Difference between revisions of "BIN-Storing data"
m |
m |
||
| (26 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | <div id=" | + | <div id="ABC"> |
| − | + | <div style="padding:5px; border:4px solid #000000; background-color:#b3dbce; font-size:300%; font-weight:400; color: #000000; width:100%;"> | |
Storing Data | Storing Data | ||
| − | + | <div style="padding:5px; margin-top:20px; margin-bottom:10px; background-color:#b3dbce; font-size:30%; font-weight:200; color: #000000; "> | |
| − | + | (Representation of data; common data formats; implementing a data model; JSON.) | |
| − | + | </div> | |
| − | |||
| − | <div | ||
| − | |||
| − | Representation of data; common data formats | ||
</div> | </div> | ||
| − | {{ | + | {{Smallvspace}} |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | <div style="padding:5px; border:1px solid #000000; background-color:#b3dbce33; font-size:85%;"> | ||
| + | <div style="font-size:118%;"> | ||
| + | <b>Abstract:</b><br /> | ||
| + | <section begin=abstract /> | ||
| + | This unit discusses options for storing and organizing data in a variety of formats, and develops R code for a protein data model, based on JSON formatted source data. | ||
| + | <section end=abstract /> | ||
| + | </div> | ||
| + | <!-- ============================ --> | ||
| + | <hr> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td style="padding:10px;"> | ||
| + | <b>Objectives:</b><br /> | ||
| + | This unit will ... | ||
| + | * Introduce principles of storing data in different formats and principles of supporting storage, maintenance, and retrieval with different tools; | ||
| + | * Implement relational data models as lists of data frames in R; | ||
| + | * Develop code for this purpose and teach how to work with such code; | ||
| + | * Find the closest homologue of Mbp1 in MYSPE. | ||
| + | </td> | ||
| + | <td style="padding:10px;"> | ||
| + | <b>Outcomes:</b><br /> | ||
| + | After working through this unit you ... | ||
| + | * can recommend alternatives for storing data, depending on the context and objectives; | ||
| + | * can create, edit and validate JSON formatted data; | ||
| + | * have practiced creating a relational database as a list of dataframes; | ||
| + | * can query the database directly, and via cross-referencing tables; | ||
| + | * have discovered the closest homologue to yeast Mbp1 in MYSPE; | ||
| + | * have added key information about this protein to the database; | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <!-- ============================ --> | ||
| + | <hr> | ||
| + | <b>Deliverables:</b><br /> | ||
| + | <section begin=deliverables /> | ||
| + | <ul> | ||
| + | <li><b>Time management</b>: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit.</li> | ||
| + | <li><b>Journal</b>: Document your progress in your [[FND-Journal|Course Journal]]. Some tasks may ask you to include specific items in your journal. Don't overlook these.</li> | ||
| + | <li><b>Insights</b>: If you find something particularly noteworthy about this unit, make a note in your [[ABC-Insights|'''insights!''' page]].</li> | ||
| + | </li>'''Your protein database''' Write a database-generating script that loads a protein database from JSON files.</li> | ||
| + | </ul> | ||
| + | <section end=deliverables /> | ||
| + | <!-- ============================ --> | ||
| + | <hr> | ||
| + | <section begin=prerequisites /> | ||
| + | <b>Prerequisites:</b><br /> | ||
| + | This unit builds on material covered in the following prerequisite units:<br /> | ||
| + | *[[BIN-Abstractions|BIN-Abstractions (Abstractions for Bioinformatics)]] | ||
| + | <section end=prerequisites /> | ||
| + | <!-- ============================ --> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | {{Smallvspace}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | {{Smallvspace}} | ||
| − | |||
| − | |||
| − | |||
| − | + | __TOC__ | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| Line 70: | Line 75: | ||
=== Evaluation === | === Evaluation === | ||
| − | < | + | This learning unit can be evaluated for a maximum of 5 marks. If you want to submit the tasks for this unit for credit: |
| − | < | + | <ol> |
| − | < | + | <li>Create a new page on the student Wiki as a subpage of your User Page.</li> |
| − | + | <li>There are a number of tasks in which you are explicitly asked you to submit code or other text for credit. Put all of these submission on this one page.</li> | |
| + | <li>When you are done with everything, go to the [https://q.utoronto.ca/courses/180416/assignments Quercus '''Assignments''' page] and open the first Learning Unit that you have not submitted yet. Paste the URL of your Wiki page into the form, and click on '''Submit Assignment'''.</li> | ||
| + | </ol> | ||
| − | + | Your link can be submitted only once and not edited. But you may change your Wiki page at any time. However only the last version before the due date will be marked. All later edits will be silently ignored. | |
| + | {{Smallvspace}} | ||
| − | |||
| − | |||
== Contents == | == Contents == | ||
| − | |||
{{Task|1= | {{Task|1= | ||
| Line 91: | Line 96: | ||
| + | Any software project requires modelling on many levels - data-flow models, logic models, user interaction models and more. But all of these ultimately rely on a '''data model''' that defines how the world is going to be represented in the computer for the project's purpose. The process of abstraction of data entities and defining their relationships can (and should) take up a major part of the project definition, often taking several iterations until you get it right. Whether your data can be completely described, consistently stored and efficiently retrieved is determined to a large part by your data model. | ||
| − | + | {{WP|Database|Databases}} can take many forms, from memories in your brain, to shoe-cartons under your bed, to software applications on your computer, or warehouse-sized data centres. Fundamentally, these all do the same thing: collect information and make it available for retrieval. | |
| − | |||
| − | |||
| − | + | Let us consider collecting information on APSES-domain transcription factors in various fungi, with the goal of being able to compare the transcription factors. Let's specify this as follows: | |
| + | |||
| + | <div class="emphasis-box">Store data on APSES-domain proteins so that we can | ||
| + | * cross reference the source databases; | ||
| + | * study if they have the same features (e.g. domains); | ||
| + | * and compare the features across species. | ||
| + | </div> | ||
| + | |||
| + | The underlying information can easily be retrieved for a protein from its RefSeq or UniProt entry. | ||
| − | |||
| − | + | ===Text files=== | |
| − | + | A first attempt to organize the data might be simply to write it down as text: | |
| − | + | name: Mbp1 | |
| + | refseq ID: NP_010227 | ||
| + | uniprot ID: P39678 | ||
| + | species: Saccharomyces cerevisiae | ||
| + | taxonomy ID: 4392 | ||
| + | sequence: | ||
| + | MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKR | ||
| + | TRILEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF | ||
| + | DFTQTDGSASPPPAPKHHHASKVDRKKAIRSASTSAIMETKRNNKKAEEN | ||
| + | QFQSSKILGNPTAAPRKRGRPVGSTRGSRRKLGVNLQRSQSDMGFPRPAI | ||
| + | PNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQPQQNNSAQFKEI | ||
| + | DLEDGLSSDVEPSQQLQQVFNQNTGFVPQQQSSLIQTQQTESMATSVSSS | ||
| + | PSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDINDKV | ||
| + | NKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPYIDAPIDPELHTAFH | ||
| + | WACSMGNLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRI | ||
| + | FQLLHETVFDIDSQSQTVIHHIVKRKSTTPSAVYYLDVVLSKIKDFSPQY | ||
| + | RIELLLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTAN | ||
| + | EIMNQQYEQMMIQNGTNQHVNSSNTDLNIHVNTNNIETKNDVNSMVIMSP | ||
| + | VSPSDYITYPSQIATNISRNIPNVVNSMKQMASIYNDLHEQHDNEIKSLQ | ||
| + | KTLKSISKTKIQVSLKTLEVLKESSKDENGEAQTNDDFEILSRLQEQNTK | ||
| + | KLRKRLIRYKRLIKQKLEYRQTVLLNKLIEDETQATTNNTVEKDNNTLER | ||
| + | LELAQELTMLQLQRKNKLSSLVKKFEDNAKIHKYRRIIREGTEMNIEEVD | ||
| + | SSLDVILQTLIANNNKNKGAEQIITISNANSHA | ||
| + | length: 833 | ||
| + | Kila-N domain: 21-93 | ||
| + | Ankyrin domains: 369-455, 505-549 | ||
| − | + | ... | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ... and save it all in one large text file and whenever you need to look something up, you just open the file, look for e.g. the name of the protein and read what's there. Or - for a more structured approach, you could put this into several files in a folder.<ref>Your operating system can help you keep the files organized. The "file system" '''is''' a database.</ref> This is a perfectly valid approach and for some applications it might not be worth the effort to think more deeply about how to structure the data, and store it in a way that it is robust and scales easily to large datasets. Alas, small projects have a tendency to grow into large projects and if you work in this way, it's almost guaranteed that you will end up doing many things by hand that could easily be automated. Imagine asking questions like: | |
| + | * How many proteins do I have? | ||
| + | * What's the sequence of the Kila-N domain? | ||
| + | * What percentage of my proteins have an Ankyrin domain? | ||
| + | * Or two ...? | ||
| − | + | Answering these questions "by hand" is possible, but tedious. | |
| − | + | ===Spreadsheets=== | |
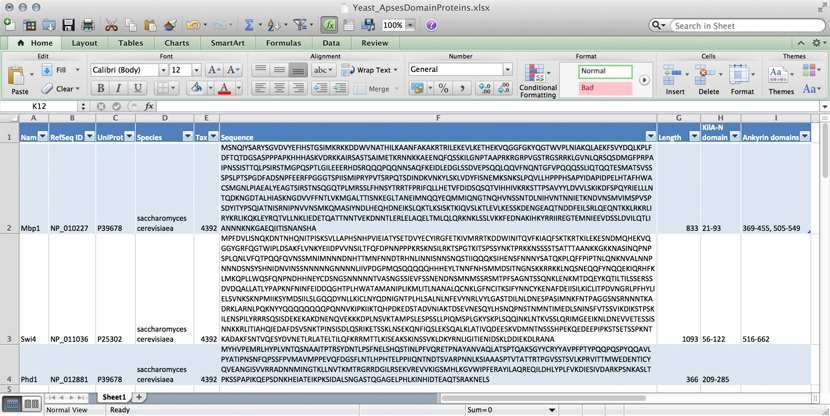
| + | [[Image:DB_Excel-spreadsheet.jpg|frame|Data for three yeast APSES domain proteins in an Excel spreadsheet.]] | ||
| − | + | Many serious researchers keep their project data in spreadsheets. Often they use Excel, or an alternative like the free [https://www.openoffice.org/product/calc.html OpenOffice Calc], or [https://www.google.ca/sheets/about/ Google Sheets], both of which are compatible with Excel and have some interesting advantages. Here, all your data is in one place, easy to edit. You can even do simple calculations - although you should '''never''' use Excel for statistics<ref>For real: Excel is miserable and often wrong on statistics, and it makes horrible, ugly plots. See [http://www.practicalstats.com/xlstats/excelstats.html here] and [http://www.burns-stat.com/documents/tutorials/spreadsheet-addiction/ here] why Excel problems are not merely cosmetic.</ref>. You could answer ''What percentage of my proteins have an Ankyrin domain?'' quite easily<ref>At the bottom of the window there is a menu that says "sum = ..." by default. This provides simple calculations on the selected range. Set the choice to "count", select all Ankyrin domain entries, and the count shows you how many cells actually have a value.</ref>. | |
| − | + | There are two major downsides to spreadsheets. For one, complex queries need programming. There is no way around this. You '''can''' program inside Excel with ''Visual Basic''. But you might as well export your data so you can work on it with a "real" programming language. The other thing is that Excel does not scale very well. Once you have more than a hundred proteins in your spreadsheet, you can see how finding anything can become tedious. | |
| − | + | However, just because Excel was built for business applications, and designed for use by office assistants, does not mean it is intrinsically unsuitable for our domain. It's important to be pragmatic, not dogmatic, when choosing tools: choose according to your real requirements. Sometimes "quick and dirty" is just fine, because quick. | |
{{Vspace}} | {{Vspace}} | ||
| − | + | ===R=== | |
| − | + | '''R''' can keep complex data in data frames and lists. If we do data analysis with '''R''', we have to load the data first. We can use any of the <code>read.table()</code> functions for structured data, read lines of raw text with <code>readLines()</code>, or slurp in entire files with <code>scan()</code>. Convenient packages exist to parse structured data like XML or JSON and import it. But we could also keep the data in an '''R''' object in the first place that we can read from disk, analyze, modify, and write back. In this case, '''R''' becomes our database engine. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <pre> | |
| − | + | # Sample construction of an R database table as a dataframe | |
| − | + | # Data for the Mbp1 protein | |
| − | + | proteins <- data.frame( | |
| − | + | name = "Mbp1", | |
| − | + | refSeq = "NP_010227", | |
| + | uniProt = "P39678", | ||
| + | species = "Saccharomyces cerevisiae", | ||
| + | taxId = "4392", | ||
| + | sequence = paste( | ||
| + | "MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKR", | ||
| + | "TRILEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF", | ||
| + | "DFTQTDGSASPPPAPKHHHASKVDRKKAIRSASTSAIMETKRNNKKAEEN", | ||
| + | "QFQSSKILGNPTAAPRKRGRPVGSTRGSRRKLGVNLQRSQSDMGFPRPAI", | ||
| + | "PNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQPQQNNSAQFKEI", | ||
| + | "DLEDGLSSDVEPSQQLQQVFNQNTGFVPQQQSSLIQTQQTESMATSVSSS", | ||
| + | "PSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDINDKV", | ||
| + | "NKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPYIDAPIDPELHTAFH", | ||
| + | "WACSMGNLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRI", | ||
| + | "FQLLHETVFDIDSQSQTVIHHIVKRKSTTPSAVYYLDVVLSKIKDFSPQY", | ||
| + | "RIELLLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTAN", | ||
| + | "EIMNQQYEQMMIQNGTNQHVNSSNTDLNIHVNTNNIETKNDVNSMVIMSP", | ||
| + | "VSPSDYITYPSQIATNISRNIPNVVNSMKQMASIYNDLHEQHDNEIKSLQ", | ||
| + | "KTLKSISKTKIQVSLKTLEVLKESSKDENGEAQTNDDFEILSRLQEQNTK", | ||
| + | "KLRKRLIRYKRLIKQKLEYRQTVLLNKLIEDETQATTNNTVEKDNNTLER", | ||
| + | "LELAQELTMLQLQRKNKLSSLVKKFEDNAKIHKYRRIIREGTEMNIEEVD", | ||
| + | "SSLDVILQTLIANNNKNKGAEQIITISNANSHA", | ||
| + | sep=""), | ||
| + | seqLen = 833, | ||
| + | KilAN = "21-93", | ||
| + | Ankyrin = "369-455, 505-549") | ||
| − | + | # add data for the Swi4 protein | |
| − | + | proteins <- rbind(proteins, | |
| − | + | data.frame( | |
| + | name = "Swi4", | ||
| + | refSeq = "NP_011036", | ||
| + | uniProt = "P25302", | ||
| + | species = "Saccharomyces cerevisiae", | ||
| + | taxId = "4392", | ||
| + | sequence = paste( | ||
| + | "MPFDVLISNQKDNTNHQNITPISKSVLLAPHSNHPVIEIATYSETDVYEC", | ||
| + | "YIRGFETKIVMRRTKDDWINITQVFKIAQFSKTKRTKILEKESNDMQHEK", | ||
| + | "VQGGYGRFQGTWIPLDSAKFLVNKYEIIDPVVNSILTFQFDPNNPPPKRS", | ||
| + | "KNSILRKTSPGTKITSPSSYNKTPRKKNSSSSTSATTTAANKKGKKNASI", | ||
| + | "NQPNPSPLQNLVFQTPQQFQVNSSMNIMNNNDNHTTMNFNNDTRHNLINN", | ||
| + | "ISNNSNQSTIIQQQKSIHENSFNNNYSATQKPLQFFPIPTNLQNKNVALN", | ||
| + | "NPNNNDSNSYSHNIDNVINSSNNNNNGNNNNLIIVPDGPMQSQQQQQHHH", | ||
| + | "EYLTNNFNHSMMDSITNGNSKKRRKKLNQSNEQQFYNQQEKIQRHFKLMK", | ||
| + | "QPLLWQSFQNPNDHHNEYCDSNGSNNNNNTVASNGSSIEVFSSNENDNSM", | ||
| + | "NMSSRSMTPFSAGNTSSQNKLENKMTDQEYKQTILTILSSERSSDVDQAL", | ||
| + | "LATLYPAPKNFNINFEIDDQGHTPLHWATAMANIPLIKMLITLNANALQC", | ||
| + | "NKLGFNCITKSIFYNNCYKENAFDEIISILKICLITPDVNGRLPFHYLIE", | ||
| + | "LSVNKSKNPMIIKSYMDSIILSLGQQDYNLLKICLNYQDNIGNTPLHLSA", | ||
| + | "LNLNFEVYNRLVYLGASTDILNLDNESPASIMNKFNTPAGGSNSRNNNTK", | ||
| + | "ADRKLARNLPQKNYYQQQQQQQQPQNNVKIPKIIKTQHPDKEDSTADVNI", | ||
| + | "AKTDSEVNESQYLHSNQPNSTNMNTIMEDLSNINSFVTSSVIKDIKSTPS", | ||
| + | "KILENSPILYRRRSQSISDEKEKAKDNENQVEKKKDPLNSVKTAMPSLES", | ||
| + | "PSSLLPIQMSPLGKYSKPLSQQINKLNTKVSSLQRIMGEEIKNLDNEVVE", | ||
| + | "TESSISNNKKRLITIAHQIEDAFDSVSNKTPINSISDLQSRIKETSSKLN", | ||
| + | "SEKQNFIQSLEKSQALKLATIVQDEESKVDMNTNSSSHPEKQEDEEPIPK", | ||
| + | "STSETSSPKNTKADAKFSNTVQESYDVNETLRLATELTILQFKRRMTTLK", | ||
| + | "ISEAKSKINSSVKLDKYRNLIGITIENIDSKLDDIEKDLRANA", | ||
| + | sep=""), | ||
| + | seqLen = 1093, | ||
| + | KilAN = "56-122", | ||
| + | Ankyrin = "516-662") | ||
| + | ) | ||
| − | + | # how many proteins? | |
| + | nrow(proteins) | ||
| − | + | #what are their names? | |
| + | proteins[,"name"] | ||
| + | # how many do not have an Ankyrin domain? | ||
| + | sum(proteins[,"Ankyrin"] == "") | ||
| + | # save it to file | ||
| + | saveRDS(proteins, file="proteinData.rds") | ||
| − | + | # delete it from memory | |
| + | rm(proteins) | ||
| − | + | # check... | |
| + | proteins # ... yes, it's gone | ||
| − | + | # read it back in: | |
| − | + | proteins <- readRDS("proteinData.rds") | |
| − | |||
| − | + | # did this work? | |
| + | sum(proteins[ , "seqLen"]) # 1926 amino acids | ||
| − | |||
| − | + | # [END] | |
| + | </pre> | ||
| − | |||
| − | ; | + | |
| − | + | The third way to use '''R''' for data is to connect it to a "real" database: | |
| − | + | *a relational database like {{WP|mySQL}}, {{WP|MariaDB}}, or {{WP|PostgreSQL}}; | |
| − | + | *an object/document database like {{WP|MongoDB}; | |
| + | * or even a graph-database like {{WP|Neo4j}}. | ||
| − | }} | + | '''R''' "drivers" are available for all of these. However all of these require installing extra software on your computer: the actual database, which runs as an independent application. If you need a rock-solid database with guaranteed integrity, multi-user support, {{WP|ACID}} transactional gurantees, industry standard performance, and scalability to even '''very''' large datasets, don't think of rolling your own solution. One of the above is the way to go. |
{{Vspace}} | {{Vspace}} | ||
| + | ===MySQL and friends=== | ||
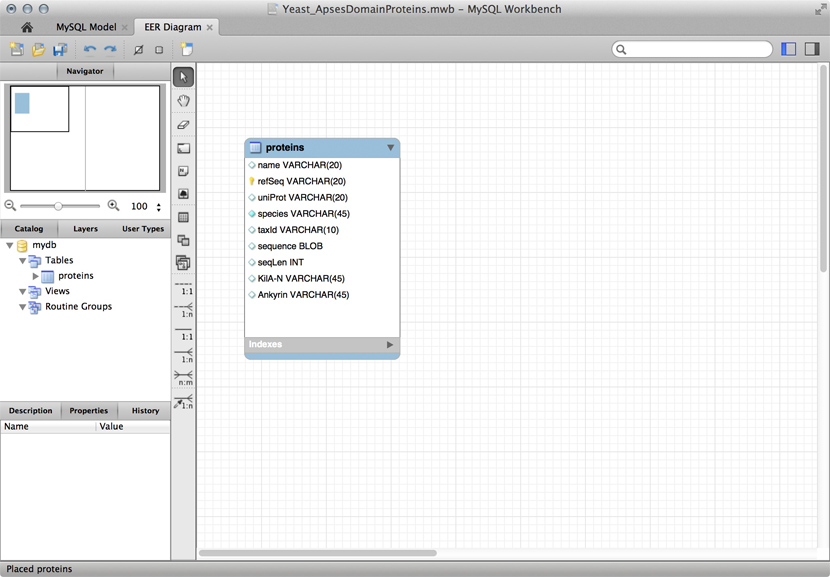
| + | [[Image:DB_MySQL-Workbench.jpg|frame|A "Schema" for a ''table'' that stores data for APSES domain proteins. This is a screenshot of the free MySQL Workbench application.]] | ||
| + | MySQL is a free, open '''relational database''' that powers some of the largest corporations as well as some of the smallest laboratories. It is based on a client-server model. The database engine runs as a daemon in the background and waits for connection attempts. When a connection is established, the '''server''' process establishes a communication session with the client. The client sends requests, and the server responds. One can do this interactively, by running the '''client''' program <tt>/usr/local/mysql/bin/mysql</tt> (on Unix systems). Or, when you are using a program such as '''R''', Python, Perl, etc. you use the appropriate method calls or functions—the driver—to establish the connection. | ||
| + | These types of databases use their own language to describe actions: {{WP|SQL|'''SQL'''}} - which handles data definition, data manipulation, and data control. | ||
| + | |||
| + | Just for illustration, the Figure above shows a table for our APSES domain protein data, built as a table in the [https://www.mysql.com/products/workbench/ MySQL workbench] application and presented as an Entity Relationship Diagram (ERD). There is only one entity though - the protein "table". The application can generate the actual code that implements this model on a SQL compliant database: | ||
| − | |||
| − | |||
| − | + | <pre> | |
| + | CREATE TABLE IF NOT EXISTS `mydb`.`proteins` ( | ||
| + | `name` VARCHAR(20) NULL, | ||
| + | `refSeq` VARCHAR(20) NOT NULL, | ||
| + | `uniProt` VARCHAR(20) NULL, | ||
| + | `species` VARCHAR(45) NOT NULL COMMENT ' ', | ||
| + | `taxId` VARCHAR(10) NULL, | ||
| + | `sequence` BLOB NULL, | ||
| + | `seqLen` INT NULL, | ||
| + | `KilA-N` VARCHAR(45) NULL, | ||
| + | `Ankyrin` VARCHAR(45) NULL, | ||
| + | PRIMARY KEY (`refSeq`)) | ||
| + | ENGINE = InnoDB | ||
| − | + | </pre> | |
| + | This looks at least as complicated as putting the model into '''R''' in the first place. Why then would we do this, if we need to load it into '''R''' for analysis anyway? There are several important reasons. | ||
| − | {{ | + | *Scalability: these systems are built to work with '''very''' large datasets and optimized for performance. In theory '''R''' has very good performance with large data objects, but not so when the data becomes larger than what the computer can keep in memory all at once. |
| + | *Concurrency: when several users need to access the data potentially at the same time, you '''must''' use a "real" database system. Handling problems of concurrent access is what they are made for. | ||
| + | *ACID compliance. {{WP|ACID}} describes four aspects that make a database robust, these are crucial for situations in which you have only partial control over your system or its input, and they would be quite laborious to implement for your hand built '''R''' data model: | ||
| + | **'''A'''tomicity: Atomicity requires that each transaction is handled "indivisibly": it either succeeds fully, with all requested elements, or not at all. | ||
| + | **'''C'''onsistency: Consistency requires that any transaction will bring the database from one valid state to another. In particular any data-validation rules have to be enforced. | ||
| + | **'''I'''solation: Isolation ensures that any concurrent execution of transactions results in the exact same database state as if transactions would have been executed serially, one after the other. | ||
| + | **'''D'''urability: The Durability requirement ensures that a committed transaction remains permanently committed, even in the event that the database crashes or later errors occur. You can think of this like an "autosave" function on every operation. | ||
| + | All the database systems I have mentioned above are ACID compliant<ref>For a list of relational Database Management Systems, see {{WP|https://en.wikipedia.org/wiki/Comparison_of_relational_database_management_systems|here}}.</ref>. | ||
| − | + | Incidentally - RStudio has inbuilt database support via the '''Connections''' tab of the top-right pane. Read more about database connections directly from RStudio [https://db.rstudio.com/rstudio/connections/ '''here''']. | |
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| − | + | ==Store Data== | |
| − | == | ||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| + | ===A protein datamodel=== | ||
| − | < | + | {{Vspace}} |
| − | < | + | <table> |
| − | + | <tr> | |
| − | < | + | <td> |
| − | |||
| − | |||
| − | + | [[File:ProteinDBschema.svg|width=500px|link=https://docs.google.com/presentation/d/13vWaVcFpWEOGeSNhwmqugj2qTQuH1eZROgxWdHGEMr0]] | |
| − | < | + | </td> |
| − | + | </tr> | |
| − | < | + | <tr> |
| − | + | <td><small> | |
| + | Entity-Relationship Diagram (ERD) for a protein data model that includes protein, taxonomy and feature annotations. Entity names are at the top of each box, attributes are listed below. If you think of an entity as an R dataframe, or a spreadsheet table, the attributes are the column names and each specific instance of an entity fills one row (or "record").All relationships link to unique primary keys and are thus 1 to (0, n). The diagram was drawn as a "Google slide" and you can [https://docs.google.com/presentation/d/13vWaVcFpWEOGeSNhwmqugj2qTQuH1eZROgxWdHGEMr0 view it on the Web] and, if you have a Google account, you can make a copy to use for your own purposes. | ||
| + | </small></td> | ||
| + | </tr> | ||
| + | </table> | ||
| − | + | {{Vspace}} | |
| − | |||
| − | + | ===Implementing the Data Model in R=== | |
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| − | + | To actually implement the data model in '''R''' we will create tables as data frames, and we will collect them in a list. We don't '''have''' to keep the tables in a list - we could also keep them as independent objects, but a list is neater, more flexible (we might want to have several of these), it reflects our intent about the model better, and doesn't require very much more typing. | |
{{Vspace}} | {{Vspace}} | ||
| − | + | {{ABC-unit|BIN-Storing_data.R}} | |
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| − | |||
| − | --- | + | == Further reading, links and resources == |
| + | *{{WP|Database normalization}} | ||
| + | <div class="reference-box">[https://www.lucidchart.com/pages/database-diagram/database-models Overview of data model types] (Lucidchart)</div> | ||
| + | == Notes == | ||
| + | <references /> | ||
{{Vspace}} | {{Vspace}} | ||
| + | |||
<div class="about"> | <div class="about"> | ||
| Line 263: | Line 375: | ||
:2017-08-05 | :2017-08-05 | ||
<b>Modified:</b><br /> | <b>Modified:</b><br /> | ||
| − | : | + | :2020-10-07 |
<b>Version:</b><br /> | <b>Version:</b><br /> | ||
| − | : | + | :1.2 |
<b>Version history:</b><br /> | <b>Version history:</b><br /> | ||
| − | *0.1 First stub | + | *1.2 Edit policy update |
| + | *1.1 Update to new marking scheme | ||
| + | *1.0.1 Add link to data model types overview | ||
| + | *1.0 First Live version | ||
| + | *0.1 First stub | ||
</div> | </div> | ||
| − | |||
| − | |||
{{CC-BY}} | {{CC-BY}} | ||
| + | [[Category:ABC-units]] | ||
| + | {{EVAL}} | ||
| + | {{LIVE}} | ||
| + | {{EVAL}} | ||
</div> | </div> | ||
<!-- [END] --> | <!-- [END] --> | ||
Latest revision as of 05:01, 10 October 2020
Storing Data
(Representation of data; common data formats; implementing a data model; JSON.)
Abstract:
This unit discusses options for storing and organizing data in a variety of formats, and develops R code for a protein data model, based on JSON formatted source data.
|
Objectives:
|
Outcomes:
|
Deliverables:
- Time management: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit.
- Journal: Document your progress in your Course Journal. Some tasks may ask you to include specific items in your journal. Don't overlook these.
- Insights: If you find something particularly noteworthy about this unit, make a note in your insights! page. Your protein database Write a database-generating script that loads a protein database from JSON files.
Prerequisites:
This unit builds on material covered in the following prerequisite units:
Contents
Evaluation
This learning unit can be evaluated for a maximum of 5 marks. If you want to submit the tasks for this unit for credit:
- Create a new page on the student Wiki as a subpage of your User Page.
- There are a number of tasks in which you are explicitly asked you to submit code or other text for credit. Put all of these submission on this one page.
- When you are done with everything, go to the Quercus Assignments page and open the first Learning Unit that you have not submitted yet. Paste the URL of your Wiki page into the form, and click on Submit Assignment.
Your link can be submitted only once and not edited. But you may change your Wiki page at any time. However only the last version before the due date will be marked. All later edits will be silently ignored.
Contents
Task:
- Read the introductory notes on concepts about storing data for bioinformatics.
Any software project requires modelling on many levels - data-flow models, logic models, user interaction models and more. But all of these ultimately rely on a data model that defines how the world is going to be represented in the computer for the project's purpose. The process of abstraction of data entities and defining their relationships can (and should) take up a major part of the project definition, often taking several iterations until you get it right. Whether your data can be completely described, consistently stored and efficiently retrieved is determined to a large part by your data model.
Databases can take many forms, from memories in your brain, to shoe-cartons under your bed, to software applications on your computer, or warehouse-sized data centres. Fundamentally, these all do the same thing: collect information and make it available for retrieval.
Let us consider collecting information on APSES-domain transcription factors in various fungi, with the goal of being able to compare the transcription factors. Let's specify this as follows:
- cross reference the source databases;
- study if they have the same features (e.g. domains);
- and compare the features across species.
The underlying information can easily be retrieved for a protein from its RefSeq or UniProt entry.
Text files
A first attempt to organize the data might be simply to write it down as text:
name: Mbp1 refseq ID: NP_010227 uniprot ID: P39678 species: Saccharomyces cerevisiae taxonomy ID: 4392 sequence: MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKR TRILEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF DFTQTDGSASPPPAPKHHHASKVDRKKAIRSASTSAIMETKRNNKKAEEN QFQSSKILGNPTAAPRKRGRPVGSTRGSRRKLGVNLQRSQSDMGFPRPAI PNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQPQQNNSAQFKEI DLEDGLSSDVEPSQQLQQVFNQNTGFVPQQQSSLIQTQQTESMATSVSSS PSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDINDKV NKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPYIDAPIDPELHTAFH WACSMGNLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRI FQLLHETVFDIDSQSQTVIHHIVKRKSTTPSAVYYLDVVLSKIKDFSPQY RIELLLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTAN EIMNQQYEQMMIQNGTNQHVNSSNTDLNIHVNTNNIETKNDVNSMVIMSP VSPSDYITYPSQIATNISRNIPNVVNSMKQMASIYNDLHEQHDNEIKSLQ KTLKSISKTKIQVSLKTLEVLKESSKDENGEAQTNDDFEILSRLQEQNTK KLRKRLIRYKRLIKQKLEYRQTVLLNKLIEDETQATTNNTVEKDNNTLER LELAQELTMLQLQRKNKLSSLVKKFEDNAKIHKYRRIIREGTEMNIEEVD SSLDVILQTLIANNNKNKGAEQIITISNANSHA length: 833 Kila-N domain: 21-93 Ankyrin domains: 369-455, 505-549
...
... and save it all in one large text file and whenever you need to look something up, you just open the file, look for e.g. the name of the protein and read what's there. Or - for a more structured approach, you could put this into several files in a folder.[1] This is a perfectly valid approach and for some applications it might not be worth the effort to think more deeply about how to structure the data, and store it in a way that it is robust and scales easily to large datasets. Alas, small projects have a tendency to grow into large projects and if you work in this way, it's almost guaranteed that you will end up doing many things by hand that could easily be automated. Imagine asking questions like:
- How many proteins do I have?
- What's the sequence of the Kila-N domain?
- What percentage of my proteins have an Ankyrin domain?
- Or two ...?
Answering these questions "by hand" is possible, but tedious.
Spreadsheets
Many serious researchers keep their project data in spreadsheets. Often they use Excel, or an alternative like the free OpenOffice Calc, or Google Sheets, both of which are compatible with Excel and have some interesting advantages. Here, all your data is in one place, easy to edit. You can even do simple calculations - although you should never use Excel for statistics[2]. You could answer What percentage of my proteins have an Ankyrin domain? quite easily[3].
There are two major downsides to spreadsheets. For one, complex queries need programming. There is no way around this. You can program inside Excel with Visual Basic. But you might as well export your data so you can work on it with a "real" programming language. The other thing is that Excel does not scale very well. Once you have more than a hundred proteins in your spreadsheet, you can see how finding anything can become tedious.
However, just because Excel was built for business applications, and designed for use by office assistants, does not mean it is intrinsically unsuitable for our domain. It's important to be pragmatic, not dogmatic, when choosing tools: choose according to your real requirements. Sometimes "quick and dirty" is just fine, because quick.
R
R can keep complex data in data frames and lists. If we do data analysis with R, we have to load the data first. We can use any of the read.table() functions for structured data, read lines of raw text with readLines(), or slurp in entire files with scan(). Convenient packages exist to parse structured data like XML or JSON and import it. But we could also keep the data in an R object in the first place that we can read from disk, analyze, modify, and write back. In this case, R becomes our database engine.
# Sample construction of an R database table as a dataframe
# Data for the Mbp1 protein
proteins <- data.frame(
name = "Mbp1",
refSeq = "NP_010227",
uniProt = "P39678",
species = "Saccharomyces cerevisiae",
taxId = "4392",
sequence = paste(
"MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKR",
"TRILEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF",
"DFTQTDGSASPPPAPKHHHASKVDRKKAIRSASTSAIMETKRNNKKAEEN",
"QFQSSKILGNPTAAPRKRGRPVGSTRGSRRKLGVNLQRSQSDMGFPRPAI",
"PNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQPQQNNSAQFKEI",
"DLEDGLSSDVEPSQQLQQVFNQNTGFVPQQQSSLIQTQQTESMATSVSSS",
"PSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDINDKV",
"NKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPYIDAPIDPELHTAFH",
"WACSMGNLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRI",
"FQLLHETVFDIDSQSQTVIHHIVKRKSTTPSAVYYLDVVLSKIKDFSPQY",
"RIELLLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTAN",
"EIMNQQYEQMMIQNGTNQHVNSSNTDLNIHVNTNNIETKNDVNSMVIMSP",
"VSPSDYITYPSQIATNISRNIPNVVNSMKQMASIYNDLHEQHDNEIKSLQ",
"KTLKSISKTKIQVSLKTLEVLKESSKDENGEAQTNDDFEILSRLQEQNTK",
"KLRKRLIRYKRLIKQKLEYRQTVLLNKLIEDETQATTNNTVEKDNNTLER",
"LELAQELTMLQLQRKNKLSSLVKKFEDNAKIHKYRRIIREGTEMNIEEVD",
"SSLDVILQTLIANNNKNKGAEQIITISNANSHA",
sep=""),
seqLen = 833,
KilAN = "21-93",
Ankyrin = "369-455, 505-549")
# add data for the Swi4 protein
proteins <- rbind(proteins,
data.frame(
name = "Swi4",
refSeq = "NP_011036",
uniProt = "P25302",
species = "Saccharomyces cerevisiae",
taxId = "4392",
sequence = paste(
"MPFDVLISNQKDNTNHQNITPISKSVLLAPHSNHPVIEIATYSETDVYEC",
"YIRGFETKIVMRRTKDDWINITQVFKIAQFSKTKRTKILEKESNDMQHEK",
"VQGGYGRFQGTWIPLDSAKFLVNKYEIIDPVVNSILTFQFDPNNPPPKRS",
"KNSILRKTSPGTKITSPSSYNKTPRKKNSSSSTSATTTAANKKGKKNASI",
"NQPNPSPLQNLVFQTPQQFQVNSSMNIMNNNDNHTTMNFNNDTRHNLINN",
"ISNNSNQSTIIQQQKSIHENSFNNNYSATQKPLQFFPIPTNLQNKNVALN",
"NPNNNDSNSYSHNIDNVINSSNNNNNGNNNNLIIVPDGPMQSQQQQQHHH",
"EYLTNNFNHSMMDSITNGNSKKRRKKLNQSNEQQFYNQQEKIQRHFKLMK",
"QPLLWQSFQNPNDHHNEYCDSNGSNNNNNTVASNGSSIEVFSSNENDNSM",
"NMSSRSMTPFSAGNTSSQNKLENKMTDQEYKQTILTILSSERSSDVDQAL",
"LATLYPAPKNFNINFEIDDQGHTPLHWATAMANIPLIKMLITLNANALQC",
"NKLGFNCITKSIFYNNCYKENAFDEIISILKICLITPDVNGRLPFHYLIE",
"LSVNKSKNPMIIKSYMDSIILSLGQQDYNLLKICLNYQDNIGNTPLHLSA",
"LNLNFEVYNRLVYLGASTDILNLDNESPASIMNKFNTPAGGSNSRNNNTK",
"ADRKLARNLPQKNYYQQQQQQQQPQNNVKIPKIIKTQHPDKEDSTADVNI",
"AKTDSEVNESQYLHSNQPNSTNMNTIMEDLSNINSFVTSSVIKDIKSTPS",
"KILENSPILYRRRSQSISDEKEKAKDNENQVEKKKDPLNSVKTAMPSLES",
"PSSLLPIQMSPLGKYSKPLSQQINKLNTKVSSLQRIMGEEIKNLDNEVVE",
"TESSISNNKKRLITIAHQIEDAFDSVSNKTPINSISDLQSRIKETSSKLN",
"SEKQNFIQSLEKSQALKLATIVQDEESKVDMNTNSSSHPEKQEDEEPIPK",
"STSETSSPKNTKADAKFSNTVQESYDVNETLRLATELTILQFKRRMTTLK",
"ISEAKSKINSSVKLDKYRNLIGITIENIDSKLDDIEKDLRANA",
sep=""),
seqLen = 1093,
KilAN = "56-122",
Ankyrin = "516-662")
)
# how many proteins?
nrow(proteins)
#what are their names?
proteins[,"name"]
# how many do not have an Ankyrin domain?
sum(proteins[,"Ankyrin"] == "")
# save it to file
saveRDS(proteins, file="proteinData.rds")
# delete it from memory
rm(proteins)
# check...
proteins # ... yes, it's gone
# read it back in:
proteins <- readRDS("proteinData.rds")
# did this work?
sum(proteins[ , "seqLen"]) # 1926 amino acids
# [END]
The third way to use R for data is to connect it to a "real" database:
- a relational database like mySQL, MariaDB, or PostgreSQL;
- an object/document database like {{WP|MongoDB};
- or even a graph-database like Neo4j.
R "drivers" are available for all of these. However all of these require installing extra software on your computer: the actual database, which runs as an independent application. If you need a rock-solid database with guaranteed integrity, multi-user support, ACID transactional gurantees, industry standard performance, and scalability to even very large datasets, don't think of rolling your own solution. One of the above is the way to go.
MySQL and friends
MySQL is a free, open relational database that powers some of the largest corporations as well as some of the smallest laboratories. It is based on a client-server model. The database engine runs as a daemon in the background and waits for connection attempts. When a connection is established, the server process establishes a communication session with the client. The client sends requests, and the server responds. One can do this interactively, by running the client program /usr/local/mysql/bin/mysql (on Unix systems). Or, when you are using a program such as R, Python, Perl, etc. you use the appropriate method calls or functions—the driver—to establish the connection.
These types of databases use their own language to describe actions: SQL - which handles data definition, data manipulation, and data control.
Just for illustration, the Figure above shows a table for our APSES domain protein data, built as a table in the MySQL workbench application and presented as an Entity Relationship Diagram (ERD). There is only one entity though - the protein "table". The application can generate the actual code that implements this model on a SQL compliant database:
CREATE TABLE IF NOT EXISTS `mydb`.`proteins` ( `name` VARCHAR(20) NULL, `refSeq` VARCHAR(20) NOT NULL, `uniProt` VARCHAR(20) NULL, `species` VARCHAR(45) NOT NULL COMMENT ' ', `taxId` VARCHAR(10) NULL, `sequence` BLOB NULL, `seqLen` INT NULL, `KilA-N` VARCHAR(45) NULL, `Ankyrin` VARCHAR(45) NULL, PRIMARY KEY (`refSeq`)) ENGINE = InnoDB
This looks at least as complicated as putting the model into R in the first place. Why then would we do this, if we need to load it into R for analysis anyway? There are several important reasons.
- Scalability: these systems are built to work with very large datasets and optimized for performance. In theory R has very good performance with large data objects, but not so when the data becomes larger than what the computer can keep in memory all at once.
- Concurrency: when several users need to access the data potentially at the same time, you must use a "real" database system. Handling problems of concurrent access is what they are made for.
- ACID compliance. ACID describes four aspects that make a database robust, these are crucial for situations in which you have only partial control over your system or its input, and they would be quite laborious to implement for your hand built R data model:
- Atomicity: Atomicity requires that each transaction is handled "indivisibly": it either succeeds fully, with all requested elements, or not at all.
- Consistency: Consistency requires that any transaction will bring the database from one valid state to another. In particular any data-validation rules have to be enforced.
- Isolation: Isolation ensures that any concurrent execution of transactions results in the exact same database state as if transactions would have been executed serially, one after the other.
- Durability: The Durability requirement ensures that a committed transaction remains permanently committed, even in the event that the database crashes or later errors occur. You can think of this like an "autosave" function on every operation.
All the database systems I have mentioned above are ACID compliant[4].
Incidentally - RStudio has inbuilt database support via the Connections tab of the top-right pane. Read more about database connections directly from RStudio here.
Store Data
A protein datamodel
|
Entity-Relationship Diagram (ERD) for a protein data model that includes protein, taxonomy and feature annotations. Entity names are at the top of each box, attributes are listed below. If you think of an entity as an R dataframe, or a spreadsheet table, the attributes are the column names and each specific instance of an entity fills one row (or "record").All relationships link to unique primary keys and are thus 1 to (0, n). The diagram was drawn as a "Google slide" and you can view it on the Web and, if you have a Google account, you can make a copy to use for your own purposes. |
Implementing the Data Model in R
To actually implement the data model in R we will create tables as data frames, and we will collect them in a list. We don't have to keep the tables in a list - we could also keep them as independent objects, but a list is neater, more flexible (we might want to have several of these), it reflects our intent about the model better, and doesn't require very much more typing.
Task:
- Open RStudio and load the
ABC-unitsR project. If you have loaded it before, choose File → Recent projects → ABC-Units. If you have not loaded it before, follow the instructions in the RPR-Introduction unit. - Choose Tools → Version Control → Pull Branches to fetch the most recent version of the project from its GitHub repository with all changes and bug fixes included.
- Type
init()if requested. - Open the file
BIN-Storing_data.Rand follow the instructions.
Note: take care that you understand all of the code in the script. Evaluation in this course is cumulative and you may be asked to explain any part of code.
Further reading, links and resources
Notes
- ↑ Your operating system can help you keep the files organized. The "file system" is a database.
- ↑ For real: Excel is miserable and often wrong on statistics, and it makes horrible, ugly plots. See here and here why Excel problems are not merely cosmetic.
- ↑ At the bottom of the window there is a menu that says "sum = ..." by default. This provides simple calculations on the selected range. Set the choice to "count", select all Ankyrin domain entries, and the count shows you how many cells actually have a value.
- ↑ For a list of relational Database Management Systems, see here.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2020-10-07
Version:
- 1.2
Version history:
- 1.2 Edit policy update
- 1.1 Update to new marking scheme
- 1.0.1 Add link to data model types overview
- 1.0 First Live version
- 0.1 First stub
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.