, # high value ... what are these? which(I > 4) MSA[,which(I > 4)] # And what is in the columns with low values? MSA[,which(I < 1.5)] # =================================================== # plot the information # (c.f. Assignment 5, see there for explanations) # =================================================== IP <- (I-min(I))/(max(I) - min(I) + 0.0001) nCol <- 15 IP <- floor(IP * nCol) + 1 spect <- colorRampPalette(c("#DD0033", "#00BB66", "#3300DD"), bias=0.6)(nCol) # lets set the information scores from single informations to grey. We # change the highest level of the spectrum to grey. #spect[nCol] <- "#CCCCCC" Icol <- vector() for (i in 1:length(I)) { Icol[i] <- spect[ IP[i] ] } plot(1,1, xlim=c(0, lAli), ylim=c(-0.5, 5) , type="n", bty="n", xlab="position in alignment", ylab="Information (bits)") # plot as rectangles: height is information and color is coded to information for (i in 1:lAli) { rect(i, 0, i+1, I[i], border=NA, col=Icol[i]) } # As you can see, some of the columns reach very high values, but they are not # contiguous in sequence. Are they contiguous in structure? We will find out in # a later assignment, when we map computed values to structure.
Calculating conservation scores
Task:
- Study this code carefully, execute it, section by section and make sure you understand all of it. Ask on the list if anything is not clear.
# BiostringsExample.R # Short tutorial on sequence alignment with the Biostrings package. # Boris Steipe for BCH441, 2013 - 2014 # setwd("~/path/to/your/R_files/") setwd("~/Documents/07.TEACHING/37-BCH441 Bioinformatics 2014/05-Materials/Assignment_5 data") # Biostrings is a package within the bioconductor project. # bioconducter packages have their own installation system, # they are normally not installed via CRAN. # First, you load the BioConductor installer... source("http://bioconductor.org/biocLite.R") # Then you can install the Biostrings package and all of its dependencies. biocLite("Biostrings") # ... and load the library. library(Biostrings) # Some basic (technical) information is available ... library(help=Biostrings) # ... but for more in depth documentation, use the # so called "vignettes" that are provided with every R package. browseVignettes("Biostrings") # In this code, we mostly use functions that are discussed in the # pairwise alignement vignette. # Read in two fasta files - you will need to edit this for YFO sacce <- readAAStringSet("mbp1-sacce.fa", format="fasta") # "USTMA" is used only as an example here - modify for YFO :-) ustma <- readAAStringSet("mbp1-ustma.fa", format="fasta") sacce names(sacce) names(sacce) <- "Mbp1 SACCE" names(ustma) <- "Mbp1 USTMA" # Example only ... modify for YFO width(sacce) as.character(sacce) # Biostrings takes a sophisticated approach to sequence alignment ... ?pairwiseAlignment # ... but the use in practice is quite simple: ali <- pairwiseAlignment(sacce, ustma, substitutionMatrix = "BLOSUM50") ali pattern(ali) subject(ali) writePairwiseAlignments(ali) p <- aligned(pattern(ali)) names(p) <- "Mbp1 SACCE aligned" s <- aligned(subject(ali)) names(s) <- "Mbp1 USTMA aligned" # don't overwrite your EMBOSS .fal files writeXStringSet(p, "mbp1-sacce.R.fal", append=FALSE, format="fasta") writeXStringSet(s, "mbp1-ustma.R.fal", append=FALSE, format="fasta") # Done.- Compare the alignments you received from the EMBOSS server, and that you computed using R. Are they approximately the same? Exactly? You did use different matrices and gap parameters, so minor differences are to be expected. But by and large you should get the same alignments.
We will now use the aligned sequences to compute a graphical display of alignment quality.
Task:
- Study this code carefully, execute it, section by section and make sure you understand all of it. Ask on the list if anything is not clear.
# aliScore.R # Evaluating an alignment with a sliding window score # Boris Steipe, October 2012. Update October 2013 setwd("~/path/to/your/R_files/") # Scoring matrices can be found at the NCBI. # ftp://ftp.ncbi.nih.gov/blast/matrices/BLOSUM62 # It is good practice to set variables you might want to change # in a header block so you don't need to hunt all over the code # for strings you need to update. # fa1 <- "mbp1-sacce.R.fal" fa2 <- "mbp1-ustma.R.fal" code1 <- "SACCE" code2 <- "USTMA" mdmFile <- "BLOSUM62.mdm" window <- 9 # window-size (should be an odd integer) # ================================================ # Read data files # ================================================ # read fasta datafiles using seqinr function read.fasta() install.packages("seqinr") library(seqinr) tmp <- unlist(read.fasta(fa1, seqtype="AA", as.string=FALSE, seqonly=TRUE)) seq1 <- unlist(strsplit(as.character(tmp), split="")) tmp <- unlist(read.fasta(fa2, seqtype="AA", as.string=FALSE, seqonly=TRUE)) seq2 <- unlist(strsplit(as.character(tmp), split="")) if (length(seq1) != length(seq2)) { print("Error: Sequences have unequal length!") } lSeq <- length(seq1) # ================================================ # Read scoring matrix # ================================================ MDM <- read.table(mdmFile, skip=6) # This is a dataframe. Study how it can be accessed: MDM MDM[1,] MDM[,1] MDM[5,5] # Cys-Cys MDM[20,20] # Val-Val MDM[,"W"] # the tryptophan column MDM["R","W"] # Arg-Trp pairscore MDM["W","R"] # Trp-Arg pairscore: pairscores are symmetric colnames(MDM) # names of columns rownames(MDM) # names of rows colnames(MDM)[3] # third column rownames(MDM)[12] # twelfth row # change the two "*" names to "-" so we can use them to score # indels of the alignment. This is a bit of a hack, since this # does not reflect the actual indel penalties (which is, as you) # remember from your lectures, calculated as a gap opening # + gap extension penalty; it can't be calculated in a pairwise # manner) EMBOSS defaults for BLODSUM62 are opening -10 and # extension -0.5 i.e. a gap of size 3 (-11.5) has approximately # the same penalty as a 3-character score of "-" matches (-12) # so a pairscore of -4 is not entirely unreasonable. colnames(MDM)[24] rownames(MDM)[24] colnames(MDM)[24] <- "-" rownames(MDM)[24] <- "-" colnames(MDM)[24] rownames(MDM)[24] MDM["Q", "-"] MDM["-", "D"] # so far so good. # ================================================ # Tabulate pairscores for alignment # ================================================ # It is trivial to create a pairscore vector along the # length of the aligned sequences. PS <- vector() for (i in 1:lSeq) { aa1 <- seq1[i] aa2 <- seq2[i] PS[i] = MDM[aa1, aa2] } PS # The same vector could be created - albeit perhaps not so # easy to understand - with the expression ... MDM[cbind(seq1,seq2)] # ================================================ # Calculate moving averages # ================================================ # In order to evaluate the alignment, we will calculate a # sliding window average over the pairscores. Somewhat surprisingly # R doesn't (yet) have a native function for moving averages: options # that are quoted are: # - rollmean() in the "zoo" package http://rss.acs.unt.edu/Rdoc/library/zoo/html/rollmean.html # - MovingAverages() in "TTR" http://rss.acs.unt.edu/Rdoc/library/TTR/html/MovingAverages.html # - ma() in "forecast" http://robjhyndman.com/software/forecast/ # But since this is easy to code, we shall implement it ourselves. PSma <- vector() # will hold the averages winS <- floor(window/2) # span of elements above/below the centre winC <- winS+1 # centre of the window # extend the vector PS with zeros (virtual observations) above and below PS <- c(rep(0, winS), PS , rep(0, winS)) # initialize the window score for the first position winScore <- sum(PS[1:window]) # write the first score to PSma PSma[1] <- winScore # Slide the window along the sequence, and recalculate sum() # Loop from the next position, to the last position that does not exceed the vector... for (i in (winC + 1):(lSeq + winS)) { # subtract the value that has just dropped out of the window winScore <- winScore - PS[(i-winS-1)] # add the value that has just entered the window winScore <- winScore + PS[(i+winS)] # put score into PSma PSma[i-winS] <- winScore } # convert the sums to averages PSma <- PSma / window # have a quick look at the score distributions boxplot(PSma) hist(PSma) # ================================================ # Plot the alignment scores # ================================================ # normalize the scores PSma <- (PSma-min(PSma))/(max(PSma) - min(PSma) + 0.0001) # spread the normalized values to a desired range, n nCol <- 10 PSma <- floor(PSma * nCol) + 1 # Assign a colorspectrum to a vector (with a bit of colormagic, # don't worry about that for now). Dark colors are poor scores, # "hot" colors are high scores spect <- colorRampPalette(c("black", "red", "yellow", "white"), bias=0.4)(nCol) # Color is an often abused aspect of plotting. One can use color to label # *quantities* or *qualities*. For the most part, our pairscores measure amino # acid similarity. That is a quantity and with the spectrum that we just defined # we associte the measured quantities with the color of a glowing piece # of metal: we start with black #000000, then first we ramp up the red # (i.e. low-energy) part of the visible spectrum to red #FF0000, then we # add and ramp up the green spectrum giving us yellow #FFFF00 and finally we # add blue, giving us white #FFFFFF. Let's have a look at the spectrum: s <- rep(1, nCol) barplot(s, col=spect, axes=F, main="Color spectrum") # But one aspect of our data is not quantitatively different: indels. # We valued indels with pairscores of -4. But indels are not simply poor alignment, # rather they are non-alignment. This means stretches of -4 values are really # *qualitatively* different. Let's color them differently by changing the lowest # level of the spectrum to grey. spect[1] <- "#CCCCCC" barplot(s, col=spect, axes=F, main="Color spectrum") # Now we can display our alignment score vector with colored rectangles. # Convert the integers in PSma to color values from spect PScol <- vector() for (i in 1:length(PSma)) { PScol[i] <- spect[ PSma[i] ] # this is how a value from PSma is used as an index of spect } # Plot the scores. The code is similar to the last assignment. # Create an empty plot window of appropriate size plot(1,1, xlim=c(-100, lSeq), ylim=c(0, 2) , type="n", yaxt="n", bty="n", xlab="position in alignment", ylab="") # Add a label to the left text (-30, 1, adj=1, labels=c(paste("Mbp1:\n", code1, "\nvs.\n", code2)), cex=0.9 ) # Loop over the vector and draw boxes without border, filled with color. for (i in 1:lSeq) { rect(i, 0.9, i+1, 1.1, border=NA, col=PScol[i]) } # Note that the numbers along the X-axis are not sequence numbers, but numbers # of the alignment, i.e. sequence number + indel length. That is important to # realize: if you would like to add the annotations from the last assignment # which I will leave as an exercise, you need to map your sequence numbering # into alignment numbering. Let me know in case you try that but need some help.
Genomics
Large scale genome sequencing and annotation has made a wealth of information available that is all related to the same biological objects: the DNA. The information however can be of very different types, it includes:
- the actual sequence
- sequence variants (SNPs and CNVs)
- conservation between related species
- genes (with introns and exons)
- mRNAs
- expression levels
- regulatory features such as transcription factor bindings sites
and much more.
Since all of this information relates to specific positions or ranges on the chromosome, displaying it alongside the chromosomal coordinates is a useful way to integrate and visualize it. We call such strips of annotation tracts and display them in genome browsers. Quite a number of such browsers exist and most work on the same principle: server hosted databases are queried through a Web interface; the resulting data is displayed graphically in a Web browser window. The large data centres each have their own browsers, but arguably the best engineered, most informative and mostly widely used one is provided by the University of California Santa Cruz (UCSC) Genome Browser Project.
Compiling the data requires a massive annotation effort, which has not been completed for all genome-sequenced species. In particular, not all of our YFOs have been included in the major model-organism annotation efforts. The general strategy for analysis of a gene in YFO is thus to map it to homologous genes in model organisms. In this assignment you will explore the UCSC genome browser and we will go through an exercise that relates fungal replication genes to human genes. We have previously focused a lot on Mbp1 homologs, but these have no clear equivalences in "higher" eukaryotes. However one of the key target genes of Mbp1 is the cell cycle protein Cdc6, which is well conserved in fungi and other eukaryotes eukaryotes and has a human homolog. Since generally speaking the annotation level for human genes is the highest, we will have a closer look at that gene.
The UCSC genome browser
The University of California Santa Cruz (UCSC) Genome Browser Project has the largest offering of annotation information. However it is strictly model-organism oriented and you will probably not find YFO among its curated genomes. Nevertheless, if you are studying eg. human genes, or yeast, the UCSC browser will probably be your first choice.
Task:
In this task you will access the UCSC genome browser view of the human Cdc6 gene. You will explore some of the very large number of tracks that are available and study the transcription factor binding region.- Navigate to the UCSC Genome Bioinformatics entry page and follow the link to the Genome Browser in the "Our tools" section.
- Click on the link to humans. Note that this is the hg38 assembly.
- Enter CDC6 into the "Position/Search Term" field and click "Go". You should get a list of entries, click on the top link, the gene on chromosome 17: CDC6 (uc002huj.2) at chr17:40287633-40304657
- Zoom out 1.5x to view the upstream regulatory region: the end of the adjacent WIPF2 gene should have just come into view on the left.
- Study the Genome Browser view of the human CDC6 homolog.
- In particular, note the extensive functional annotations of DNA and the alignments of vertebrate syntenic regions that allow detailed genomic comparisons.
- Distinguish between exon and intron sequence.
- Note that the mammal Conservation track has high values for all of the exons, but not only for exons.
- Find more information on the "Layered H3K27Ac" tract.
- Note the large number of available tracks that have been integrated into this view. Most of them are switched off. Find the Regulation section, and follow the link to the "ORegAnno" information to see what that is about. Note that you can switch individual annotations on or off on this page, as well as set the display format for all of the results. Select the check-box only for "transcription factor binding site" to be on, select the "Display mode" to full and click submit.
- Study this information and note:
- There is a cluster of TFBS just upstream of the transcription initiation site.
- This cluster coincides with the highest H3K27Ac density.
- If you <control>-click (right-click?) on the top orange bar of this cluster, a contextual menu opens from which you can access the details page for OREG1791811 in a new window. Follow the link to the RBL2 transcription factor via ENST00000379935 ... from where you can access transcript and gene and expression and protein family and GO and all other information.
- Go back to the Genome Browser and set the ORegAnno tract to "pack" and click "refresh".
- Slide the SNP track to just beneath the RefSeq genes track that contains the introns and exons. You will notice that one of the SNPs is green, and two are red. Why? Set the "Common SNPs" track display mode to "pack" and click "refresh".
Based on this kind of information, it should be straightforward to identify human transcription factors that potentially regulate human Cdc6 and determine - via sequence comparisons - whether any of them are homologous to any of the yeast transcription factors or factors in YFO. Through a detailed analysis of existing systems, their regulatory components and the conservation of regulation, one can in principle establish functional equivalences across large evolutionary distances.
Task:
Finally:- Print this page, but print the first page only.
- With a red pen, mark and label the following four items on your print-out:
- The first exon of CDC6.
- The chromosomal coordinates of the current view.
- The binding sites for the transcription factors that bind to the CDC6 promoter.
- The locations of the missense-variant SNPs.
- Write your name and Student number on this page and bring it to class to hand it in on Tuesday.
Links and resources
Wang et al. (2013) A brief introduction to web-based genome browsers. Brief Bioinformatics 14:131-43. (pmid: 22764121) [ PubMed ] [ DOI ] Genome browser provides a graphical interface for users to browse, search, retrieve and analyze genomic sequence and annotation data. Web-based genome browsers can be classified into general genome browsers with multiple species and species-specific genome browsers. In this review, we attempt to give an overview for the main functions and features of web-based genome browsers, covering data visualization, retrieval, analysis and customization. To give a brief introduction to the multiple-species genome browser, we describe the user interface and main functions of the Ensembl and UCSC genome browsers using the human alpha-globin gene cluster as an example. We further use the MSU and the Rice-Map genome browsers to show some special features of species-specific genome browser, taking a rice transcription factor gene OsSPL14 as an example.
Sloan et al. (2016) ENCODE data at the ENCODE portal. Nucleic Acids Res 44:D726-32. (pmid: 26527727) [ PubMed ] [ DOI ] The Encyclopedia of DNA Elements (ENCODE) Project is in its third phase of creating a comprehensive catalog of functional elements in the human genome. This phase of the project includes an expansion of assays that measure diverse RNA populations, identify proteins that interact with RNA and DNA, probe regions of DNA hypersensitivity, and measure levels of DNA methylation in a wide range of cell and tissue types to identify putative regulatory elements. To date, results for almost 5000 experiments have been released for use by the scientific community. These data are available for searching, visualization and download at the new ENCODE Portal (www.encodeproject.org). The revamped ENCODE Portal provides new ways to browse and search the ENCODE data based on the metadata that describe the assays as well as summaries of the assays that focus on data provenance. In addition, it is a flexible platform that allows integration of genomic data from multiple projects. The portal experience was designed to improve access to ENCODE data by relying on metadata that allow reusability and reproducibility of the experiments.
Pazin (2015) Using the ENCODE Resource for Functional Annotation of Genetic Variants. Cold Spring Harb Protoc 2015:522-36. (pmid: 25762420) [ PubMed ] [ DOI ] This article illustrates the use of the Encyclopedia of DNA Elements (ENCODE) resource to generate or refine hypotheses from genomic data on disease and other phenotypic traits. First, the goals and history of ENCODE and related epigenomics projects are reviewed. Second, the rationale for ENCODE and the major data types used by ENCODE are briefly described, as are some standard heuristics for their interpretation. Third, the use of the ENCODE resource is examined. Standard use cases for ENCODE, accessing the ENCODE resource, and accessing data from related projects are discussed. Although the focus of this article is the use of ENCODE data, some of the same approaches can be used with data from other projects.
ENCODE Project Consortium (2011) A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9:e1001046. (pmid: 21526222) [ PubMed ] [ DOI ] The mission of the Encyclopedia of DNA Elements (ENCODE) Project is to enable the scientific and medical communities to interpret the human genome sequence and apply it to understand human biology and improve health. The ENCODE Consortium is integrating multiple technologies and approaches in a collective effort to discover and define the functional elements encoded in the human genome, including genes, transcripts, and transcriptional regulatory regions, together with their attendant chromatin states and DNA methylation patterns. In the process, standards to ensure high-quality data have been implemented, and novel algorithms have been developed to facilitate analysis. Data and derived results are made available through a freely accessible database. Here we provide an overview of the project and the resources it is generating and illustrate the application of ENCODE data to interpret the human genome.
Zarrei et al. (2015) A copy number variation map of the human genome. Nat Rev Genet 16:172-83. (pmid: 25645873) [ PubMed ] [ DOI ] A major contribution to the genome variability among individuals comes from deletions and duplications - collectively termed copy number variations (CNVs) - which alter the diploid status of DNA. These alterations may have no phenotypic effect, account for adaptive traits or can underlie disease. We have compiled published high-quality data on healthy individuals of various ethnicities to construct an updated CNV map of the human genome. Depending on the level of stringency of the map, we estimated that 4.8-9.5% of the genome contributes to CNV and found approximately 100 genes that can be completely deleted without producing apparent phenotypic consequences. This map will aid the interpretation of new CNV findings for both clinical and research applications.
Footnotes and references
- ↑ This is not strictly true in all cases: some algorithms measure similarity through an alignment-free approach, for example by comparing structural features, or domain annotations. These methods are less sensitive, but important when sequences are so highly diverged that no meaningful sequence alignment can be produced.
- ↑ "indel": insertion / deletion – a difference in sequence length between two aligned sequences that is accommodated by gaps in the alignment. Since we can't tell from the comparison of two sequences whether such a change was introduced by insertion into or deletion from the ancestral sequence, we join both into a portmanteau.
- ↑
Russ et al. (2016) Genome Sequence of Spizellomyces punctatus. Genome Announc 4:. (pmid: 27540072) [ PubMed ] [ DOI ] Spizellomyces punctatus is a basally branching chytrid fungus that is found in the Chytridiomycota phylum. Spizellomyces species are common in soil and of importance in terrestrial ecosystems. Here, we report the genome sequence of S. punctatus, which will facilitate the study of this group of early diverging fungi.
- ↑ That directory also contains sourcecode to generate the PAM matrices. This may be of interest if you ever want to produce scoring matrices from your own datasets.
- ↑ That directory also contains sourcecode to generate the PAM matrices. This may be of interest if you ever want to produce scoring matrices from your own datasets.
- ↑ While this seems like an unnecessary limitation, given that we could easily write such code to transform to-upper when looking up values in the MDM, perhaps it is meant as an additional sanity check that we haven't inadvertently included text in the sequence that does not belong there, such as the FASTA header line perhaps.
- ↑ Note that RBMs are usually orthologues, but the definition of orthologue and RBM is not the same. Most importantly, many orthologues are not RBMs. We will explore this more when we discuss phylogenetic inference.
- ↑ One such case we encountered involved a protein that has a corrupted annotation for the DNA binding domain. It appears to be the correct orthologue, but it only has the second highest BLAST score.
- ↑ The -myceta are well supported groups above the Class rank. See Leotiomyceta for details and references.
Ask, if things don't work for you!
- If anything about this page is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- Review Netiquette for the course mailing list.
- Read How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example.
Data Sequence Structure Phylogeny Function