BIN-FUNC-Domain annotation
Domain Annotation
(Domain discovery by multiple sequence alignment; HMMER algorithm; Domain databases: Pfam, SMART, CDART; Annotation of sequences)
Abstract:
This unit introduces the observation that evolution composes higher-order functions from domains that are folding units, functional units, and units of inheritance. It then covers some of the databases and services that support discovery and analysis of domains, and guides through an exercise in domain annotation.
|
Objectives:
|
Outcomes:
|
Deliverables:
Prerequisites:
This unit builds on material covered in the following prerequisite units:
Contents
Evaluation
Evaluation: NA
Contents
Task:
- Read the introductory notes on how domain annotations support the annotation of gene function.
Pfam
The Pfam protein domain family database is a large, curated collection of domain definitions and doamin annotations. Domains are discovered from multiple sequecne alignments, and represented as Hidden Markov Models (a probabilistic technique that can define a probability that a given sequence is part of a family the model was trained on).
Task:
- Retrieve the MYSPE UniProt ID from your Journal, or by issuing the following R commands:
pName <- sprintf("MBP1_%s", biCode(MYSPE))
sel <- which(myDB$protein$name == pName)
myDB$protein$UniProtID[sel]- Navigate to the Pfam database.
- Search for your Mbp1 protein by entering the ID into the search field and clicking Go.
- Study the annotations. Download the annotations as a JSON file and name it Mbp1MYSPE_Pfam.JSON.
- Are all expected domains present? (APSES or KilA-N domain, disordered segments, Ankyrin domains, coiled coil, more ?)
- "disorder", "low complexity", and "coiled coil" annotations are not based on alignments, but on sequence analysis algorithms. Visit Finn et al. (2014) and read how these regions are defined[1].
- Study the (well curated!) family annotations for:
SMART domain annotation
The SMART database at the EMBL in Heidelberg integrates a number of feature detection tools including Pfam domain annotation and its own, HMM based SMART domain database. You can search by sequence, or by accession number and retrieve domain annotations and more.
SMART search
Task:
- Access the SMART database at http://smart.embl-heidelberg.de/
- Click the link to access SMART in the normal mode.
- Paste the MYSPE Mbp1 UniProtID into the Sequence ID or ACC field.
- Check all the boxes for:
- outlier homologues (also including homologues in the PDB structure database)
- PFAM domains (domains defined by sequence similarity in the PFAM database)
- signal peptides (using the Gunnar von Heijne's SignalP 4.0 server at the Technical University in Lyngby, Denmark)
- internal repeats (using the programs ariadne and prospero at the Wellcome Trust Centre for Human Genetics at Oxford University, England)
- Click on Sequence SMART to run the search and annotation. (In case you get an error like: "Sorry, your entry seems to have no SMART domain ...", try again with the actual sequence instead of the accession number.)
Study the results.
- Study the family annotations for:
- Note that neither the domain definitions on the sequence nor the descriptions are identical to Pfam.
- Note down the following information so you can enter the annotation in the protein database for MYSPE:
- From the section on "Confidently predicted domains ..."
- The start and end coordinates of the KilA-N domain (...according to SMART, not Pfam, in case the two differ).
- All start and end coordinates of low complexity segments
- All start and end coordinates of ANK (Ankyrin) domains
- Start and end coordinates of coiled coil domain(s) I expect only one.
- Start and end coordinates of AT hook domain(s) I expect some but not all not all Mbp1 orthologues have one.
- From the section on "Features NOT shown ..."
- All start and end coordinates of low complexity segments for which the Reason is "overlap".
- Any start and end coordinates of overlapping coiled coil segments.
- I expect all other annotations - besides the overlapping KilA-N domain defined by Pfam - to arise from the succession of ankyrin domains that the proteins have, both Pfam_ANK.. domains, as well as internal repeats, OR to be excluded because they did not exceed the significance threshold. However, if there are other features I have not mentioned here, please let me know.
- From the section on "Outlier homologues ..."
- Start and end coordinates of a PDB:1SW6|B annotation (if you have one): this is a region of sequence similarity to a protein for which the 3D structural coordinate are known.
- Of course there should also be annotations to the structure of 1BM8 / 1MB1 and/or 1L3G - all of which are structures of the Mbp1 APSES domain that we have already annotated as an"APSES fold" feature previously. And there will be BLAST annotations to Ankyrin domains. We will not annotate these separately either.
- From the section on "Confidently predicted domains ..."
- Follow the links to the database entries for the information so you know what these domains and features are.
Next we'll enter the features into our database, so we can compare them with the annotations that I have prepared from SMART annotations of Mbp1 orthologues from the ten reference fungi.
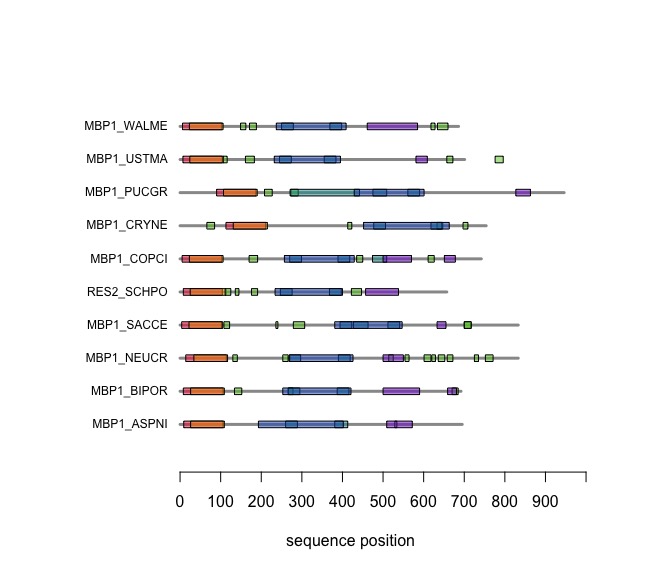
Visual comparison of domain annotations in R
The versatile plotting functions of R allow us to compare domain annotations. The distribution of segments that are annotated as "low-complexity, presumably disordered, is particularly interesting: these are functional features that are often not alignable since there is no selective pressure on sequence similarity but they may have arisen from convergent evolution or diverged while maintaining average composition, not specific sequence. Sequence alignment is after all based on amino acid pair scores that have been optimized to detect amino acids that behave similarly in the same context of folded proteins.
In the following code tutorial, we create a plot similar to the CDD and SMART displays. It is based on the SMART domain annotations of the reference species in our protein database.
Task:
- Open RStudio and load the
ABC-unitsR project. If you have loaded it before, choose File → Recent projects → ABC-Units. If you have not loaded it before, follow the instructions in the RPR-Introduction unit. - Choose Tools → Version Control → Pull Branches to fetch the most recent version of the project from its GitHub repository with all changes and bug fixes included.
- Type
init()if requested. - Open the file
BIN-FUNC-Domain_annotation.Rand follow the instructions.
Note: take care that you understand all of the code in the script. Evaluation in this course is cumulative and you may be asked to explain any part of code.
After you have worked through this code, your plot should look similar to this one:
CDART
The CDART database (Conserved Domain Architecture Retrieval Tool) finds proteins that have a similar domain architecture as a query. This has the potential to find homologous and functionally related proteins that are far todissimilar to be detected with sequence similarity searches.
Task:
- Retrieve the MYSPE Refseq ID from your Journal or by issuing the following R commands:
pName <- sprintf("MBP1_%s", biCode(MYSPE))
sel <- which(myDB$protein$name == pName)
myDB$protein$RefSeqID[sel]- Navigate to CDART.
- Paste your Mbp1 protein ID and click Submit.
- Note that the first page of the (very long!) results list shows proteisn that contain both KilA-N and Ankyrin domains. However a few other domains are found as well (Atrophin, GNVR, SMV_N), and this raises the intriguing possibility that the MBP1_MYSPE protein might contain some or all of these as well, although the sequence similarity may be to low to detect this outright.
- Study the domain annotations for
Further reading, links and resources
| El-Gebali et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47:D427-D432. (pmid: 30357350) |
| Letunic & Bork (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493-D496. (pmid: 29040681) |
| Geer et al. (2002) CDART: protein homology by domain architecture. Genome Res 12:1619-23. (pmid: 12368255) |
| Yang et al. (2020) NCBI's Conserved Domain Database and Tools for Protein Domain Analysis. Curr Protoc Bioinformatics 69:e90. (pmid: 31851420) |
Notes
- ↑ "Disorder" comes from IUPred predictions, "low complexity regions" are predicted by SEG, and "coiled coils" are predicted according to a scale developed by Rob Russell and Rune Linding; their coils server appears defunct but a similar algorithm (due to A. Lupas) is hosted at SIB.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2020-09-24
Version:
- 1.1
Version history:
- 1.1 2020 Updates, fixed Pfam/SMART annotation mix-up
- 1.0 Live version
- 0.1 First stub, import of 2016 material
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.