Difference between revisions of "BIN-FUNC-Domain annotation"
m (Boris moved page BIN-ALI-Domains by sequence to BIN-FUNC-Domain annotation without leaving a redirect) |
m |
||
| Line 1: | Line 1: | ||
<div id="BIO"> | <div id="BIO"> | ||
<div class="b1"> | <div class="b1"> | ||
| − | + | Domain Annotation | |
</div> | </div> | ||
| Line 8: | Line 8: | ||
<div class="keywords"> | <div class="keywords"> | ||
<b>Keywords:</b> | <b>Keywords:</b> | ||
| − | Domain discovery by multiple sequence alignment | + | Domain discovery by multiple sequence alignment; HMMER algorithm; Domain databases: Pfam, SMART, CDART; Annotation of sequences |
</div> | </div> | ||
| Line 19: | Line 19: | ||
| − | {{ | + | {{DEV}} |
{{Vspace}} | {{Vspace}} | ||
| Line 27: | Line 27: | ||
<div id="ABC-unit-framework"> | <div id="ABC-unit-framework"> | ||
== Abstract == | == Abstract == | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "abstract" --> |
... | ... | ||
| Line 35: | Line 35: | ||
== This unit ... == | == This unit ... == | ||
=== Prerequisites === | === Prerequisites === | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "prerequisites" --> |
<!-- included from "ABC-unit_components.wtxt", section: "notes-prerequisites" --> | <!-- included from "ABC-unit_components.wtxt", section: "notes-prerequisites" --> | ||
You need to complete the following units before beginning this one: | You need to complete the following units before beginning this one: | ||
| Line 44: | Line 44: | ||
=== Objectives === | === Objectives === | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "objectives" --> |
... | ... | ||
| Line 51: | Line 51: | ||
=== Outcomes === | === Outcomes === | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "outcomes" --> |
... | ... | ||
| Line 58: | Line 58: | ||
=== Deliverables === | === Deliverables === | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "deliverables" --> |
<!-- included from "ABC-unit_components.wtxt", section: "deliverables-time_management" --> | <!-- included from "ABC-unit_components.wtxt", section: "deliverables-time_management" --> | ||
*<b>Time management</b>: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit. | *<b>Time management</b>: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit. | ||
| Line 70: | Line 70: | ||
=== Evaluation === | === Evaluation === | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "evaluation" --> |
<!-- included from "ABC-unit_components.wtxt", section: "eval-none" --> | <!-- included from "ABC-unit_components.wtxt", section: "eval-none" --> | ||
<b>Evaluation: NA</b><br /> | <b>Evaluation: NA</b><br /> | ||
| Line 81: | Line 81: | ||
<div id="BIO"> | <div id="BIO"> | ||
== Contents == | == Contents == | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "contents" --> |
| − | ... | + | |
| + | |||
| + | {{Task|1= | ||
| + | *Read the introductory notes on {{ABC-PDF|BIN-FUNC-Domain_annotation|how domain annotations support the annotation of gene function}}. | ||
| + | }} | ||
| + | |||
| + | |||
| + | == SMART domain annotation == | ||
| + | |||
| + | |||
| + | The [http://smart.embl-heidelberg.de/ SMART database] at the EMBL in Heidelberg integrates a number of feature detection tools including Pfam domain annotation and its own, HMM based SMART domain database. You can search by sequence, or by accession number and retrieve domain annotations and more. | ||
| + | |||
| + | |||
| + | ===SMART search=== | ||
| + | |||
| + | {{task|1= | ||
| + | # Access the [http://smart.embl-heidelberg.de/ '''SMART database'''] at http://smart.embl-heidelberg.de/ | ||
| + | # Click the lick to access SMART in the '''normal''' mode. | ||
| + | # Paste the YFO Mbp1 UniProtKB Accession number into the '''Sequence ID or ACC''' field. If you were not able to find a UniProt ID, paste the sequence instead. | ||
| + | # Check all the boxes for: | ||
| + | ## '''outlier homologues''' (also including homologues in the PDB structure database) | ||
| + | ## '''PFAM domains''' (domains defined by sequence similarity in the PFAM database) | ||
| + | ## '''signal peptides''' (using the Gunnar von Heijne's SignalP 4.0 server at the Technical University in Lyngby, Denmark) | ||
| + | ## '''internal repeats''' (using the programs ''ariadne'' and ''prospero'' at the Wellcome Trust Centre for Human Genetics at Oxford University, England) | ||
| + | # Click on '''Sequence SMART''' to run the search and annotation. <small>(In case you get an error like: "Sorry, your entry seems to have no SMART domain ...", try again with the actual sequence instead of the accession number.)</small> | ||
| + | |||
| + | Study the results. | ||
| + | |||
| + | # Note down the following information so you can enter the annotation in the protein database for YFO: | ||
| + | ## From the section on "Confidently predicted domains ..." | ||
| + | ### The start and end coordinates of the '''KilA-N''' domain <small>(...according to SMART, not Pfam, in case the two differ)</small>. | ||
| + | ### All start and end coordinates of '''low complexity segments''' | ||
| + | ### All start and end coordinates of '''ANK''' (Ankyrin) domains | ||
| + | ### Start and end coordinates of '''coiled coil''' domain(s) <small>I expect only one.</small> | ||
| + | ### Start and end coordinates of '''AT hook''' domain(s) <small>I expect at most one - not all Mbp1 orthologues have one.</small> | ||
| + | ## From the section on "Features NOT shown ..." | ||
| + | ### All start and end coordinates of '''low complexity segments''' for which the ''Reason'' is "overlap". | ||
| + | ### Any start and end coordinates of overlapping '''coiled coil''' segments. | ||
| + | ### <small>I expect all other annotations - besides the overlapping KilA-N domain defined by Pfam - to arise from the succession of ankyrin domains that the proteins have, both '''Pfam_ANK..''' domains, as well as internal repeats. However, if there are other features I have not mentioned here, feel encouraged to let me know.</small> | ||
| + | ## From the section on "Outlier homologues ..." | ||
| + | ### Start and end coordinates of a '''PDB:1SW6{{!}}B''' annotation (if you have one): this is a region of sequence similarity to a protein for which the 3D structural coordinate are known. | ||
| + | ### <small>Of course there should also be annotations to the structure of 1BM8 / 1MB1 and/or 1L3G - all of which are structures of the Mbp1 APSES domain that we have already annotated as an"APSES fold" feature previously. And there will be BLAST annotations to Ankyrin domains. We will not annotate these separately either.</small> | ||
| + | # Follow the links to the database entries for the information so you know what these domains and features are. | ||
| + | |||
| + | }} | ||
| + | |||
| + | Next we'll enter the features into our database, so we can compare them with the annotations that I have prepared from SMART annotations of Mbp1 orthologues from the ten reference fungi. | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | === Visual comparison of domain annotations in '''R''' === | ||
| + | |||
| + | The versatile plotting functions of '''R''' allow us to compare domain annotations. The distribution of segments that are annotated as "low-complexity, presumably disordered, is particularly interesting: these are functional features that are often not associated with sequence similarity but may have arisen from convergent evolution. Those would not be detectable through sequence alignment - which is after all based on amino acid pair scores and therefore context independent. | ||
| + | |||
| + | In the following code tutorial, we create a plot similar to the CDD and SMART displays. It is based on the SMART domain annotations of the six-fungal reference species for the course. | ||
| + | |||
| + | |||
| + | |||
| + | {{task|1 = | ||
| + | |||
| + | * Return to your RStudio session. | ||
| + | * Make sure you have saved <code>myDB</code> as instructed previously. Then quit the program, restart, and re-open the project via the '''File''' → '''Recent projects ...''' menu. This is to clear out-of-date assignments and functions from the workspace. | ||
| + | * Do not type <code>init()</code> yet, but '''pull''' the most recent version of files from github. Then type <code>init()</code>. | ||
| + | * Study and work through the code in the <code>SMART domain annotations</code> section of the <code>BCH441_A04.R</code> script. This includes entering your domain and other feature annotations into the database. | ||
| + | * At the end of the script, print out your plot of the domain annotations for MB1_YFO and the reference proteins. Bring this plot with you for the next quiz. | ||
| + | * Can this plot be improved? What would you do differently to maximize its utility from an information-design point of view? | ||
| + | |||
| + | }} | ||
| + | |||
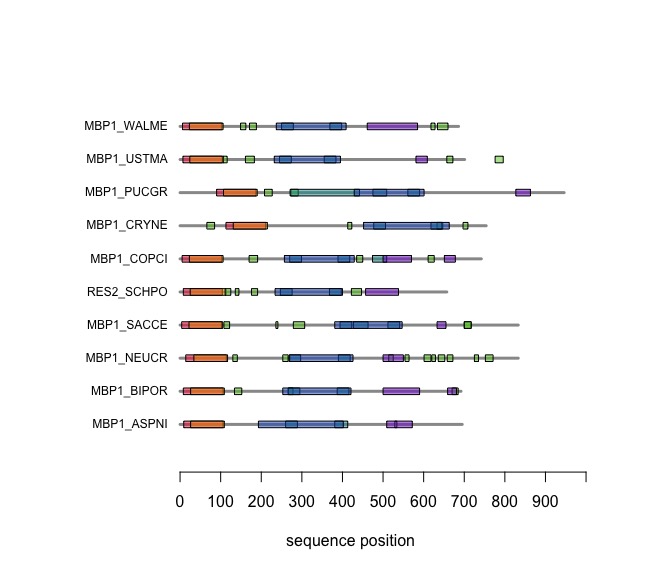
| + | When you execute the code, your plot should look similar to this one: | ||
| + | |||
| + | [[Image:DomainAnnotations.jpg|frame|none|SMART domain annotations for Mbp1 proteins for the ten reference fungi. | ||
| + | ]] | ||
| + | |||
| + | A note on the '''R''' code up to this point: You will find that we have been writing a lot of nested expressions for selections that join data from multiple tables of our data model. When I teach '''R''' workshops for graduate students, postdocs and research fellows, I find that the single greatest barrier in their actual research work is the preparation of data for analysis: filtering, selecting, cross-referencing, and integrating data from different sources. By now, I hope you will have acquired a somewhat robust sense for achieving this. You can imagine that there are ways to simplify those tasks with functions you write, or special resources from a variety of different packages you cab install. But the "pedestrian" approach we have been taking in our scripts has the advantage of working from a very small number of principles, with very few syntactic elements. | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | {{task|1= | ||
| + | |||
| + | ; Optional - care to share? | ||
| + | |||
| + | # Copy one of the list definitions for Mbp1 domains and edit it with the appropriate values for your own annotations. | ||
| + | # Test that you can add the YFO annotation to the plot. | ||
| + | # Submit your validated code block to the [http://biochemistry.utoronto.ca/steipe/abc/students/index.php/BCH441_2014_Assignment_4_domain_annotations '''Student Wiki here''']. The goal is to compile an overview of all species we are studying in class. | ||
| + | # If your working annotation block is in the Wiki before noontime on Wednesday, you will be awarded a 10% bonus on the quiz. | ||
| + | }} | ||
| + | --> | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | |||
| + | |||
{{Vspace}} | {{Vspace}} | ||
| Line 96: | Line 188: | ||
== Notes == | == Notes == | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "notes" --> |
<!-- included from "ABC-unit_components.wtxt", section: "notes" --> | <!-- included from "ABC-unit_components.wtxt", section: "notes" --> | ||
<references /> | <references /> | ||
| Line 106: | Line 198: | ||
<div id="ABC-unit-framework"> | <div id="ABC-unit-framework"> | ||
== Self-evaluation == | == Self-evaluation == | ||
| − | <!-- included from "../components/BIN- | + | <!-- included from "../components/BIN-FUNC-Domain_annotation.components.wtxt", section: "self-evaluation" --> |
<!-- | <!-- | ||
=== Question 1=== | === Question 1=== | ||
Revision as of 04:26, 31 August 2017
Domain Annotation
Keywords: Domain discovery by multiple sequence alignment; HMMER algorithm; Domain databases: Pfam, SMART, CDART; Annotation of sequences
Contents
This unit is under development. There is some contents here but it is incomplete and/or may change significantly: links may lead to nowhere, the contents is likely going to be rearranged, and objectives, deliverables etc. may be incomplete or missing. Do not work with this material until it is updated to "live" status.
Abstract
...
This unit ...
Prerequisites
You need to complete the following units before beginning this one:
Objectives
...
Outcomes
...
Deliverables
- Time management: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit.
- Journal: Document your progress in your course journal.
- Insights: If you find something particularly noteworthy about this unit, make a note in your insights! page.
Evaluation
Evaluation: NA
- This unit is not evaluated for course marks.
Contents
Task:
- Read the introductory notes on how domain annotations support the annotation of gene function.
SMART domain annotation
The SMART database at the EMBL in Heidelberg integrates a number of feature detection tools including Pfam domain annotation and its own, HMM based SMART domain database. You can search by sequence, or by accession number and retrieve domain annotations and more.
SMART search
Task:
- Access the SMART database at http://smart.embl-heidelberg.de/
- Click the lick to access SMART in the normal mode.
- Paste the YFO Mbp1 UniProtKB Accession number into the Sequence ID or ACC field. If you were not able to find a UniProt ID, paste the sequence instead.
- Check all the boxes for:
- outlier homologues (also including homologues in the PDB structure database)
- PFAM domains (domains defined by sequence similarity in the PFAM database)
- signal peptides (using the Gunnar von Heijne's SignalP 4.0 server at the Technical University in Lyngby, Denmark)
- internal repeats (using the programs ariadne and prospero at the Wellcome Trust Centre for Human Genetics at Oxford University, England)
- Click on Sequence SMART to run the search and annotation. (In case you get an error like: "Sorry, your entry seems to have no SMART domain ...", try again with the actual sequence instead of the accession number.)
Study the results.
- Note down the following information so you can enter the annotation in the protein database for YFO:
- From the section on "Confidently predicted domains ..."
- The start and end coordinates of the KilA-N domain (...according to SMART, not Pfam, in case the two differ).
- All start and end coordinates of low complexity segments
- All start and end coordinates of ANK (Ankyrin) domains
- Start and end coordinates of coiled coil domain(s) I expect only one.
- Start and end coordinates of AT hook domain(s) I expect at most one - not all Mbp1 orthologues have one.
- From the section on "Features NOT shown ..."
- All start and end coordinates of low complexity segments for which the Reason is "overlap".
- Any start and end coordinates of overlapping coiled coil segments.
- I expect all other annotations - besides the overlapping KilA-N domain defined by Pfam - to arise from the succession of ankyrin domains that the proteins have, both Pfam_ANK.. domains, as well as internal repeats. However, if there are other features I have not mentioned here, feel encouraged to let me know.
- From the section on "Outlier homologues ..."
- Start and end coordinates of a PDB:1SW6|B annotation (if you have one): this is a region of sequence similarity to a protein for which the 3D structural coordinate are known.
- Of course there should also be annotations to the structure of 1BM8 / 1MB1 and/or 1L3G - all of which are structures of the Mbp1 APSES domain that we have already annotated as an"APSES fold" feature previously. And there will be BLAST annotations to Ankyrin domains. We will not annotate these separately either.
- From the section on "Confidently predicted domains ..."
- Follow the links to the database entries for the information so you know what these domains and features are.
Next we'll enter the features into our database, so we can compare them with the annotations that I have prepared from SMART annotations of Mbp1 orthologues from the ten reference fungi.
Visual comparison of domain annotations in R
The versatile plotting functions of R allow us to compare domain annotations. The distribution of segments that are annotated as "low-complexity, presumably disordered, is particularly interesting: these are functional features that are often not associated with sequence similarity but may have arisen from convergent evolution. Those would not be detectable through sequence alignment - which is after all based on amino acid pair scores and therefore context independent.
In the following code tutorial, we create a plot similar to the CDD and SMART displays. It is based on the SMART domain annotations of the six-fungal reference species for the course.
Task:
- Return to your RStudio session.
- Make sure you have saved
myDBas instructed previously. Then quit the program, restart, and re-open the project via the File → Recent projects ... menu. This is to clear out-of-date assignments and functions from the workspace. - Do not type
init()yet, but pull the most recent version of files from github. Then typeinit(). - Study and work through the code in the
SMART domain annotationssection of theBCH441_A04.Rscript. This includes entering your domain and other feature annotations into the database. - At the end of the script, print out your plot of the domain annotations for MB1_YFO and the reference proteins. Bring this plot with you for the next quiz.
- Can this plot be improved? What would you do differently to maximize its utility from an information-design point of view?
When you execute the code, your plot should look similar to this one:
A note on the R code up to this point: You will find that we have been writing a lot of nested expressions for selections that join data from multiple tables of our data model. When I teach R workshops for graduate students, postdocs and research fellows, I find that the single greatest barrier in their actual research work is the preparation of data for analysis: filtering, selecting, cross-referencing, and integrating data from different sources. By now, I hope you will have acquired a somewhat robust sense for achieving this. You can imagine that there are ways to simplify those tasks with functions you write, or special resources from a variety of different packages you cab install. But the "pedestrian" approach we have been taking in our scripts has the advantage of working from a very small number of principles, with very few syntactic elements.
Further reading, links and resources
Notes
Self-evaluation
If in doubt, ask! If anything about this learning unit is not clear to you, do not proceed blindly but ask for clarification. Post your question on the course mailing list: others are likely to have similar problems. Or send an email to your instructor.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2017-08-05
Version:
- 0.1
Version history:
- 0.1 First stub
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.