BIO Assignment Week 8
Assignment for Week 7
Predictions: Homology Modeling
| < Assignment 6 | Assignment 8 > |
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz.
Contents
Introduction
- How could the search for ultimate truth have revealed so hideous and visceral-looking an object?
-
- Max Perutz (on his first glimpse of the Hemoglobin structure)
Where is the hidden beauty in structure, and where, the "ultimate truth"? In the previous assignments we have studied sequence conservation in APSES family domains and we have discovered homologues in all fungal species. This is an ancient protein family that had already duplicated to several paralogues at the time the cenancestor of all fungi lived, more than 600,000,000 years ago, in the Vendian period of the Proterozoic era of Precambrian times.
In order to understand how specific residues in the sequence contribute to the putative function of the protein, and why and how they are conserved throughout evolution, we would need to study an explicit molecular model of an APSES domain protein, bound to its cognate DNA sequence. Explanations of a protein's observed properties and functions can't rely on the general fact that it binds DNA, we need to consider details in terms of specific residues and their spatial arrangement. In particular, it would be interesting to correlate the conservation patterns of key residues with their potential to make specific DNA binding interactions. Unfortunately, no APSES domain structures in complex with bound DNA has been solved up to now, and the experimental evidence we have considered in Assignment 2 (Taylor et al., 2000) is not sufficient to unambiguously define the details of how a DNA double helix might be bound. Moreover, at least two distinct modes of DNA binding are known for proteins of the winged-helix superfamily, of which the APSES domain is a member.
In this and the following assignment you will (1) construct a molecular model of the APSES domain from the Mbp1 orthologue in your assigned species, (2) identify similar structures of distantly related domains for which protein-DNA complexes are known, (3) assemble a hypothetical complex structure and (4) consider whether the available evidence allows you to distinguish between different modes of ligand binding.
For the following, please remember the following terminology:
- Target
- The protein that you are planning to model.
- Template
- The protein whose structure you are using as a guide to build the model.
- Model
- The structure that results from the modeling process. It has the Target sequence and is similar to the Template structure.
A brief overview article on the construction and use of homology models is linked to the resource section at the bottom of this page. That section also contains links to other sites and resources you might find useful or interesting.
Preparation
Target sequence
The first step of homology modelling is to determine which sequence to model. We have determined the putative orthologue with conserved function in YFO by reciprocal best match with saccharomyces cervisiae Mbp1. Your sequence was initially found with an APSES domain search in YFO and the alignments with the yeast sequence are straightforward for the most part.
There are two exceptions however: the alignment of ASPFU gene XP_754232 and the CAPCO gene XP_007722875 both are missing part of the domin's N-terminus. This is odd, because this may imply the APSES domain of these genes might not be properly folded. When such surprising results of alignement occurr, you must consider whether there could be an error in the published sequence, perhaps stemming from an erroneous gene model. This is not absolutely germane to this assignment, so I have placed the process into the collapsible section below - optional reading. However it may be useful for you to understand what the issue is here and how to address it.
- Correcting the ASPFU Mbp1 gene model.
An alignment of APSES domain sequence shows the shortened N-terminus of the ASPFU and the CAPCOprotein, relative to SACCE and e.g. the closely related aspergillus nidulans, ASPNI:
APSES domains: Mbp1_SACCE QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAA... Mbp1_ASPNI NVYSATYSSVPVYEFKIGTDSVMRRRSDDWINATHILKVA... Mbp1_ASPFU ----------------------MRRRGDDWINATHILKVA... Mbp1_CAPCO ----------------------MRRRSDDWVNATHILKVA...
We analyse this for the ASPFU gene.
Working from the possibility that this may be a gene model error - e.g. a false translational start, a frameshift due to a sequencing error, or an erroneously modelled intron, we check whether the translation of the genomic sequence supports the presence of the expected amino acids. This is easily done running TBLASTN - BLASTing the protein query against the six reading frames of the ASPFU genome. We find the following:
Aspergillus fumigatus Af293 chromosome 3, whole genome shotgun sequence
Sequence ID: ref|NC_007196.1|Length: 4079167Number of Matches: 2
[...]
Query 10 VDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILE ...
V VYEF S+M+R+ DDW+NATHILK A F K RTRILE ...
Sbjct 3691193 VPVYEFKVDGESVMRRRGDDWINATHILKVAGFDKPARTRILE ...
Indeed, there is sequence upstream of the gene's published translation start that matches well with our query! But where is the correct translation start? For that we need to look at the actual nucleotide sequence and translate it. Remember: BLAST is a local sequence alignment algorithm and it won't retrieve everything that matches to our query, just the best matching segment. ASPFU chromosome 3 is over 4 megabases large, so let us try to obtain only the region we are actually interested in: downstream of bases 3691193, lets say 3691100 (make sure this offset is divisible by three, to stay in the same reading frame) and upstream to, say, 3691372.
- At the NCBI genome project site we search for aspergillus fumigatus.
- At the aspergillus fumigatus genome project site we click on chromosome 3 to access the map viewer.
- Hovering over the Download/View sequence link shows us how an URL to access sequence data is structured:
http://www.ncbi.nlm.nih.gov/projects/mapview/seq_reg.cgi?taxid=746128&chr=3&from=1&to=4079167
- We can easily adapt this to the sequence range we need ...
- ... and follow: http://www.ncbi.nlm.nih.gov/nuccore/NC_007196.1?from=3691003&to=3691243&report=fasta to yield:
>gi|71025130:3691003-3691243 Aspergillus fumigatus Af293 chromosome 3, whole genome shotgun sequence ACGGTTTGCGGAGACGGGCATTATGGCGGCGGTGGATTTCTCAAAAATCTATTCTGCTACATACAGCAGC GTAAGTCTCTTCTAATTGCGTATCTCTGTTTTCCCTACAGCCTCAAATTTTCCCCAATGCCTCTTTCCAT CCATTTTGCCCCTTCCTTCGCCGCGAAGCCAATCTAACGCAGTTCAATAGGTTCCAGTTTACGAGTTCAA AGTCGATGGCGAAAGTGTTATGCGCCGACGA
- To translate this, we navigate to any of the EMBOSS tools servers and use "remap" - we want to see the translation matched to the nucleotide sequence. We turn restriction sites off, translate all three forward frames and paste and manually align the SACCE Mbp1 sequence into the output to see what we expect and what we got. I have selected only the frame(s) that actually give a match, and I have pasted the homologous CAPCO and SACCE sequences (lower case) to demonstrate their similarity:
ASPFU ACGGTTTGCGGAGACGGGCATTATGGCGGCGGTGGATTTCTCAAAAATCTATTCTGCTACATACAGCAGC
ASPFU R F A E T G I M A A V D F S K I Y S A T Y S S
CAPCO m - a f d - k e i y s a t y s n
SACCE m s - - - - n q i y s a r y s g
ASPFU GTAAGTCTCTTCTAATTGCGTATCTCTGTTTTCCCTACAGCCTCAAATTTTCCCCAATGCCTCTTTCCAT
ASPFU V S L F * ...
CAPCO v a - - ...
SACCE v d - - ...
ASPFU CCATTTTGCCCCTTCCTTCGCCGCGAAGCCAATCTAACGCAGTTCAATAGGTTCCAGTTTACGAGTTCAA
... V Y E F K
CAPCO ... v y e l k
SACCE ... v y e f i
ASPFU AGTCGATGGCGAAAGTGTTATGCGCCGACGAGGCGATGATTGGATCAATGCTACACATATTCTTAAA
ASPFU V D G E S V M R R R G D D W I N A T H I L K ...
CAPCO v a g d h i m r r r s d d w v n a t h i l k ...
SACCE h s t g s i m k r k k d d w v n a t h i l k ...
- This clearly shows us that there is N-terminal sequence that ought to be added to the gene model, upstream of the reported translational start of MRRR.... The sequences thus most likely begin as follows:
ASPFU MAAVDFSKIYSATYSSVSLFVYEFKVDGE-----SVMRRRGDDWINATHILK... CAPCO ma-fd-keiysatysnva--vyelkvagd-----himrrrsddwvnathilk... SACCE ms----nqiysarysgvd--ysgvdvyefihstgsimkrkkddwvnathilk...
The fact that the truncated N-terminus appears in both closely related genes and species suggests that what we see here is a mis-annotated intron. The take-home lesson is: if your retrieved protein sequence does not conform to your expectations, it may be worthwhile to follow up with the actual nucleotide sequence.
Template choice and template sequence
The SWISS-MODEL server provides several different options for constructing homology models. The easiest option requires only a target sequence as input. In this mode the program will automatically choose suitable templates and create an input alignment. I would argue however that that is not the best way to use such a service: template choice and alignment both may be significantly influenced by biochemical reasoning, and an automated algorithm cannot make the necessary decisions. Should you use a structure of reduced resolution that however has a ligand bound? Should you move an indel from an active site to a loop region even though the sequence similarity score might be less? Questions like that may yield answers that are counter to the best choices an automated algorithm could make. But Swiss Model is flexible and allows us to upload an explicit alignment between target and template. Please note: the model you will produce is "easy" - the sequence similarity is high and there are no indels to consider, the automated mode would have done just as well. But the strategy we pursue here is suitable also for much more difficult problems. The automated strategy probably is not.
Template choice is the first step. Often more than one related structure can be found in the PDB. We have touched on principles of selecting template structures in the lectures; please refer to the template choice principles page on this Wiki where I have reviewed the principles and discussed more details and alternatives. One can either search the PDB itself through its Advanced Search interface; for example one can search for sequence similarity with a BLAST search, or search for structural similarity by accessing structures according to their CATH or SCOP classification. But the BLAST search is probably the method of choice: after all, the most important measure of the probability of success for homology modeling is sequence similarity.
In Assignment 3, you have defined the extent of the APSES domain in yeast Mbp1. In Assignment 6, you have used PSI-BLAST to search for APSES domains in YFO. In Assignment 7 you have confirmed by Reciprocal Best Match which of these APSES domain sequences is the closest related orthologue to yeast Mbp1. This sequence is the best candidate for having a conserved function similar to yeast Mbp1. Therefore, this sequence is the one you will model: it is called the target for the homology modeling procedure. In the same assignment you have also computed a multiple sequence alignment that includes the sequence of Mbp1 with YFO.
Defining a template means finding a PDB coordinate set that has sufficient sequence similarity to your target that you can build a model based on that template. In Assignment 2 you have used a keyword search at the PDB to find "Mbp1" structures - but some of these structures were not homologs: keyword searches are notoriously unreliable. To find suitable PDB structures, we will perform a BLAST search at the PDB instead.
Task:
- Retrieve your YFO Mbp1-like APSES domain sequence. You can find the domain boundaries for the yeast protein in the Mbp1 annotation reference page, and you can get the aligned sequence from your Jalview alignment, or simply recompute it with the
needleprogram of the EMBOSS suite. This YFO sequence is your target sequence. - Navigate to the PDB.
- Click on Advanced to enter the advanced search interface.
- Open the menu to Choose a Query Type:
- Find the Sequence features section and choose Sequence (BLAST...)
- Paste your target sequence into the Sequence field, select not to mask low-complexity regions and Submit Query. Since the E-value is set rather high by default, you will get a number of low-confidence hits as well as the actual homologs, these have very low E-values.
All hits that are homologs are potentially suitable templates, but some are more suitable than others. Consider how the coordinate sets differ and which features would make each more or less suitable for creating a homology model: you should consider ...
- sequence similarity to your target
- size of expected model (= length of alignment)
- presence or absence of ligands
- experimental method and quality of the data set
Sequence similarity is the most important, but we can have the PDB tabulate the other features concisely for this task.
- There is a menu to create Reports: - select customizable table.
- Select (at least) the following information items:
- Structure Summary
- Experimental Method
- Sequence
- Chain Length
- Ligands
- Ligand Name
- Biological details
- Macromolecule Name
- refinement Details
- Resolution
- R Work
- R free
- click: Create report.
Unfortunately you don't get the E-values into the report, and those should strongly influence your final decision. However in our case the sequences and therefore the E-values of the top three hits are all the same. Neither of the structures has a bound DNA ligand, but the experimental methods and structure quality are different. Two of the sequences have a longer chain-length ... but those are only disordered residues (otherwise these would be better suited templates; regrettably, you'd need to check that in the real world, there is no automatic tool to evaluate disorder and its effects on template choice). In my opinion that leaves pretty much only one unambiguous choice: 1BM8. In case you don't agree, please let me know.
- Finally
- Click on the 1BM8 ID to navigate to the structure page for the template and save the FASTA sequence to your computer. This is the template sequence.
Sequence numbering
It is not straightforward at all how to number sequence in such a project. A "natural" numbering starts with the start-codon of the full length protein and goes sequentially from there. However, this does not map exactly to other numbering schemes we have encountered. As you know the first residue of the APSES domain (as defined by CDD) is not Residue 1 of the Mbp1 protein. The first residue of the 1BM8 FASTA file (one of the related PDB structures) is the fourth residue of the Mbp1 protein. The first residue in the structure is GLN 3, therefore Q is the first residue in a FASTA sequence derived from the cordinate section of the PDB file (the ATOM records. In the 1MB1 structure, the original N-terminal amino acids are present in the molecule, therefore they are present in the FASTA file which starts with MSNQIY..., but they are disordered in the structure and no coordinates are present for M and S. A sequence derived explicitly from the coordinates is therefore different from the reported FASTA sequence, which is really bad because that is what the modeling program has to work with ... and so on. It can get complicated. You need to remember: a sequence number is not absolute, but assigned in a particular context and you need to be careful how to do this.
Fortunately, the numbering for the residues in the coordinate section of our target structure corresponds not to its FASTA sequence, but to the numbering of the gene. Otherwise we would need to renumber the sequence (e.g. by using the bio3D R package). If we would not do this, the sequence numbers in the model might not correspond to the sequence numbers of our target.
The input alignment
The sequence alignment between target and template is the single most important factor that determines the quality of your model. No comparative modeling process will repair an incorrect alignment; it is useful to consider a homology model rather like a three-dimensional map of a sequence alignment rather than a structure in its own right. In a homology modeling project, typically the largest amount of time should be spent on preparing the best possible alignment. Even though automated servers like the SwissModel server will align sequences and select template structures for you, it would be unwise to use these just because they are convenient. You should take advantage of the much more sophisticated alignment methods available. Analysis of wrong models can't be expected to produce right results.
The best possible alignment is usually constructed from a multiple sequence alignment that includes at least the target and template sequence and other related sequences as well. The additional sequences are an important aid in identifying the correct placement of insertions and deletions. Your alignment should have been carefully reviewed by you and wherever required, manually adjusted to move insertions or deletions between target and template out of the secondary structure elements of the template structure.
In most of the Mbp1 orthologues, we do not observe indels in the APSES domain regions. Evolutionary pressure on the APSES domains has selected against indels in the more than 600 million years these sequences have evolved independently in their respective species. To obtain an alignment between the template sequence and the target sequence from your species, proceed as follows.
Task:
Choose on of the following options to align your target and template sequence.
- In Jalview...
- Load your Jalview project with aligned APSES domain sequences or recreate it from the Mbp1 orthologue sequences from the Mbp1 protein orthologs page that I prepared for Assignment 7. Include the sequence of your template protein and re-align.
- Delete all sequence you no longer need, i.e. keep only the APSES domains of the target (from your species) and the template (from the PDB) and choose Edit → Remove empty columns. This is your input alignment.
- Choose File→Output to textbox→FASTA to obtain the aligned sequences. They should both have exactly the same length, i.e. N- or C- termini have to be padded by hyphens if the original sequences had different length. Save the sequences in a text-file.
- Using a different MSA program
- Copy the FASTA formatted sequences of the Mbp1 proteins in the reference species from the Reference APSES domain page.
- Access e.g. the MSA tools page at the EBI.
- Paste the Mbp1 sequence set, your target sequence and the template sequence into the input form.
- Run the alignment and save the output.
- Using the EMBOSS explorer
- Use the
needletool for the alignment ... but remember that pairwise alignments will only be suitable in case the alignment is absolutely unambiguous (such as here) . If there are any indels, an MSA will give much more reliable information.
- By hand
APSES domains are strongly conserved and have few if any indels. You could also simply align by hand.
- Copy the CLUSTAL formatted reference alignment of the Mbp1 proteins in the reference species from the Reference APSES domain page.
- Open a new file in a text editor.
- Paste the Mbp1 sequence set, your target sequence and the template sequence into the file.
- Align by hand, replace all spaces with hyphens and save the output.
Whatever method you use: the result should be a two sequence alignment in multi-FASTA format, that was constructed from a number of supporting sequences and that contains your aligned target and template sequence. This is your input alignment for the homology modeling server. For a Schizosaccharomyces pombe model, which I am using as an example here, it looks like this:
>1BM8_A QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF >Mbp1_SCHPO 2-100 NP_593032 AVHVAVYSGVEVYECFIKGVSVMRRRRDSWLNATQILKVADFDKPQRTRV LERQVQIGAHEKVQGGYGKYQGTWVPFQRGVDLATKYKVDGIMSPILSL
=Creating an Ankyrin domain alignment
APSES domains are relatively easy to identify and annotate but we have had problems with the ankyrin domains in Mbp1 homologues. Both CDD as well as SMART have identified such domains, but while the domain model was based on the same Pfam profile for both, and both annotated approximately the same regions, the details of the alignments and the extent of the predicted region was different.
Mbp1 forms heterodimeric complexes with a homologue, Swi6. Swi6 does not have an APSES domain, thus it does not bind DNA. But it is similar to Mbp1 in the region spanning the ankyrin domains and in 1999 Foord et al. published its crystal structure (1SW6). This structure is a good model for Ankyrin repeats in Mbp1. For details, please refer to the consolidated Mbp1 annotation page I have prepared.
In what follows, we will use the program JALVIEW - a Java based multiple sequence alignment editor to load and align sequences and to consider structural similarity between yeast Mbp1 and its closest homologue in your organism.
In this part of the assignment,
- You will load sequences that are most similar to Mbp1 into an MSA editor;
- You will add sequences of ankyrin domain models;
- You will perform a multiple sequence alignment;
- You will try to improve the alignment manually;
Jalview, loading sequences
Geoff Barton's lab in Dundee has developed an integrated MSA editor and sequence annotation workbench with a number of very useful functions. It is written in Java and should run on Mac, Linux and Windows platforms without modifications.
| Waterhouse et al. (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189-91. (pmid: 19151095) |
|
[ PubMed ] [ DOI ] UNLABELLED: Jalview Version 2 is a system for interactive WYSIWYG editing, analysis and annotation of multiple sequence alignments. Core features include keyboard and mouse-based editing, multiple views and alignment overviews, and linked structure display with Jmol. Jalview 2 is available in two forms: a lightweight Java applet for use in web applications, and a powerful desktop application that employs web services for sequence alignment, secondary structure prediction and the retrieval of alignments, sequences, annotation and structures from public databases and any DAS 1.53 compliant sequence or annotation server. AVAILABILITY: The Jalview 2 Desktop application and JalviewLite applet are made freely available under the GPL, and can be downloaded from www.jalview.org. |
We will use this tool for this assignment and explore its features as we go along.
Task:
- Navigate to the Jalview homepage click on Download, install Jalview on your computer and start it. A number of windows that showcase the program's abilities will load, you can close these.
- Prepare homologous Mbp1 sequences for alignment:
- Open the Reference Mbp1 orthologues (all fungi) page. (This is the list of Mbp1 orthologs I mentioned above.)
- Copy the FASTA sequences of the reference proteins, paste them into a text file (TextEdit on the Mac, Notepad on Windows) and save the file; you could give it an extension of
.fa–but you don't have to. - Check whether the sequence for YFO is included in the list. If it is, fine. If it is not, retrieve it from NCBI, paste it into the file and edit the header like the other sequences. If the wrong sequence from YFO is included, replace it and let me know.
- Return to Jalview and select File → Input Alignment → from File and open your file. A window with sequences should appear.
- Copy the sequences for ankyrin domain models (below), click on the Jalview window, select File → Add sequences → from Textbox and paste them into the Jalview textbox. Paste two separate copies of the CD00204 consensus sequence and one copy of 1SW6.
- When all the sequences are present, click on Add.
Jalview now displays all the sequences, but of course this is not yet an alignment.
- Ankyrin domain models
>CD00204 ankyrin repeat consensus sequence from CDD NARDEDGRTPLHLAASNGHLEVVKLLLENGADVNAKDNDGRTPLHLAAKNGHLEIVKLLL EKGADVNARDKDGNTPLHLAARNGNLDVVKLLLKHGADVNARDKDGRTPLHLAAKNGHL
>1SW6 from PDB - unstructured loops replaced with xxxx GPIITFTHDLTSDFLSSPLKIMKALPSPVVNDNEQKMKLEAFLQRLLFxxxxSFDSLLQE VNDAFPNTQLNLNIPVDEHGNTPLHWLTSIANLELVKHLVKHGSNRLYGDNMGESCLVKA VKSVNNYDSGTFEALLDYLYPCLILEDSMNRTILHHIIITSGMTGCSAAAKYYLDILMGW IVKKQNRPIQSGxxxxDSILENLDLKWIIANMLNAQDSNGDTCLNIAARLGNISIVDALL DYGADPFIANKSGLRPVDFGAG
Computing alignments
try two MSA's algorithms and load them in Jalview. Locally: which one do you prefer? Modify the consensus. Annotate domains.
The EBI has a very convenient page to access a number of MSA algorithms. This is especially convenient when you want to compare, e.g. T-Coffee and Muscle and MAFFT results to see which regions of your alignment are robust. You could use any of these tools, just paste your sequences into a Webform, download the results and load into Jalview. Easy.
But even easier is to calculate the alignments directly from Jalview. available. (Not today. Bummer.)
No. Claculate an external alignment and import.
- Calculate a MAFFT alignment using the Jalview Web service option
Task:
- In Jalview, select Web Service → Alignment → MAFFT with defaults.... The alignment is calculated in a few minutes and displayed in a new window.
- Calculate a MAFFT alignment when the Jalview Web service is NOT available
Task:
- In Jalview, select File → Output to Textbox → FASTA
- Copy the sequences.
- Navigate to the MAFFT Input form at the EBI.
- Paste your sequences into the form.
- Click on Submit.
- Close the Jalview sequence window and either save your MAFFT alignment to file and load in Jalview, or simply 'File → Input Alignment → from Textbox, paste and click New Window.
In any case, you should now have an alignment.
Task:
- Choose Colour → Hydrophobicity and → by Conservation. Then adjust the slider left or right to see which columns are highly conserved. You will notice that the Swi6 sequence that was supposed to align only to the ankyrin domains was in fact aligned to other parts of the sequence as well. This is one part of the MSA that we will have to correct manually and a common problem when aligning sequences of different lengths.
Editing ankyrin domain alignments
A good MSA comprises only columns of residues that play similar roles in the proteins' mechanism and/or that evolve in a comparable structural context. Since the alignment reflects the result of biological selection and conservation, it has relatively few indels and the indels it has are usually not placed into elements of secondary structure or into functional motifs. The contiguous features annotated for Mbp1 are expected to be left intact by a good alignment.
A poor MSA has many errors in its columns; these contain residues that actually have different functions or structural roles, even though they may look similar according to a (pairwise!) scoring matrix. A poor MSA also may have introduced indels in biologically irrelevant positions, to maximize spurious sequence similarities. Some of the features annotated for Mbp1 will be disrupted in a poor alignment and residues that are conserved may be placed into different columns.
Often errors or inconsistencies are easy to spot, and manually editing an MSA is not generally frowned upon, even though this is not a strictly objective procedure. The main goal of manual editing is to make an alignment biologically more plausible. Most comonly this means to mimize the number of rare evolutionary events that the alignment suggests and/or to emphasize conservation of known functional motifs. Here are some examples for what one might aim for in manually editing an alignment:
- Reduce number of indels
From a Probcons alignment: 0447_DEBHA ILKTE-K-T---K--SVVK ILKTE----KTK---SVVK 9978_GIBZE MLGLN-PGLKEIT--HSIT MLGLNPGLKEIT---HSIT 1513_CANAL ILKTE-K-I---K--NVVK ILKTE----KIK---NVVK 6132_SCHPO ELDDI-I-ESGDY--ENVD ELDDI-IESGDY---ENVD 1244_ASPFU ----N-PGLREIC--HSIT -> ----NPGLREIC---HSIT 0925_USTMA LVKTC-PALDPHI--TKLK LVKTCPALDPHI---TKLK 2599_ASPTE VLDAN-PGLREIS--HSIT VLDANPGLREIS---HSIT 9773_DEBHA LLESTPKQYHQHI--KRIR LLESTPKQYHQHI--KRIR 0918_CANAL LLESTPKEYQQYI--KRIR LLESTPKEYQQYI--KRIR
Gaps marked in red were moved. The sequence similarity in the alignment does not change considerably, however the total number of indels in this excerpt is reduced to 13 from the original 22
- Move indels to more plausible position
From a CLUSTAL alignment: 4966_CANGL MKHEKVQ------GGYGRFQ---GTW MKHEKVQ------GGYGRFQ---GTW 1513_CANAL KIKNVVK------VGSMNLK---GVW KIKNVVK------VGSMNLK---GVW 6132_SCHPO VDSKHP-----------QID---GVW -> VDSKHPQ-----------ID---GVW 1244_ASPFU EICHSIT------GGALAAQ---GYW EICHSIT------GGALAAQ---GYW
The two characters marked in red were swapped. This does not change the number of indels but places the "Q" into a a column in which it is more highly conserved (green). Progressive alignments are especially prone to this type of error.
- Conserve motifs
From a CLUSTAL alignment: 6166_SCHPO --DKRVA---GLWVPP --DKRVA--G-LWVPP XBP1_SACCE GGYIKIQ---GTWLPM GGYIKIQ--G-TWLPM 6355_ASPTE --DEIAG---NVWISP -> ---DEIA--GNVWISP 5262_KLULA GGYIKIQ---GTWLPY GGYIKIQ--G-TWLPY
The first of the two residues marked in red is a conserved, solvent exposed hydrophobic residue that may mediate domain interactions. The second residue is the conserved glycine in a beta turn that cannot be mutated without structural disruption. Changing the position of a gap and insertion in one sequence improves the conservation of both motifs.
The Ankyrin domains are quite highly diverged, the boundaries not well defined and not even CDD, SMART and SAS agree on the precise annotations. We expect there to be alignment errors in this region. Nevertheless we would hope that a good alignment would recognize homology in that region and that ideally the required indels would be placed between the secondary structure elements, not in their middle. But judging from the sequence alignment alone, we cannot judge where the secondary structure elements ought to be. You should therefore add the following "sequence" to the alignment; it contains exactly as many characters as the Swi6 sequence above and annotates the secondary structure elements. I have derived it from the 1SW6 structure
>SecStruc 1SW6 E: strand t: turn H: helix _: irregular _EEE__tt___ttt______EE_____t___HHHHHHHHHHHHHHHH_xxxx_HHHHHHH HHHH_t_____t_____t____HHHHHHH__tHHHHHHHHH____t___tt____HHHHH HH__HHHH___HHHHHHHHHHHHHEE_t____HHHHHHHHH__t__HHHHHHHHHHHHHH HHHHHH__EEE_xxxx_HHHHHt_HHHHHHH______t____HHHHHHHH__HHHHHHHH H____t____t____HHHH___
To proceed:
- Manually align the Swi6 sequence with yeast Mbp1
- Bring the Secondary structure annotation into its correct alignment with Swi6
- Bring both CDD ankyrin profiles into the correct alignment with yeast Mbp1
Proceed along the following steps:
Task:
- Add the secondary structure annotation to the sequence alignment in Jalview. Copy the annotation, select File → Add sequences → from Textbox and paste the sequence.
- Select Help → Documentation and read about Editing Alignments, Cursor Mode and Key strokes.
- Click on the yeast Mbp1 sequence row to select the entire row. Then use the cursor key to move that sequence down, so it is directly above the 1SW6 sequence. Select the row of 1SW6 and use shift/mouse to move the sequence elements and edit the alignment to match yeast Mbp1. Refer to the alignment given in the Mbp1 annotation page for the correct alignment.
- Align the secondary structure elements with the 1SW6 sequence: Every character of 1SW6 should be matched with either E, t, H, or _. The result should be similar to the Mbp1 annotation page. If you need to insert gaps into all sequences in the alignment, simply drag your mouse over all row headers - movement of sequences is constrained to selected regions, the rest is locked into place to prevent inadvertent misalignments. Remember to save your project from time to time: File → save so you can reload a previous state if anything goes wrong and can't be fixed with Edit → Undo.
- Finally align the two CD00204 consensus sequences to their correct positions (again, refer to the Mbp1 annotation page).
- You can now consider the principles stated above and see if you can improve the alignment, for example by moving indels out of regions of secondary structure if that is possible without changing the character of the aligned columns significantly. Select blocks within which to work to leave the remaining alignment unchanged. So that this does not become tedious, you can restrict your editing to one Ankyrin repeat that is structurally defined in Swi6. You may want to open the 1SW6 structure in VMD to define the boundaries of one such repeat. You can copy and paste sections from Jalview into your assignment for documentation or export sections of the alignment to HTML (see the example below).
Editing ankyrin domain alignments - Sample
This sample was created by
- Editing the alignments as described above;
- Copying a block of aligned sequence;
- Pasting it To New Alignment;
- Colouring the residues by Hydrophobicity and setting the colour saturation according to Conservation;
- Choosing File → Export Image → HTML and pasting the resulting HTML source into this Wikipage.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Aligned sequences before editing. The algorithm has placed gaps into the Swi6 helix
LKWIIANand the four-residue gaps before the block of well aligned sequence on the right are poorly supported.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Aligned sequence after editing. A significant cleanup of the frayed region is possible. Now there is only one insertion event, and it is placed into the loop that connects two helices of the 1SW6 structure.
Final analysis
Task:
- Compare the distribution of indels in the ankyrin repeat regions of your alignments.
- Review whether the indels in this region are concentrated in segments that connect the helices, or if they are more or less evenly distributed along the entire region of similarity.
- Think about whether the assertion that indels should not be placed in elements of secondary structure has merit in your alignment.
- Recognize that an indel in an element of secondary structure could be interpreted in a number of different ways:
- The alignment is correct, the annotation is correct too: the indel is tolerated in that particular case, for example by extending the length of an α-helix or β-strand;
- The alignment algorithm has made an error, the structural annotation is correct: the indel should be moved a few residues;
- The alignment is correct, the structural annotation is wrong, this is not a secondary structure element after all;
- Both the algorithm and the annotation are probably wrong, but we have no data to improve the situation.
(NB: remember that the structural annotations have been made for the yeast protein and might have turned out differently for the other proteins...)
You should be able to analyse discrepancies between annotation and expectation in a structured and systematic way. In particular if you notice indels that have been placed into an annotated region of secondary structure, you should be able to comment on whether the location of the indel has strong support from aligned sequence motifs, or whether the indel could possibly be moved into a different location without much loss in alignment quality.
- Considering the whole alignment and your experience with editing, you should be able to state whether the position of indels relative to structural features of the ankyrin domains in your organism's Mbp1 protein is reliable. That would be the result of this task, in which you combine multiple sequence and structural information.
- You can also critically evaluate database information that you have encountered:
- Navigate to the CDD annotation for yeast Mbp1.
- You can check the precise alignment boundaries of the ankyrin domains by clicking on the (+) icon to the left of the matching domain definition.
- Confirm that CDD extends the ankyrin domain annotation beyond the 1SW6 domain boundaries. Given your assessment of conservation in the region beyond the structural annotation: do you think that extending the annotation is reasonable also in YFO's protein? Is there evidence for this in the alignment of the CD00204 consensus with well aligned blocks of sequence beyond the positions that match Swi6?
Homology model
SwissModel
Access the Swissmodel server at http://swissmodel.expasy.org and click on Start Modelling. Then, under the Supported Inputs, click on Target-Template Alignment.
Task:
- Paste your alignment for target and model into the form field. Click on the question mark next to "Supported Inputs" if you are not sure about the format. SwissModel will analyse the sequences and ask you to identify target and template. The YFO sequence is your target. The 1BM8 sequence is the template.
- Click Validate Target Template Alignment and check that the returned alignment is correct.
- Click Build Model to start the modeling process.
- The resulting page returns information about the resulting model. Mouse over the Model 01, open the PDB file and save the coordinates to your computer. Read the information on what is being returned by the server (click on the question mark icon). Study the quality measures.
- Also save:
- The output page as pdf (for reference)
- The modeling report (as pdf)
Model analysis
The PDB file
Task:
Open your model coordinates in a text-editor (make sure you view the PDB file in a fixed-width font (like "courier") so all the columns line up correctly) and consider the following questions:
- What is the residue number of the first residue in the model? What should it be, based on the alignment? If the putative DNA binding region was reported to be residues 50-74 in the Mbp1 protein, which residues of your model correspond to that region?
R code: renumbering the model
As you have seen, SwissModel numbers the first residue "1" and does not keep the numbering of the template. We should renumber the model so we can compare the model and the template with the same residue numbers. Fortunately there is a very useful R package that will help us with that.
Task:
- Navigate to the bio3D home page. bio3d is not available for installation via CRAN, but needs to be installed from source. Instructions for the different platforms are here http://thegrantlab.org/bio3d/tutorials/installing-bio3d Follow the instructions and install bio3d for R on your platform.
- Explore and execute the following R script. I am assuming that your model is in your working directory, change paths and filenames as required.
# renumberPDB.R
# This is a simple renumbering script that uses the bio3D
# package. We simply set the first residue number to what it
# should be and renumber all residues based on the first one.
# The script assumes your input PDBfile is in your working
# directory.
# To run this, you must have installed the bio3D R package; instructions
# are here: http://thegrantlab.org/bio3d/tutorials/installing-bio3d
setwd("~/my/working/directory")
PDBin <- "YFO_model.pdb"
PDBout <- "YFO_model_ren.pdb"
first <- 4 # residue number that the first residue should have
# ================================================
# Read coordinate file
# ================================================
# read PDB file using bio3D function read.pdb()
library(bio3d)
pdb <- read.pdb(PDBin) # read the PDB file into a list
pdb # examine the information
pdb$atom[1,] # get information for the first atom
# you can explore ?read.pdb and study the examples.
# ================================================
# Change residue numbers
# ================================================
resNum <- as.numeric(pdb$atom[,"resno"]) # get residue numbers for all atoms
resNum <- resNum + (first - resNum[1]) # calculate offset
pdb$atom[,"resno"] <- resNum # replace old numbers with new
pdb$atom[1,] # check result
# ================================================
# Write output to file
# ================================================
write.pdb(pdb=pdb,file=PDBout)
# Done. Open the PDB file you have written in a text editor and confirm
# that this has worked.
First visualization
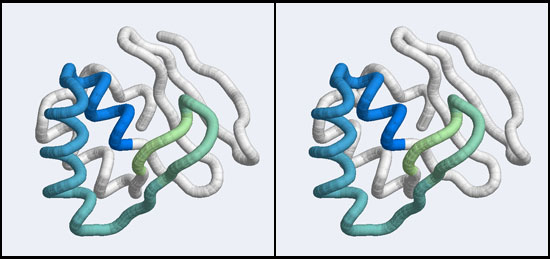
Since a homology model inherits its structural details from the template, your model of the YFO sequence should look very similar to the original 1BM8 structure.
Task:
- Start Chimera and load the model coordinates that you have just renumbered.
- From the PDB, also load the template structure. (Use File → Fetch by ID ...)
- In the Favourites → Model Panel window you can switch between the two molecules.
- Hide the ribbon and choose backbone only → full. You will note that the backbone of the two structures is virtually identical.
- Next, choose Actions → Atoms/Bonds → show to display display the two molecules in a stick style and note how the sidechains have been modeled. Note especially how sidechain coordinates have been guessed, where the template had shorter sidechains than the target. It may be more clear if you hide H-atoms: Select → Chemistry → Element → H and Actions → Atoms/Bonds → hide
- Display only residue 50 to 74 to focus on the putative helix-turn-helix domain. Choose Favourites → Sequence, select the residues for one model, then Select → Invert (selected model) and Actions → Atoms/Bonds → hide.
- Study the result. A model of the HTH domain of YFO Mbp1.
Coloring the model by energy
SwissModel calculates energies for each residue of the model with a molecular mechanics forcefield. The SwissModel modeling summary page contains a plot of these energies as a function of sequence number like. The values - between 0.0 and 1.0 - are stored in the PDB files B-factor field.
Task:
- Back in Chimera, use the model panel to close the 1BM8 structure.
- Choose Tools → Depiction → Render by attribute and select attributes of atoms, Attribute: bfactor, check color atoms and click OK.
- Study the result: It seems that residues in the core of the protein have better energies than residues at the surface. Why could that be the case?
Study the options of this window a bit, rendering by attribute is a powerful way to store and depict all manners of information with the molecule. Simply write a little R script that uses bio3D to replace the B-factor or occupancy values with any value you might be interested in: energies, conservation scores, information ... whatever. The rewnder this property to map it on the 3D structure of your molecule. If you want to experience with this a bit, you could apply the information scores from the previous assignment to your model, using a script that is easy to derive from the renumbering R-script you have studied above.
Introduction
One of the really interesting questions we can discuss with reference to our homology model is how sequence variation might result in changed DNA recognition sites, and then lead to changed cognate DNA binding sequences. In order to address this, we would need to generate a plausible structural model for how DNA is bound to APSES domains.
Since there is currently no software available that would reliably model such a complex from first principles[1], we will base a model of a bound complex on homology modelling as well. This means we need to find a similar structure for which the position of bound DNA is known, then superimpose that structure with our model. This places the DNA molecule into the spatial context of the model we are studying. However, you may remember from the third assignment that the APSES domains in fungi seem to be a relatively small family. And there is no structure available of an APSES domain-DNA complex. How can we find a coordinate set of a structurally similar protein-DNA complex?
This assignment is based on the homology model you built. You will (1) identify similar structures of distantly related domains for which protein-DNA complexes are known, (2) assemble a hypothetical complex structure and (3) consider whether the available evidence allows you to distinguish between different modes of ligand binding,
Modeling a DNA ligand
Finding a similar protein-DNA complex
Remember that homologous sequences can have diverged to the point where their sequence similarity is no longer recognizable, however their structure may be quite well conserved. Thus if we could find similar structures in the PDB, these might provide us with some plausible hypotheses for how DNA is bound by APSES domains. We thus need a tool similar to BLAST, but not for the purpose of sequence alignment, but for structure alignment. A kind of BLAST for structures. Just like with sequence searches, we might not want to search with the entire protein, if we are interested in is a subdomain that binds to DNA. Attempting to match all structural elements in addition to the ones we are actually interested in is likely to make the search less specific - we would find false positives that are similar to some irrelevant part of our structure. However, defining too small of a subdomain would also lead to a loss of specificity: in the extreme it is easy to imagine that the search for e.g. a single helix would retrieve very many hits that would be quite meaningless.
At the NCBI, VAST is provided as a search tool for structural similarity search.
Task:
- Navigate to the VAST search interface page.
- Enter
1bm8as the PDB ID to search for and click Go. - Follow the link to Related Structures.
- Study the result.
You will see that VAST finds more than 3,000 partially similar structures, but it would be almost impossibly tedious to manually search through the list for structures of protein DNA complexes that are similar to the interacting core of the APSES domain. It turns out that our search is not specific enough in two ways: we have structural elements in our PDB file that are unnecessary for the question at hand, and thus cause the program to find irrelevant matches. But, if we constrain ourselves to just a single helix and strand (i.e. the 50-74 subdomain that has been implicated in DNA binding, the search will become too non-specific. Also we have no good way to retrieve functional information from these hits: which ones are DNA-binding proteins, that bind DNA through residues of this subdomain and for which the structure of a complex has been solved? It seems we need to define our question more precisely.
Task:
- Open VMD and load the 1BM8 structure or your YFO homology model.
- Display the backbone as a Trace (of CA atoms) and color by Index
- In the sequence viewer, highlight residues 50 to 74.
- In the representations window, find the yellow representation (with Color ID 4) that the sequence viewer has generated. Change the Drawing Method to NewCartoon.
- Now (using stereo), study the topology of the region. Focus on the helix at the N-terminus of the highlighted subdomain, it is preceded by a turn and another helix. This first helix makes interactions with the beta hairpin at the C-terminal end of the subdomain and is thus important for the orientation of these elements. (This is what is referred to as a helix-turn-helix motif, or HtH motif, it is very common in DNA-binding proteins.)
- Holding the shift key in the alignment viewer, extend your selection until you cover all of the first helix, and the residues that contact the beta hairpin. I think that the first residue of interest here is residue 33.
- Again holding the shift key, extend the selection at the C-terminus to include the residues of the beta hairpin to where they contact the helix at the N-terminus. I think that the last residue of interest here is residue 79.
- Study the topology and arrangement of this compact subdomain. It contains the DNA-binding elements and probably most of the interactions that establish its three-dimensional shape. This subdomain even has a name: it is a winged helix DNA binding motif, a member of a very large family of DNA-binding domains. I have linked a review by Gajiwala and Burley to the end of this page; note that their definition of a canonical winged helix motif is a bit larger than what we have here, with an additional helix at the N-terminus and a second "wing". )
Armed with this insight, we can attempt again to find meaningfully similar structures. At the EBI there are a number of very well designed structure analysis tools linked off the Structural Analysis page. As part of its MSD Services, PDBeFold provides a convenient interface for structure searches for our purpose
Task:
- Navigate to the PDBeFold search interface page.
- Enter
1bm8for the PDB code and choose Select range from the drop down menu. Select the residues you have defined above. - Note that you can enter the lowest acceptable match % separately for query and target. This means: what percentage of secondary structure elements would need to be matched in either query or target to produce a hit. Keep that value at 80 for our query, since we would want to find structures with almost all of the elements of the winged helix motif. Set the match to 10 % for the target, since we are interested in such domains even if they happen to be small subdomains of large proteins.
- Keep the Precision at normal. Precision and % query match could be relaxed if we wanted to find more structures.
- Finally click on: Submit your query.
- On the results page, click on the index number (in the left-hand column) of the top hit that is not one of our familiar Mbp1 structures to get a detailed view of the result. Most likely this is
1wq2:a, an enzyme. Click on View Superposed. This will open a window with the structure coordinates superimposed in the Jmol molecular viewer. Control-click anywhere in the window area to open a menu of viewing options. Select Style → Stereographic → Wall-eyed viewing. Select Trace as the rendering. Then study the superposition. You will note that the secondary structure elements match quite well, but does this mean we have a DNA-binding domain in this sulfite reductase?
All in all this appears to be well engineered software! It gives you many options to access result details for further processing. I think this can be put to very good use. But for our problem, we would have to search through too many structures because, once again, we can't tell which ones of the hits are DNA binding domains, especially domains for which the structure of a complex has been solved.
APSES domains represent one branch of the tree of helix-turn-helix (HTH) DNA binding modules. (A review on HTH proteins is linked from the resources section at the bottom of this page). Winged Helix domains typically bind their cognate DNA with a "recognition helix" which precedes the beta hairpin and binds into the major groove; additional stabilizing interactions are provided by the edge of a beta-strand binding into the minor groove. This is good news: once we have determined that the APSES domain is actually an example of a larger group of transcription factors, we can compare our model to a structure of a protein-DNA complex. Superfamilies of such structural domains are compiled in the CATH database. Unfortunately CATH itself does not provide information about whether the structures have been determined as complexes. But we can search the PDB with CATH codes and restrict the results to complexes. Essentially, this should give us a list of all winged helix domains for which the structure of complexes with DNA have been determined. This works as follows:
Task:
- For reference, access CATH domain superfamily 1.10.10.10; this is the CATH classification code we will use to find protein-DNA complexes. Click on Superfamily Superposition to get a sense of the structural core of the winged helix domain.
- Navigate to the PDB home page and follow the link to Advanced Search
- In the options menu for Choose a Query Type select Structure Features → CATH Classification Browser. A window will open that allows you to navigate down through the CATH tree. You can view the Class/Architecture/Topology names on the CATH page linked above. Click on the triangle icons (not the text) for Mainly Alpha → Orthogonal Bundle → ARC repressor mutant, subunit A then click on the link to winged helix repressor DNA binding domain. Or, just enter "winged helix" into the search field. This subquery should match more than 550 coordinate entries.
- Click on the (+) button behind Add search criteria to add an additional query. Select the option Structure Features → Macromolecule type. In the option menus that pop up, select Contains Protein→Yes, Contains DNA→Yes, Contains RNA→Ignore, Contains DNA/RNA hybrid→Ignore. This selects files that contain Protein-DNA complexes.
- Check the box below this subquery to Remove Similar Sequences at 90% identity and click on Submit Query. This query should retrieve more than 100 complexes.
- Scroll down to the beginning of the list of PDB codes and locate the Reports menu. Under the heading View select Gallery. This is a fast way to obtain an overview of the structures that have been returned. Adjust the number of Results to see all 100 images and choose Options→Resize medium.
- Finally we have a set of winged-helix domain/DNA complexes, for comparison. Scroll through the gallery and study how the protein binds DNA.
First of all you may notice that in fact not all of the structures are really different, despite having requested only to retrieve dissimilar sequences, and not all images show DNA. This appears to be a deficiency of the algorithm. But you can also easily recognize how in most of the the structures the recognition helix inserts into the major groove of B-DNA (eg. 1BC8, 1CF7) and the wing - if clearly visible at all in the image - appears to make accessory interactions with the DNA backbone.. There is one exception: the structure 1DP7 shows how the human RFX1 protein binds DNA in a non-canonical way, through the beta-strands of the "wing". This is interesting since it suggests there is more than one way for winged helix domains to bind to DNA. We can therefore use structural superposition of your homology model and two of the winged-helix proteins to decide whether the canonical or the non-canonical mode of DNA binding seems to be more plausible for Mbp1 orthologues.
Preparation and superposition of a canonical complex
The structure we shall use as a reference for the canonical binding mode is the Elk-1 transcription factor.
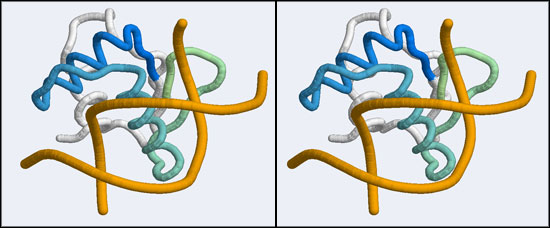
The 1DUX coordinate-file contains two protein domains and two B-DNA dimers in one asymmetric unit. For simplicity, you should delete the second copy of the complex from the PDB file. (Remember that PDB files are simply text files that can be edited.)
Task:
- Find the 1DUX structure in the image gallery and open the 1DUX structure explorer page in a separate window. Download the coordinates to your computer.
- Open the coordinate file in a text-editor (TextEdit or Notepad - NOT MS-Word!) and delete the coordinates for chains
D,EandF; you may also delete allHETATMrecords and theMASTERrecord. Save the file with a different name, e.g. 1DUX_monomer.pdb . - Open VMD and load your homology model. Turn off the axes, display the model as a Tube representation in stereo, and color it by Index. Then load your edited 1DUX file, display this coordinate set in a tube representation as well, and color it by ColorID in some color you like. It is important that you can distinguish easily which structure is which.
- You could use the Extensions→Analysis→RMSD calculator interface to superimpose the two strutcures IF you would know which residues correspond to each other. Sometimes it is useful to do exactly that: define exact correspondences between residue pairs and superimpose according to these selected pairs. For our purpose it is much simpler to use the Multiseq tool (and the structures are simple and small enough that the STAMP algorithm for structural alignment can define corresponding residue pairs automatically). Open the multiseq extension window, select the check-boxes next to both protein structures, and open the Tools→Stamp Structural Alignment interface.
- In the "'Stamp Alignment Options'" window, check the radio-button for Align the following ... Marked Structures and click on OK.
- In the Graphical Representations window, double-click on all "NewCartoon" representations for both molecules, to undisplay them.
- You should now see a superimposed tube model of your homology model and the 1DUX protein-DNA complex. You can explore it, display side-chains etc. and study some of the details of how a transcription factor recognizes and binds to its cognate DNA sequence. However, remember that your model's side-chain orientations have not been determined experimentally but inferred from the template, and that the template's structure was determined in the absence of bound DNA ligand.
- Orient and scale your superimposed structures so that their structural similarity is apparent, and the recognition helix can be clearly seen inserting into the DNA major groove. You may want to keep a copy of the image for future reference. Consider which parts of the structure appear to superimpose best. Note whether it is plausible that your model could bind a B-DNA double-helix in this orientation.
Preparation and superposition of a non-canonical complex
The structure displaying a non-canonical complex between a winged-helix domain and its cognate DNA binding site is the human Regulatory Factor X.
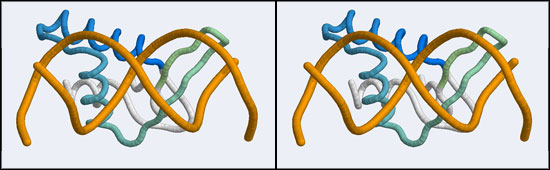
Before we can work with this however, we have to fix an annoying problem. If you download and view the 1DP7 structure in VMD, you will notice that there is only a single strand of DNA! Where is the second strand of the double helix? It is not in the coordinate file, because it happens to be exactly equivalent to the frist starnd, rotated around a two-fold axis of symmetry in the crystal lattice. We need to download and work with the so-called Biological Assembly instead. But there is a problem related to the way the PDB stores replicates in biological assemblies. The PDB generates the additional chains as copies of the original and delineates them with MODEL and ENDMDL records, just like in a multi-structure NMR file. The chain IDs and the atom numbers are the same as the original. The PDB file thus contains the same molecule in two different orientations, not two independent molecules. This is an important difference regarding how such molecules are displayed by VMD. If you try to use the biological unit file of the PDB, VMD does not recognize that there is a second molecule present and displays only one chain. And that looks exactly like the one we have seen before. We have to edit the file, extract the second DNA molecule, change its chain ID and then append it to the original 1DP7 structure[2]...
Task:
- On the structure explorer page for 1DP7, select the option Download Files → PDB File.
- Also select the option Download Files → Biological Assembly.
- Uncompress the biological assembly file.
- Open the file in a text editor.
- Delete everything except the second DNA molecule. This comes after the
MODEL 2line and has chain ID D. Keep theTERandENDlines. Save this with a new filename (e.g.1DP7_DNAonly.pdb). - Also delete all
HETATMrecords forHOH,PEGandEDO, as well as the entire second protein chain and theMASTERrecord. The resulting file should only contain the DNA chain and its copy and one protein chain. Save the file with a new name, eg.1DP7_BDNA.PDB. - Use a similar procedure as BIO_Assignment_Week_8#R code: renumbering the model in the last assignment to change the chain ID.
PDBin <- "1DP7_DNAonly.pdb"
PDBout <- "1DP7_DNAnewChain.pdb"
pdb <- read.pdb(PDBin)
pdb$atom[,"chain"] <- "E"
write.pdb(pdb=pdb,file=PDBout)- Use your text-editor to open both the
1DP7.pdbstructure file and the1DP7_DNAnewChain.pdb. Copy the DNA coordinates, paste them into the original file before theENDline and save. - Open the edited coordinate file with VMD. You should see one protein chain and a B-DNA double helix. (Actually, the BDNA helix has a gap, because the R-library did not read the BRDU nucleotide as DNA). Switch to stereo viewing and spend some time to see how amazingly beautiful the complementarity between the protein and the DNA helix is (you might want to display protein and nucleic in separate representations and color the DNA chain by Position → Radial for clarity) ... in particular, appreciate how not all positively charged side chains contact the phosphate backbone, but some pnetrate into the helix and make detailed interactions with the nucleobases!
- Then clear all molecules
- In VMD, open Extensions→Analysis→MultiSeq. When you run MultiSeq for the first time, you will be asked for a directory in which to store metadata. You can use the default, or a directory of your choice; you may subsequently skip all steps that ask you to install "required" databases locally since we will not need them for this task.
- Choose File→Import Data, browse to your directory and load one by one:
- -Your model;
- -The 1DUX complex;
- -The 1DP7 complex.
- Mark all three protein chains by selecting the checkbox next to their name and choose Tools→ STAMP structural alignment.
- Align the Marked Structures, choose a scanscore of 2 and scanslide of 5. Also choose Slow scan. You may have to play around with the setting to get the molecules to superimpose: but the can be superimposed quite well - at least the DNA-binding helices and the wings should line up.
- In the graphical representations window, double-click on the cartoon representations that multiseq has generated to undisplay them, also undisplay the Tube representation of 1DUX. Then create a Tube representation for 1DP7, and select a Color by ColorID (a different color that you like). The resulting scene should look similar to the one you have created above, only with 1DP7 in place of 1DUX and colored differently.
- Orient and scale your superimposed structures so that their structural similarity is apparent, and the differences in binding elements is clear. Perhaps visualizing a solvent accessible surface of the DNA will help understand the spatial requirements of the complex formation. You may want to keep a copy of the image for future reference. Note whether it is plausible that your model could bind a B-DNA double-helix in the "alternative" conformation.
Interpretation
Task:
- Spend some time studying the complex.
- Recapitulate in your mind how we have arrived at this comparison, in particular, how this was possible even though the sequence similarity between these proteins is low - none of these winged helix domains came up as a result of our previous BLAST search in the PDB.
- You should clearly think about the following question: considering the position of the two DNA helices relative to the YFO structural model, which binding mode appears to be more plausible for protein-DNA interactions in the YFO Mbp1 APSES domains? Is it the canonical, or the non-canonical binding mode? Is there evidence that allows you to distinguish between the two modes?
- Before you quit VMD, save the "state" of your session so you can reload it later. We will look at residue conservation once we have built phylogenetic trees. In the main VMD window, choose File→Save State....
Links and resources
| Altenhoff & Dessimoz (2012) Inferring orthology and paralogy. Methods Mol Biol 855:259-79. (pmid: 22407712) |
|
[ PubMed ] [ DOI ] The distinction between orthologs and paralogs, genes that started diverging by speciation versus duplication, is relevant in a wide range of contexts, most notably phylogenetic tree inference and protein function annotation. In this chapter, we provide an overview of the methods used to infer orthology and paralogy. We survey both graph-based approaches (and their various grouping strategies) and tree-based approaches, which solve the more general problem of gene/species tree reconciliation. We discuss conceptual differences among the various orthology inference methods and databases, and examine the difficult issue of verifying and benchmarking orthology predictions. Finally, we review typical applications of orthologous genes, groups, and reconciled trees and conclude with thoughts on future methodological developments. |
- PDB file format (see the Coordinate Section if you are unsure about chain identifiers)
- Wikipedia on Structural Superposition (although the article is called "Structural Alignment")
- Reference sequences
Footnotes and references
- ↑ Rosetta may get the structure approximately right, Autodock may get the complex approximately right, but the coordinate changes involved in induced fit makes the result unreliable - and we have no good way to validate whether the predicted complex is correct.
- ↑ My apologies if this is tedious. But in the real world, we encounter such problems a lot and I would be remiss not to use this opportunity to let you practice how to fix the issue that could otherwise be a roadblock in a project of yours.
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.
| < Assignment 6 | Assignment 8 > |