Lecture 09
Jump to navigation
Jump to search
Update Warning! This page has not been revised yet for the 2007 Fall term. Some of the slides may be reused, but please consider the page as a whole out of date as long as this warning appears here.
(Previous lecture) ... (Next lecture)
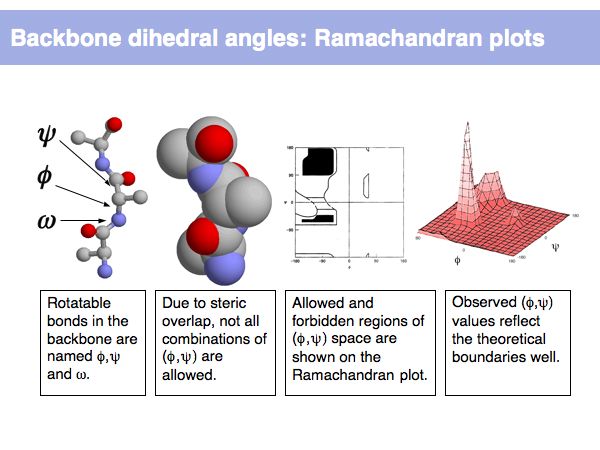
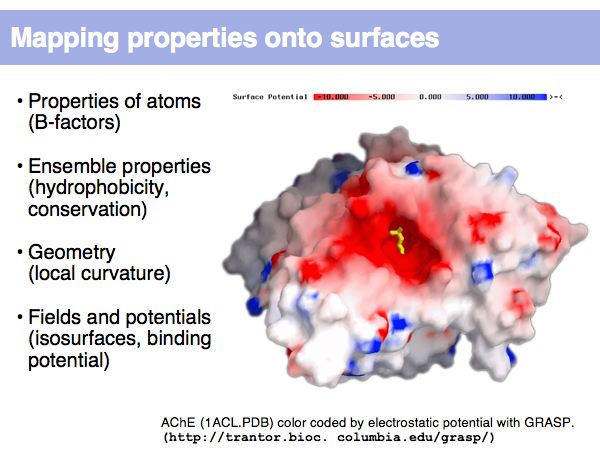
Structure Analysis
...
Add:
- Summary points
- Exercises
- Further reading
Lecture Slides
Slide 001
Slide 002
Slide 003
Slide 004
Slide 005

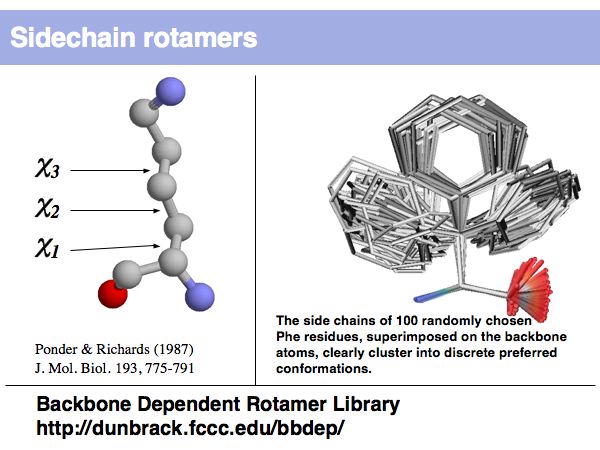
Lecture 09, Slide 005
Rotamers are low-energy conformations of side-chain dihedral angles. Only a small number of rotamer states and combinations are significantly populated in natural proteins. This tremendously simplifies protein structure modelling and prediction problems. However it also guides analysis, e.g. in enzyme active sites the rotamers often exist in strained, rare conformations.
Rotamers are low-energy conformations of side-chain dihedral angles. Only a small number of rotamer states and combinations are significantly populated in natural proteins. This tremendously simplifies protein structure modelling and prediction problems. However it also guides analysis, e.g. in enzyme active sites the rotamers often exist in strained, rare conformations.
Slide 006
Slide 007
Slide 008

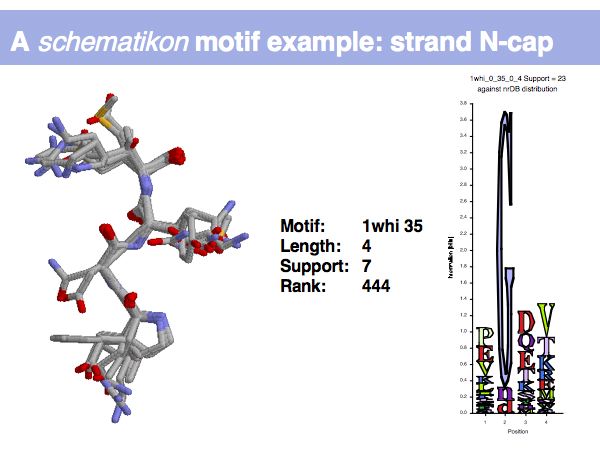
Lecture 09, Slide 008
Motifs represent (presumably) low-energy patterns of conformations. They can be discovered in structure datasets by looking for patterns that recur more frequently than expected by random chance. In almost all cases, they have significant statistical propensities to favour particular amino acids in particular positions.
Motifs represent (presumably) low-energy patterns of conformations. They can be discovered in structure datasets by looking for patterns that recur more frequently than expected by random chance. In almost all cases, they have significant statistical propensities to favour particular amino acids in particular positions.