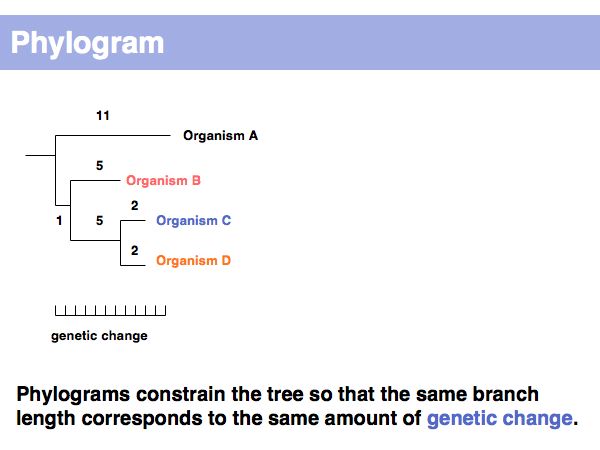
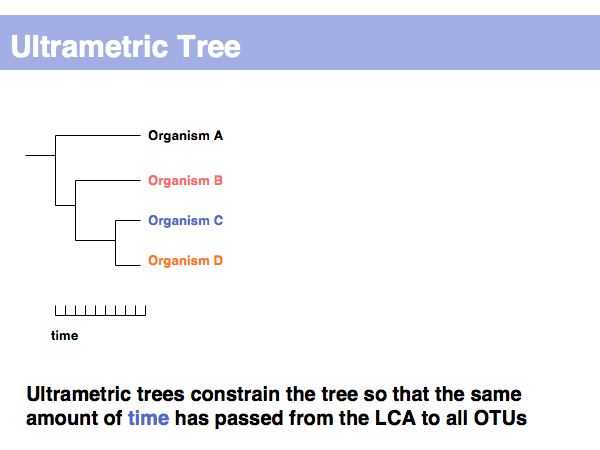
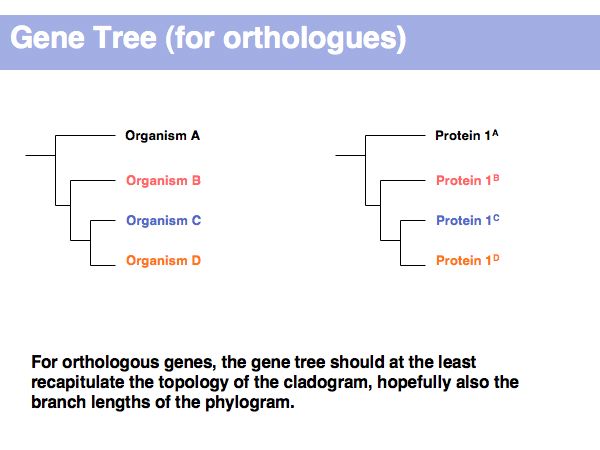
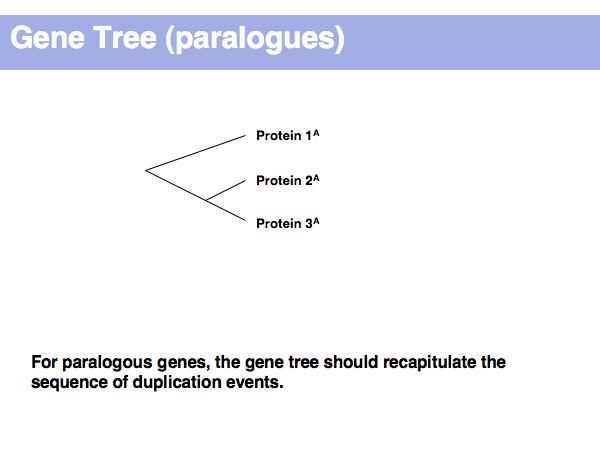
Lecture 17
(Previous lecture) ... (Next lecture)
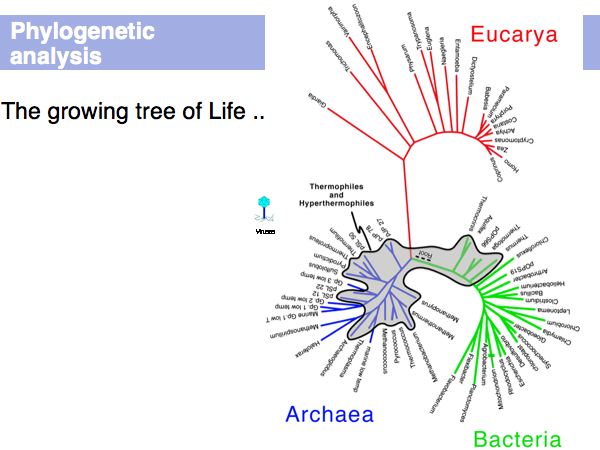
Phylogenetic Analysis
...
Add:
- Summary points
- Exercises
- Further reading
Lecture Slides
Slide 001
Slide 002
Slide 003
Slide 004
Slide 005
Slide 006
Slide 007
Slide 008
Slide 009
Slide 010
Slide 011
Slide 012
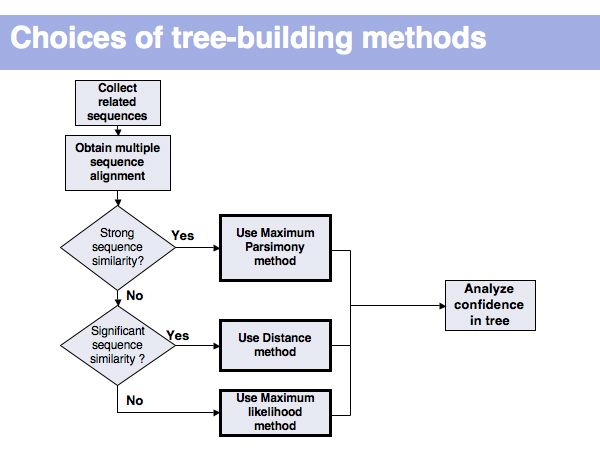
Lecture 17, Slide 012
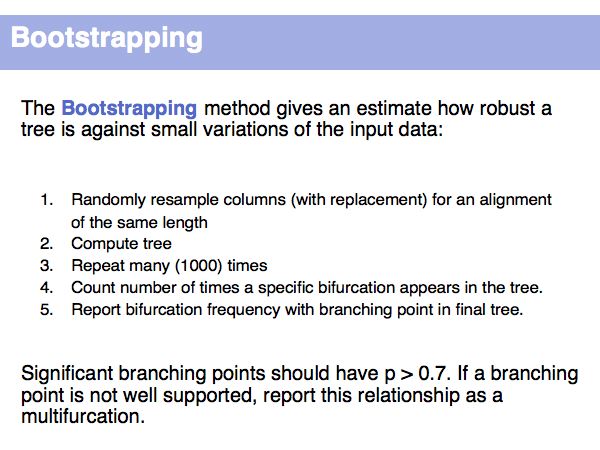
Usually, analysis of confidence implies a "bootstrapping" procedure: rerun the analysis many times with partial data and analyze which features of the tree (branching order -> topology!) are well conserved, and which ones depend critically on unreliable features of the input data.
Usually, analysis of confidence implies a "bootstrapping" procedure: rerun the analysis many times with partial data and analyze which features of the tree (branching order -> topology!) are well conserved, and which ones depend critically on unreliable features of the input data.