Difference between revisions of "Application Exam Questions"
| Line 101: | Line 101: | ||
<br> | <br> | ||
| + | ==2003 - Phylogenetic analysis== | ||
| + | |||
| + | You have studied the relationship between GlnRS and GluRS throughout your assignments (in 2003). An important contribution to this topic was made in September of 2003, with the discovery of a GluRS isozyme with intriguing specificity in ''H. pylori''. It appears that GluRS-1 aminoacylates tRNAGlu while GluRS-2 specifically aminoacylates tRNAGln. Is this a missing link in the evolution of laterally transferred GlxRS ? Is it an alternative route to independent evolution of GlnRS activity ? Or is it just a refinement of the GluRS → GatABC pathway, perhaps for better metabolic regulation ? Here is some of the evidence: | ||
| + | | ||
| + | | ||
| + | |||
| + | [[Image:PhylogeneticTreeGlnRS.jpg|frame|none|(After: Skouloubris S, DePouplana LR, DeReuse H, Hendrickson TL (2003) A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. ''PNAS'' '''100''':11297-11302.) Phylogenetic tree of GluRS sequences in proteobacteria. Species with ORFs similar to helicobacter GluRS-1 resp. GluRS-2 are boxed; those few not in the box have been identified with the suffix "-1" resp. "-2". Bacteria with a GlnRS ORF are labelled with a black diamond. GlnRS sequences are more distantly related to any of these GluRS than all of them with each other. ]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Briefly explain the principles of this phylogenetic tree in terms of what can be observed, what must be inferred and how this tree integrates this information.''' | ||
| + | *'''What do the numbers in the figure mean ?''' | ||
| + | *'''Briefly state how the principle of maximum parsimony is applied to construct phylogenetic trees such as this one.''' | ||
| + | *'''Briefly discuss the evidence in the tree for (1) independent events of horizontal gene transfer into bacteria, and (2) gene duplication leading to GluRS-1 and -2.''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
==2002== | ==2002== | ||
Revision as of 15:24, 11 December 2006
- Bioinformatics and computational biology are huge and growing areas of active research with a dynamic array of subspecialities - phylogenetic analysis has advanced in leaps and bounds with the availablity of molecular data, genomic-, transcriptomic-, proteomic and other cross-sectional views are slowly beginning to unravel some of the intricacies of the cell's inner workings, predictive models for molecular medicine and bioengineering may help shape our society's future. These tasks are what motivates bioinformatics worldwide, seeking to develp novel ways to reason about biology.
2003 - Consequences of Homology
A bacterial genome has been sequenced. Comparison with tRNA synthetase COGs has failed to turn up an annotation for a Glutaminyl tRNA synthetase gene. Assume that the genome sequence is complete, has been assembled without frameshift errors and all gene models are correct. The sequence for Glutaminyl tRNA synthetase has not simply been overlooked. Answer the following questions briefly.
- What is a COG ?
- State two reasons why the endpoint of a biochemical pathway could be present in an organism but a gene on the pathway would fail to be found in a COG analysis.
- Briefly(!) state what wet-lab experiments you would propose to confirm (or disprove) the hypotheses you have stated above ?
Comment
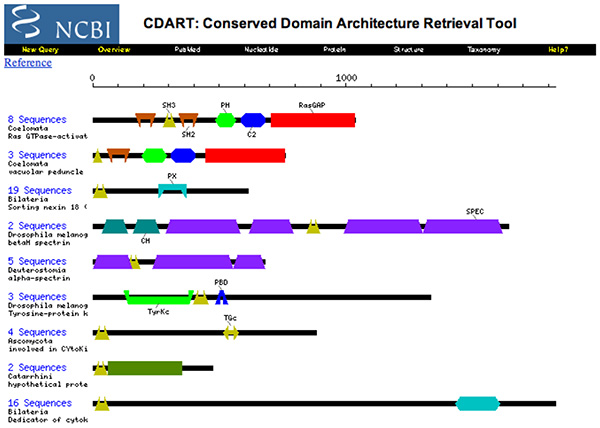
2003 - Domains and Homology
- Briefly define homologs, orthologs and paralogs.
- Are the proteins depicted above homologs ?
- Briefly explain the contents, implications and use of this CDART entry.
(By now, "contents, implications and use" would be too vague and general for my taste for an exam question.)
2003 - Homology
The COGs database employs a definition of orthologs that is based on reciprocal highest similarity.
- Briefly explain the principle and how it distinguishes orthologs from paralogs.
You have built a homology model based on the pairwise alignment of a C. elegans Glutaminyl-tRNA synthetase sequence to a template that you have identified with a BLAST search. Upon analysis of your model structure, you note that the presumed ATP-binding site of the model appears to be blocked by an arginine sidechain.
- Briefly define the key steps involved in building a homology model.
- Based on these steps, identify two probable sources of the discrepancy between the model and the function of the protein.
- Suggest a bioinformatics strategy to improve your model, given the problems you have identified above.
2003 - Expression Analysis
In order to study the coordination of gene expression in the yeast cell cycle, you have created a synchronized, growing culture of Saccharomyces cerevisiaea cells and harvested mRNA at ten successive time points along one replication cycle. You now plan to perform two-color microarray (spotted array) experiments.
- Describe the principle and key steps of this experiment, from mRNA samples to a series of scanned images.
- Describe the key steps involved in going from a scanned image to expression profiles of individual genes.
- After clustering genes according to similarity of expression profiles, you will have sets of genes with correlated expression profiles. You hypothesize that these may be coregulated genes. What bioinformatics procedure(s) can you suggest to help you annotate shared functions for your clusters of genes ?
- What bioinformatics procedure(s) can you suggest to pursue the question whether these genes may be coregulated ?
2003 - Integrated processes
Read the following abstract:
- Structure of TCTP reveals unexpected relationship with guanine nucleotidefree chaperones
- Paul Thaw, Nicola J. Baxter, Andrea M. Hounslow, Clive Price, Jonathan P. Waltho and C. Jeremy Craven: Nature Struct Biol 8: 701–704 (2001)
- The translationally controlled tumor-associated proteins (TCTPs) are a highly conserved and abundantly expressed family of eukaryotic proteins that are implicated in both cell growth and the human acute allergic response but whose intracellular biochemical function has remained elusive. We report here the solution structure of the TCTP from Schizosaccharomyces pombe, which, on the basis of sequence homology, defines the fold of the entire family. We show that TCTPs form a structural superfamily with the Mss4/Dss4 family of proteins, which bind to the GDP/GTP free form of Rab proteins (members of the Ras superfamily) and have been termed guanine nucleotide-free chaperones (GFCs). Mss4 also acts as a relatively inefficient guanine nucleotide exchange factor (GEF). We further show that the Rab protein binding site on Mss4 coincides with the region of highest sequence conservation in the TCTP family. This is the first link to any other family of proteins that has been established for the TCTP family and suggests the presence of a GFC/GEF at extremely high abundance in eukaryotic cells.
This abstract reports several pieces of data and mentions several pieces of prior information.
- List the most important data and information entities and databases that have been used in this study.
- Summarize on approximately one-half page or less the essential steps of how these entities were related to each other in this study. You may use any representation that is reasonable such as pseudocode, a flowchart, or other type of sketch.
Note that you are not required to understand the biochemical processes that are described here, nor are you required to comment on the cell-biological implications. One of the key steps has been underlined by me - you must understand how such a conclusion can be drawn in the situation that is described. You are to summarize the flow of data: the entities that are being referred to, and the experimental and computational procedures.
2003 - Phylogenetic analysis
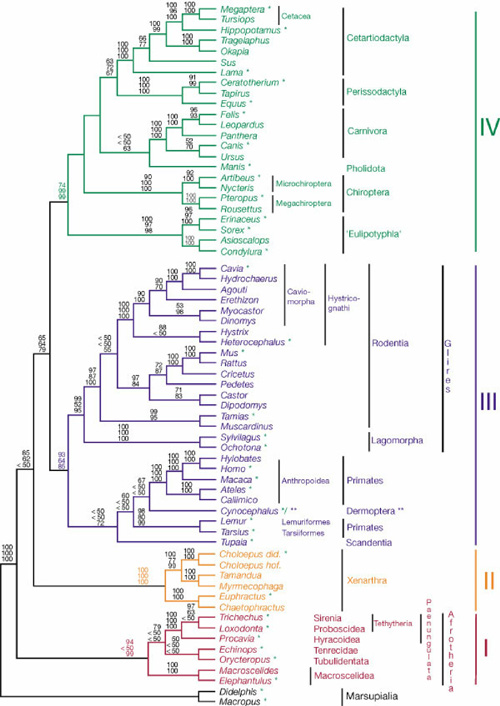
- Briefly explain the principles of this graph in terms of what can be observed, what must be inferred and how this graph integrates this information.
- What do the numbers among the branches mean ?
- Which parts of this graph correspond to the following terms:
- Clade
- Bifurcation
- Root
- Briefly state the principle of maximum parsimony and briefly explain how it is applied in principle to construct graphs of the type shown above.
2003 - Phylogenetic analysis
You have studied the relationship between GlnRS and GluRS throughout your assignments (in 2003). An important contribution to this topic was made in September of 2003, with the discovery of a GluRS isozyme with intriguing specificity in H. pylori. It appears that GluRS-1 aminoacylates tRNAGlu while GluRS-2 specifically aminoacylates tRNAGln. Is this a missing link in the evolution of laterally transferred GlxRS ? Is it an alternative route to independent evolution of GlnRS activity ? Or is it just a refinement of the GluRS → GatABC pathway, perhaps for better metabolic regulation ? Here is some of the evidence:
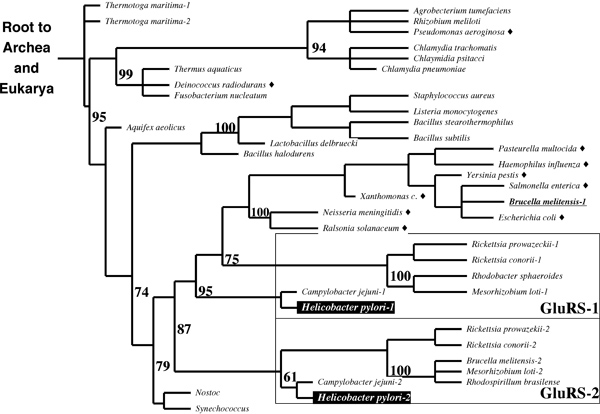
- Briefly explain the principles of this phylogenetic tree in terms of what can be observed, what must be inferred and how this tree integrates this information.
- What do the numbers in the figure mean ?
- Briefly state how the principle of maximum parsimony is applied to construct phylogenetic trees such as this one.
- Briefly discuss the evidence in the tree for (1) independent events of horizontal gene transfer into bacteria, and (2) gene duplication leading to GluRS-1 and -2.
2002
Comment