Difference between revisions of "BIN-PHYLO-Tree analysis"
m (Created page with "<div id="BIO"> <div class="b1"> Title </div> {{Vspace}} <div class="keywords"> <b>Keywords:</b> Species trees, gene trees and the importance of naming, Speciat...") |
m |
||
| Line 19: | Line 19: | ||
| − | {{ | + | {{DEV}} |
{{Vspace}} | {{Vspace}} | ||
| Line 82: | Line 82: | ||
== Contents == | == Contents == | ||
<!-- included from "../components/BIN-PHYLO-Tree_analysis.components.wtxt", section: "contents" --> | <!-- included from "../components/BIN-PHYLO-Tree_analysis.components.wtxt", section: "contents" --> | ||
| − | ... | + | |
| + | |||
| + | {{Task|1= | ||
| + | *Read the introductory notes on {{ABC-PDF|BIN-PHYLO-Tree_analysis|analysing phylogenetic trees}}. | ||
| + | }} | ||
| + | |||
| + | |||
| + | ===Analysis=== | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | Here are two principles that will help you make sense of the tree. | ||
| + | |||
| + | |||
| + | A: '''A gene that is present in an ancestral species is inherited in all descendant species'''. The gene has to be observed in all OTUs, unless its has been lost (which is a rare event). | ||
| + | |||
| + | B: '''Paralogous genes in an ancestral species should give rise to monophyletic subtrees for each of the paralogues, in all descendants'''; this means: if the MRCA of a branch has e.g. three genes, we would expect three copies of that branch below this node, one for each of the three genes. Each of these subtrees should recapitulate the reference phylogenetic tree of the species, up to the branchpoint of their MRCA. The precise relationships may not be readily apparent, due to the noise and limited resolution we saw above, but the gene ought to be '''somewhere''' in the tree and you can often assume that it is closest to where it ought to be if the topology was correct. In this way you try to reconcile your expectations with your observations - preferably with as small a number of changes as possible. | ||
| + | |||
| + | With these two simple principles (draw them out on a piece of paper if they do not seem obvious to you), you can probably pry your tree apart quite nicely. A few colored pencils and a printout of the tree will help. I would start by identifying all of the Mbp1 RBMs in the tree. | ||
| + | |||
| + | Here is a bit of code that you can use to colour the labels of the Mbp1 RBMs: | ||
| + | |||
| + | <source lang="R"> | ||
| + | |||
| + | # You have previously defined the names for Mbp1 RBMs in | ||
| + | # the vector apsMbp1Names. You can use these to check | ||
| + | # which of the tree tipLabels are in that vector and | ||
| + | # then color them red in the plot. | ||
| + | |||
| + | # You'll need to replace <TREE> with whatever you called | ||
| + | # your full tree with all APSES domain proteins. | ||
| + | |||
| + | #First, have a look at the tip labels in your tree: | ||
| + | <TREE>$tip.label | ||
| + | |||
| + | # We'll create a vector of black colours of the same length | ||
| + | # as the tip label vector: | ||
| + | tipColors = rep("#000000", Ntip(<TREE>)) | ||
| + | |||
| + | # ... then we replace each one for which the label is | ||
| + | # in apsMbp1Names with "#BB0000" (red) | ||
| + | tipColors[<TREE>$tip.label %in% apsMbp1Names] <- "#BB0000" | ||
| + | |||
| + | #inspect: | ||
| + | tipColors | ||
| + | |||
| + | # ... and then we plot: | ||
| + | plot(<TREE>, tip.color=tipColors, | ||
| + | cex=0.7, root.edge=TRUE, no.margin=TRUE) | ||
| + | |||
| + | |||
| + | </source> | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | |||
| + | ===The APSES domains of the MRCA=== | ||
| + | {{vspace}} | ||
| + | |||
| + | Note: A common confusion about cenancestral genes (MRCA = Most Recent Common Ancestor) arises from the fact that by far not all expected genes are present in the OTUs. Some will have been lost, some will have been incorrectly annotated in their genome (frameshifts!) and not been found with PSI-BLAST, some may have diverged beyond recognizability. In general you have to ask: '''given the species represented in a subclade, what is the last common ancestor of that branch'''? The expectation is that '''all''' descendants of that ancestor should be represented in that branch '''unless''' one of the above reasons why a gene might be absent would apply. Eg. if a branch contains species from ''Basidiomycota'' '''and''' ''Ascomycota'', this means that its MRCA was the ancestor of all fungi. | ||
| + | |||
| + | |||
| + | {{task|1= | ||
| + | |||
| + | |||
| + | * Consider the APSES domain proteins of the fungal cenancestor. What evidence do you see in the tree that identifies them. Note that the hallmark of a clade that originated in the cenancestor is that it contains species from '''all''' subsequent major branches of the species tree. How many of these proteins are there? What arer the names of their SACCE descendants? | ||
| + | |||
| + | }} | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ===The APSES domains of YFO=== | ||
| + | {{vspace}} | ||
| + | |||
| + | You have identified the APSES domain genes of the fungal cenancestor above. Accordingly, this defines the number of APSES protein genes the ancestor to YFO had. Identify the sequence of duplications and/or gene loss in your organism through which YFO has ended up with the APSES domains it possesses today. | ||
| + | |||
| + | {{task|1= | ||
| + | |||
| + | # Print the tree to a single sheet of paper. | ||
| + | # Mark the clades for the genes of the cenancestor. | ||
| + | # Label all subsequent branchpoints that affect the gene tree for YFO with either '''"D"''' (for duplication) or '''"S"''' (for speciation). Remember that specific speciation events can appear more than once in a tree. Identify such events. | ||
| + | # '''Bring this sheet with you to the quiz on Tuesday. Your annotated printout will be worth half of the phylogeny quiz marks.''' | ||
| + | |||
| + | }} | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ==Bonus: when did it happen?== | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | A very cool resource is [http://www.timetree.org/ '''Timetree'''] - a tool that allows you to estimate divergence times between species. For example, the speciation event that separated the main branches of the fungi - i.e. the time when the fungal cenacestor lived - is given by the divergence time of ''Schizosaccharomyces pombe'' and ''Saccharomyces cerevisiaea'': 761,000,000 years ago. For comparison, these two fungi are therefore approximately as related to each other as '''you''' are ... | ||
| + | |||
| + | A) to the rabbit?<br> | ||
| + | B) to the opossum?<br> | ||
| + | C) to the chicken?<br> | ||
| + | D) to the rainbow trout?<br> | ||
| + | E) to the warty sea squirt?<br> | ||
| + | F) to the bumblebee?<br> | ||
| + | G) to the earthworm?<br> | ||
| + | H) to the fly agaric?<br> | ||
| + | |||
| + | Check it out - the question will be on the quiz. | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | |||
| + | ==Identifying Orthologs== | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | In the last assignment we discovered homologs to ''S. cerevisiae'' Mbp1 in YFO. Some of these will be orthologs to Mbp1, some will be paralogs. Some will have similar function, some will not. We discussed previously that genes that evolve under continuously similar evolutionary pressure should be most similar in sequence, and should have the most similar "function". | ||
| + | |||
| + | In this assignment we will define the YFO gene that is the most similar ortholog to ''S. cerevisiae'' Mbp1, and perform a multiple sequence alignment with it. | ||
| + | |||
| + | Let us briefly review the basic concepts. | ||
| + | |||
| + | <div style="padding: 2px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> | ||
| + | |||
| + | <br> | ||
| + | ;All related genes are homologs. | ||
| + | </div> | ||
| + | |||
| + | |||
| + | Two central definitions about the mutual relationships between related genes go back to Walter Fitch who stated them in the 1970s: | ||
| + | <div style="padding: 2px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> | ||
| + | |||
| + | <br> | ||
| + | ;Orthologs have diverged after speciation. | ||
| + | |||
| + | ;Paralogs have diverged after duplication. | ||
| + | </div> | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
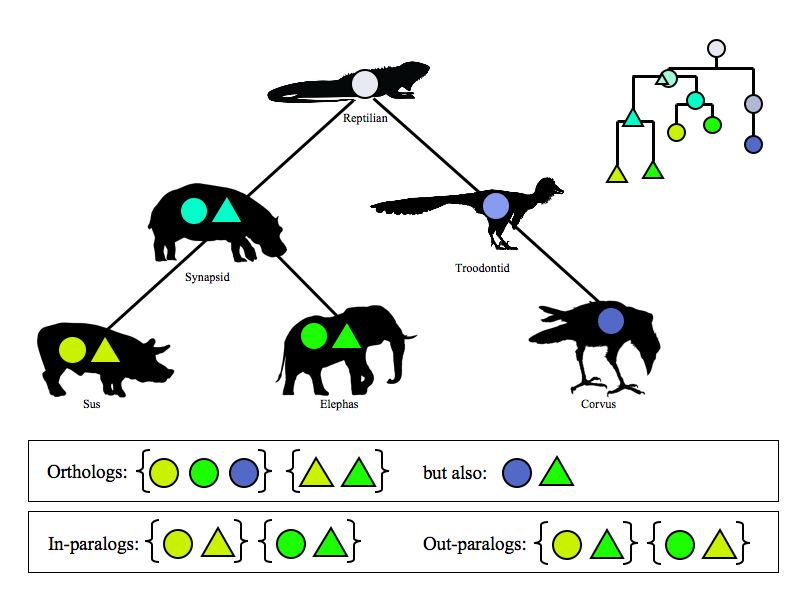
| + | [[Image:OrthologParalog.jpg|frame|none|'''Hypothetical evolutionary tree.''' A single gene evolves through two speciation events and one duplication event. A duplication occurs during the evolution from reptilian to synapsid. It is easy to see how this pair of genes (paralogs) in the ancestral synapsid gives rise to two pairs of genes in pig and elephant, respectively. All ''circle'' genes are mutually orthologs, they form a "cluster of orthologs". All genes within one species are mutual paralogs–they are so called ''in-paralogs''. The ''circle'' gene in pig and the ''triangle'' gene in the elephant are so-called ''out-paralogs''. Somewhat counterintuitively, the ''triangle'' gene in the pig and the ''circle'' gene in the raven are also orthologs - but this has to be, since the last common ancestor diverged by '''speciation'''. | ||
| + | |||
| + | The "phylogram" on the right symbolizes the amount of evolutionary change as proportional to height difference to the "root". It is easy to see how a bidirectional BLAST search will only find pairs of most similar orthologs. If applied to a group of species, bidirectional BLAST searches will find clusters of orthologs only (except if genes were lost, or there are anomalies in the evolutionary rate.)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
{{Vspace}} | {{Vspace}} | ||
| Line 88: | Line 229: | ||
== Further reading, links and resources == | == Further reading, links and resources == | ||
| − | + | ||
| − | + | {{#pmid: 26323765}} | |
| − | + | {{#pmid: 22114356}} | |
| + | {{#pmid: 19190756}} | ||
| + | |||
| + | Also: [http://www.nature.com/scitable/topicpage/reading-a-phylogenetic-tree-the-meaning-of-41956 Nature-Scitable (2008): '''Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups'''] | ||
| + | |||
{{Vspace}} | {{Vspace}} | ||
Revision as of 04:31, 31 August 2017
Title
Keywords: Species trees, gene trees and the importance of naming, Speciation and duplication signatures
Contents
This unit is under development. There is some contents here but it is incomplete and/or may change significantly: links may lead to nowhere, the contents is likely going to be rearranged, and objectives, deliverables etc. may be incomplete or missing. Do not work with this material until it is updated to "live" status.
Abstract
...
This unit ...
Prerequisites
You need to complete the following units before beginning this one:
Objectives
...
Outcomes
...
Deliverables
- Time management: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit.
- Journal: Document your progress in your course journal.
- Insights: If you find something particularly noteworthy about this unit, make a note in your insights! page.
Evaluation
Evaluation: NA
- This unit is not evaluated for course marks.
Contents
Task:
- Read the introductory notes on analysing phylogenetic trees.
Analysis
Here are two principles that will help you make sense of the tree.
A: A gene that is present in an ancestral species is inherited in all descendant species. The gene has to be observed in all OTUs, unless its has been lost (which is a rare event).
B: Paralogous genes in an ancestral species should give rise to monophyletic subtrees for each of the paralogues, in all descendants; this means: if the MRCA of a branch has e.g. three genes, we would expect three copies of that branch below this node, one for each of the three genes. Each of these subtrees should recapitulate the reference phylogenetic tree of the species, up to the branchpoint of their MRCA. The precise relationships may not be readily apparent, due to the noise and limited resolution we saw above, but the gene ought to be somewhere in the tree and you can often assume that it is closest to where it ought to be if the topology was correct. In this way you try to reconcile your expectations with your observations - preferably with as small a number of changes as possible.
With these two simple principles (draw them out on a piece of paper if they do not seem obvious to you), you can probably pry your tree apart quite nicely. A few colored pencils and a printout of the tree will help. I would start by identifying all of the Mbp1 RBMs in the tree.
Here is a bit of code that you can use to colour the labels of the Mbp1 RBMs:
# You have previously defined the names for Mbp1 RBMs in
# the vector apsMbp1Names. You can use these to check
# which of the tree tipLabels are in that vector and
# then color them red in the plot.
# You'll need to replace <TREE> with whatever you called
# your full tree with all APSES domain proteins.
#First, have a look at the tip labels in your tree:
<TREE>$tip.label
# We'll create a vector of black colours of the same length
# as the tip label vector:
tipColors = rep("#000000", Ntip(<TREE>))
# ... then we replace each one for which the label is
# in apsMbp1Names with "#BB0000" (red)
tipColors[<TREE>$tip.label %in% apsMbp1Names] <- "#BB0000"
#inspect:
tipColors
# ... and then we plot:
plot(<TREE>, tip.color=tipColors,
cex=0.7, root.edge=TRUE, no.margin=TRUE)
The APSES domains of the MRCA
Note: A common confusion about cenancestral genes (MRCA = Most Recent Common Ancestor) arises from the fact that by far not all expected genes are present in the OTUs. Some will have been lost, some will have been incorrectly annotated in their genome (frameshifts!) and not been found with PSI-BLAST, some may have diverged beyond recognizability. In general you have to ask: given the species represented in a subclade, what is the last common ancestor of that branch? The expectation is that all descendants of that ancestor should be represented in that branch unless one of the above reasons why a gene might be absent would apply. Eg. if a branch contains species from Basidiomycota and Ascomycota, this means that its MRCA was the ancestor of all fungi.
Task:
- Consider the APSES domain proteins of the fungal cenancestor. What evidence do you see in the tree that identifies them. Note that the hallmark of a clade that originated in the cenancestor is that it contains species from all subsequent major branches of the species tree. How many of these proteins are there? What arer the names of their SACCE descendants?
The APSES domains of YFO
You have identified the APSES domain genes of the fungal cenancestor above. Accordingly, this defines the number of APSES protein genes the ancestor to YFO had. Identify the sequence of duplications and/or gene loss in your organism through which YFO has ended up with the APSES domains it possesses today.
Task:
- Print the tree to a single sheet of paper.
- Mark the clades for the genes of the cenancestor.
- Label all subsequent branchpoints that affect the gene tree for YFO with either "D" (for duplication) or "S" (for speciation). Remember that specific speciation events can appear more than once in a tree. Identify such events.
- Bring this sheet with you to the quiz on Tuesday. Your annotated printout will be worth half of the phylogeny quiz marks.
Bonus: when did it happen?
A very cool resource is Timetree - a tool that allows you to estimate divergence times between species. For example, the speciation event that separated the main branches of the fungi - i.e. the time when the fungal cenacestor lived - is given by the divergence time of Schizosaccharomyces pombe and Saccharomyces cerevisiaea: 761,000,000 years ago. For comparison, these two fungi are therefore approximately as related to each other as you are ...
A) to the rabbit?
B) to the opossum?
C) to the chicken?
D) to the rainbow trout?
E) to the warty sea squirt?
F) to the bumblebee?
G) to the earthworm?
H) to the fly agaric?
Check it out - the question will be on the quiz.
Identifying Orthologs
In the last assignment we discovered homologs to S. cerevisiae Mbp1 in YFO. Some of these will be orthologs to Mbp1, some will be paralogs. Some will have similar function, some will not. We discussed previously that genes that evolve under continuously similar evolutionary pressure should be most similar in sequence, and should have the most similar "function".
In this assignment we will define the YFO gene that is the most similar ortholog to S. cerevisiae Mbp1, and perform a multiple sequence alignment with it.
Let us briefly review the basic concepts.
- All related genes are homologs.
Two central definitions about the mutual relationships between related genes go back to Walter Fitch who stated them in the 1970s:
- Orthologs have diverged after speciation.
- Paralogs have diverged after duplication.
Further reading, links and resources
| Szöllősi et al. (2015) Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos Trans R Soc Lond., B, Biol Sci 370:20140335. (pmid: 26323765) |
|
[ PubMed ] [ DOI ] Although the role of lateral gene transfer is well recognized in the evolution of bacteria, it is generally assumed that it has had less influence among eukaryotes. To explore this hypothesis, we compare the dynamics of genome evolution in two groups of organisms: cyanobacteria and fungi. Ancestral genomes are inferred in both clades using two types of methods: first, Count, a gene tree unaware method that models gene duplications, gains and losses to explain the observed numbers of genes present in a genome; second, ALE, a more recent gene tree-aware method that reconciles gene trees with a species tree using a model of gene duplication, loss and transfer. We compare their merits and their ability to quantify the role of transfers, and assess the impact of taxonomic sampling on their inferences. We present what we believe is compelling evidence that gene transfer plays a significant role in the evolution of fungi. |
| Ebersberger et al. (2012) A consistent phylogenetic backbone for the fungi. Mol Biol Evol 29:1319-34. (pmid: 22114356) |
|
[ PubMed ] [ DOI ] The kingdom of fungi provides model organisms for biotechnology, cell biology, genetics, and life sciences in general. Only when their phylogenetic relationships are stably resolved, can individual results from fungal research be integrated into a holistic picture of biology. However, and despite recent progress, many deep relationships within the fungi remain unclear. Here, we present the first phylogenomic study of an entire eukaryotic kingdom that uses a consistency criterion to strengthen phylogenetic conclusions. We reason that branches (splits) recovered with independent data and different tree reconstruction methods are likely to reflect true evolutionary relationships. Two complementary phylogenomic data sets based on 99 fungal genomes and 109 fungal expressed sequence tag (EST) sets analyzed with four different tree reconstruction methods shed light from different angles on the fungal tree of life. Eleven additional data sets address specifically the phylogenetic position of Blastocladiomycota, Ustilaginomycotina, and Dothideomycetes, respectively. The combined evidence from the resulting trees supports the deep-level stability of the fungal groups toward a comprehensive natural system of the fungi. In addition, our analysis reveals methodologically interesting aspects. Enrichment for EST encoded data-a common practice in phylogenomic analyses-introduces a strong bias toward slowly evolving and functionally correlated genes. Consequently, the generalization of phylogenomic data sets as collections of randomly selected genes cannot be taken for granted. A thorough characterization of the data to assess possible influences on the tree reconstruction should therefore become a standard in phylogenomic analyses. |
| Marcet-Houben & Gabaldón (2009) The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome. PLoS ONE 4:e4357. (pmid: 19190756) |
|
[ PubMed ] [ DOI ] A recurrent topic in phylogenomics is the combination of various sequence alignments to reconstruct a tree that describes the evolutionary relationships within a group of species. However, such approach has been criticized for not being able to properly represent the topological diversity found among gene trees. To evaluate the representativeness of species trees based on concatenated alignments, we reconstruct several fungal species trees and compare them with the complete collection of phylogenies of genes encoded in the Saccharomyces cerevisiae genome. We found that, despite high levels of among-gene topological variation, the species trees do represent widely supported phylogenetic relationships. Most topological discrepancies between gene and species trees are concentrated in certain conflicting nodes. We propose to map such information on the species tree so that it accounts for the levels of congruence across the genome. We identified the lack of sufficient accuracy of current alignment and phylogenetic methods as an important source for the topological diversity encountered among gene trees. Finally, we discuss the implications of the high levels of topological variation for phylogeny-based orthology prediction strategies. |
Also: Nature-Scitable (2008): Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups
Notes
Self-evaluation
If in doubt, ask! If anything about this learning unit is not clear to you, do not proceed blindly but ask for clarification. Post your question on the course mailing list: others are likely to have similar problems. Or send an email to your instructor.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2017-08-05
Version:
- 0.1
Version history:
- 0.1 First stub
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.