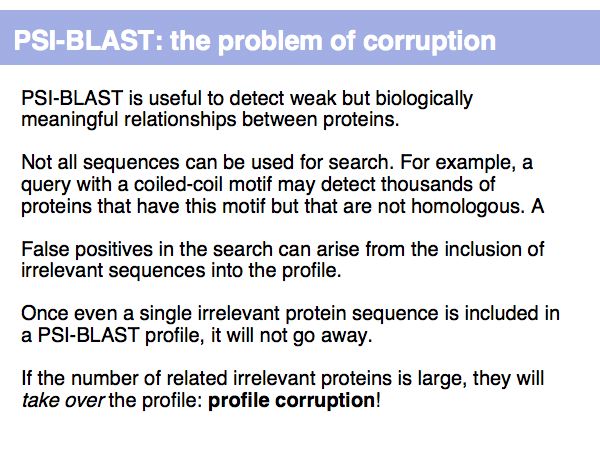
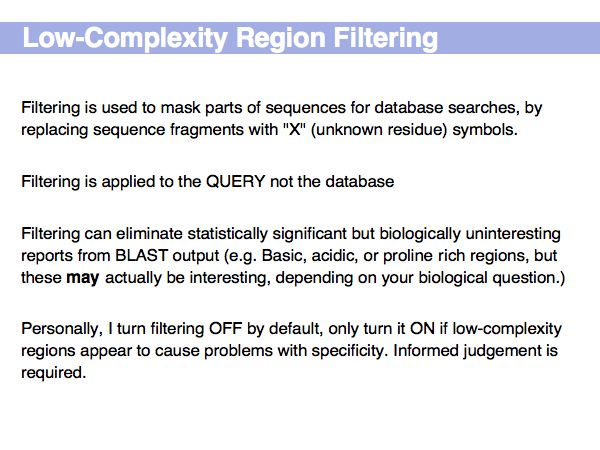
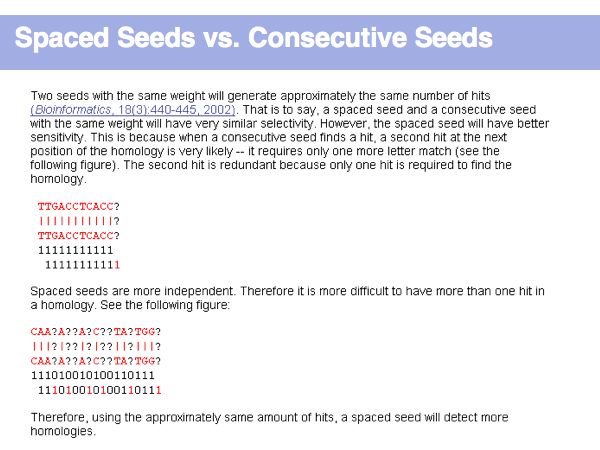
Lecture 16
Jump to navigation
Jump to search

Update Warning! This page has not been revised yet for the 2007 Fall term. Some of the slides may be reused, but please consider the page as a whole out of date as long as this warning appears here.
(Previous lecture) ... (Next lecture)
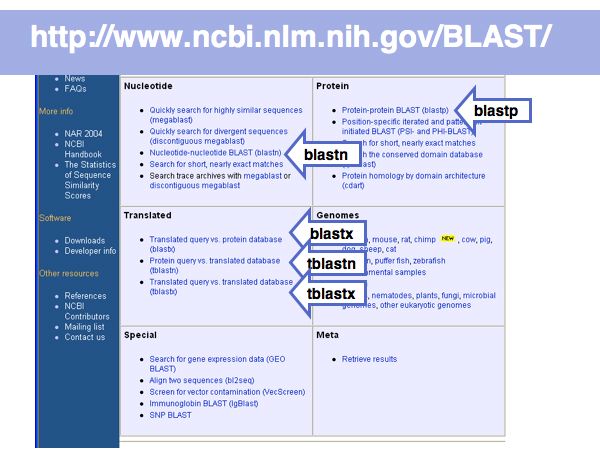
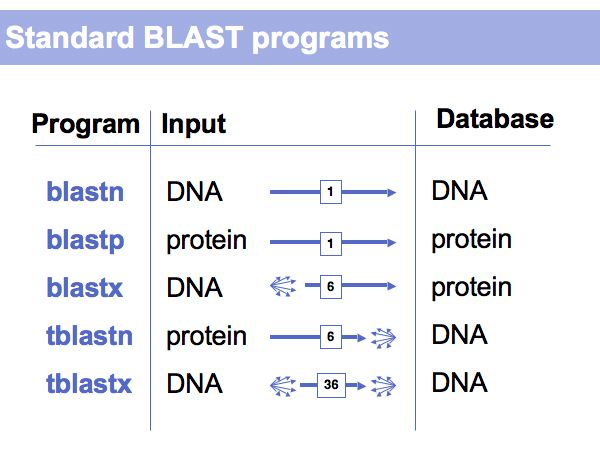
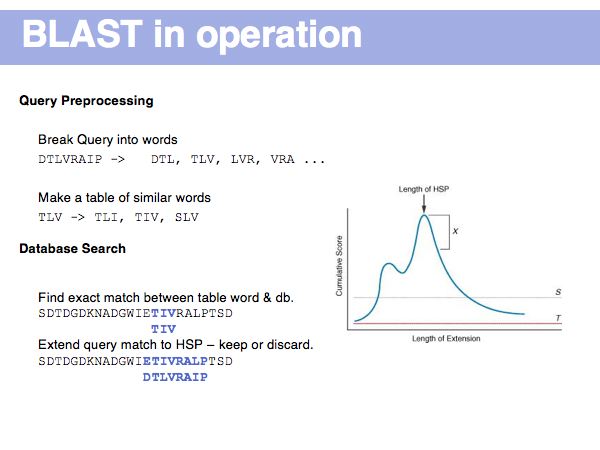
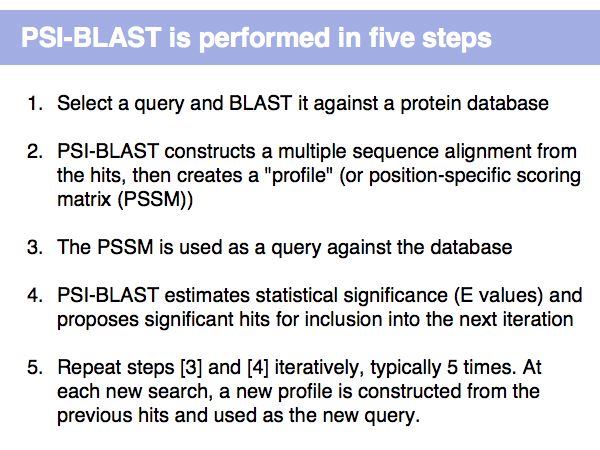
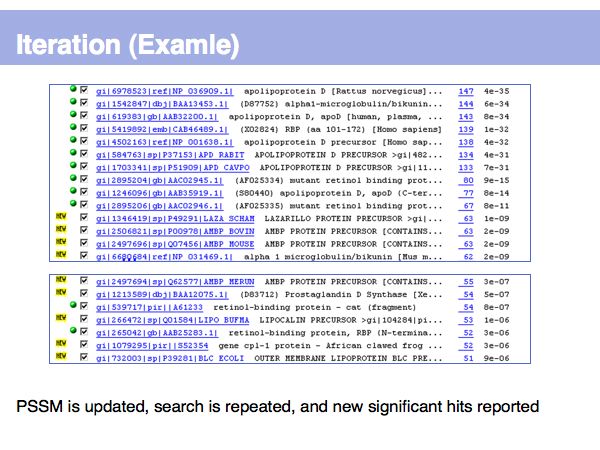
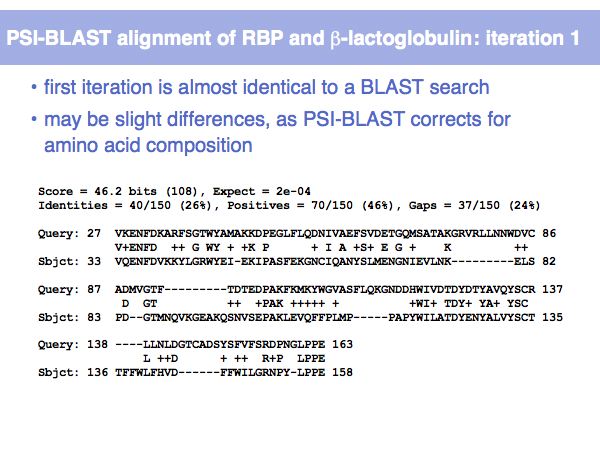
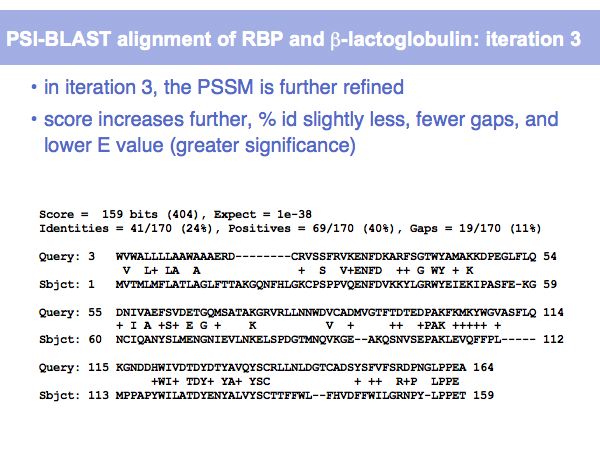
Fast Sequence Database Searches (BLAST)
...
Add:
- Summary points
- Exercises
- Further reading
Lecture Slides
Slide 001
Slide 002
Slide 003
Slide 004
Slide 005
Slide 006
Slide 007
Slide 008
Slide 009
Slide 010
Slide 011

Lecture 16, Slide 011
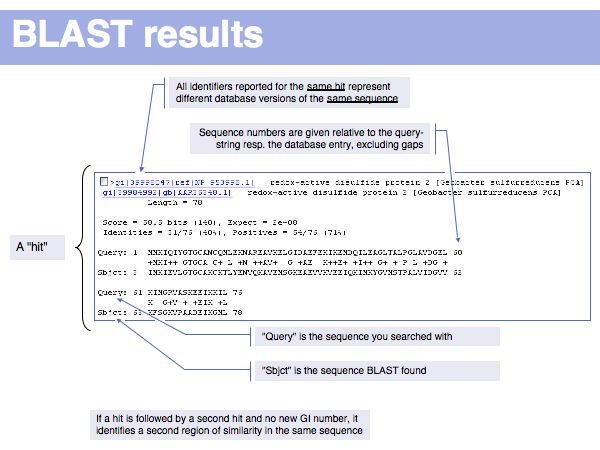
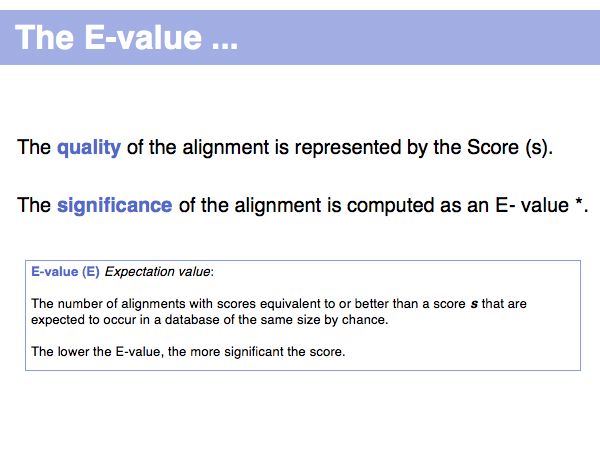
(*) Computing E-values is possible for HSPs since the statistcs of gap-less alignments are analytically tractable. Similar conclusions in general cannot be drawn from gapped alignments. Note the E-values are not a statement about the retrieved hit, but a statement about an expected distribution of scores. Or, to rephrase this, a poor e-value does not mean that your hit is not a homologue, but it means that at that score an irrelevant sequence has a a high chance of scoring well due to chance similarities.
(*) Computing E-values is possible for HSPs since the statistcs of gap-less alignments are analytically tractable. Similar conclusions in general cannot be drawn from gapped alignments. Note the E-values are not a statement about the retrieved hit, but a statement about an expected distribution of scores. Or, to rephrase this, a poor e-value does not mean that your hit is not a homologue, but it means that at that score an irrelevant sequence has a a high chance of scoring well due to chance similarities.
Slide 012

Lecture 16, Slide 012
(*) Note that restricting by organism is not a restriction on the search, but a restriction on the list of results that are being reported. The search takes just as long. Many databases of model-organism genome projects offer BLAST searches on their specific data. These may be faster.
(*) Note that restricting by organism is not a restriction on the search, but a restriction on the list of results that are being reported. The search takes just as long. Many databases of model-organism genome projects offer BLAST searches on their specific data. These may be faster.
Slide 013

Lecture 16, Slide 013
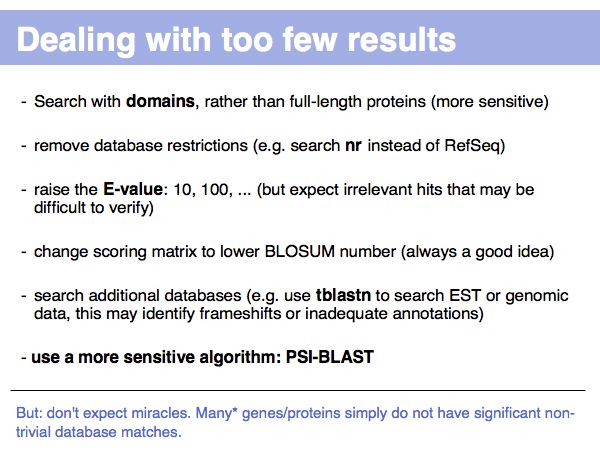
(*) How many? That depends. Unknown genes (or "ORFans") may comprise a significant (albeit diminishing) fraction of genomes. See Siew&Fischer(2003) and the ORFan database
(*) How many? That depends. Unknown genes (or "ORFans") may comprise a significant (albeit diminishing) fraction of genomes. See Siew&Fischer(2003) and the ORFan database