Difference between revisions of "BIO Assignment Week 7"
m |
m |
||
| (33 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<div id="BIO"> | <div id="BIO"> | ||
<div class="b1"> | <div class="b1"> | ||
| − | Assignment for Week | + | Assignment for Week 7<br /> |
| − | <span style="font-size: 70%"> | + | <span style="font-size: 70%">Phylogenetic Analysis</span> |
</div> | </div> | ||
| + | <table style="width:100%;"><tr> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:left; font-size:80%;">[[BIO_Assignment_Week_6|< Assignment 6]]</td> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:right; font-size:80%;">[[BIO_Assignment_Week_8|Assignment 8 >]]</td> | ||
| + | </tr></table> | ||
{{Template:Inactive}} | {{Template:Inactive}} | ||
| − | Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz. | + | <!-- Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz. --> |
| Line 13: | Line 17: | ||
| − | + | {{vspace}} | |
| − | == | + | |
| + | |||
| + | |||
| + | <div class="quote-box"> | ||
| + | {{Vspace}} | ||
| + | |||
| + | ;Nothing in Biology makes sense except in the light of evolution. | ||
| + | :''Theodosius Dobzhansky'' | ||
| + | </div> | ||
| + | |||
| + | ... but does evolution make sense in the light of biology? | ||
| + | |||
| + | {{Vspace}} | ||
| + | ---- | ||
| + | {{Vspace}} | ||
| + | |||
| + | <div class="colmask doublepage"> | ||
| + | <div class="colleft"> | ||
| + | <div class="col1"> | ||
| + | <!-- Column 1 start --> | ||
| + | |||
| + | As we have seen in the previous assignments, the Mbp1 transcription factor has homologues in all other fungi, yet there is not always a clear one-to-one mapping between members of a family in distantly related species. It appears that various systems of APSES domain transcription factors have evolved independently. Of course this bears directly on our notion of function - what it means to say that two genes in different organisms have the "same" function. In case two organisms both have an orthologous gene for the same, distinct function, calling these functions "the same" may be warranted. But what if that gene has duplicated in one species, and the two paralogues now perform different, related functions in one organism? Theses two are still orthologues to both their homologues in the other species, but now we expect functionally significant residues to have adapted to the new - and possibly distinct - roles of each paralogue. In order to be able to even ask such questions, we need to make the evolutionary history of gene families explicit. This is the domain of '''phylogenetic analysis'''. We can ask questions like: how many paralogues did the cenancestor of a clade possess? Which of these underwent additional duplications in the phylogenesis of the organism I am studying? Did any genes get lost? And - adding additional biological insight to the picture - did the observed duplications lead to the "invention" of new biological systems? When was that? And perhaps even: how did the species benefit from this event? | ||
| + | |||
| + | We will develop this kind of analysis in this assignment. In the previous assignment you have established which gene in your species is the reciprocally most closely related orthologue to yeast Mbp1 (with ''reciprocal best match'') and you have identified the full complement of APSES domain genes in your assigned organism (as a result of your PSI-BLAST search). In this assignment, we will analyse these genes' evolutionary relationship and compare it to the evolutionary relationship of other fungal APSES domains. The goal is to define families of related transcription factors and their evolutionary history. All APSES domain annotations are now available in your protein "database". Now we will attempt to compute the phylogram for these proteins. The goal is to identify orthologues and paralogues. <!-- Optionally, you will look at structural and functional conservation of residues. Future: add ankyrin domains to APSES domains. --> | ||
| + | |||
| + | A number of excellent tools for phylogenetic analysis exist; ''general purpose packages'' include the (free) [http://evolution.genetics.washington.edu/phylip.html '''PHYLIP'''] package, the [http://www.megasoftware.net/ '''MEGA''' package] and the (commercial) PAUP* package. Of these, only MEGA is still under active development, although PHYLIP still functions perfectly (except for problems with graphical windows under Mac OS 10.6). ''Specialized tools'' for tree-building include Treepuzzle or Mr. Bayes. This assignment is constructed around programs that are available in PHYLIP, however you are welcome to use other tools that fulfill a similar purpose if you wish. In this field, researchers consider trees that have been built with ML (maximum likelihood) methods to be more reliable than trees that are built with parsimony methods, or distance methods such as NJ (Neighbor Joining). However ML methods are also much more compute-intensive. Just like with multiple sequence alignments, some algorithms will come closer to guessing the truth and others will not and usually it is hard to tell which is the more trustworthy of two diverging results. The prudent researcher tries out alternatives and forms her own opinion. Specifically, we may usually assume results that converge when computed with different algorithms, to be more reliable than those that depend strongly on a particular algorithm, parameters, or details of input data. | ||
| + | |||
| + | <!-- Column 1 end --> | ||
| + | </div> | ||
| + | <div class="col2"> | ||
| + | <!-- Column 2 start --> | ||
| + | In this assignment, we will take a computational shortcut, (something you should not do in real life). We will skip establishing the reliability of the tree with a bootstrap procedure, i.e. repeat the tree-building a hundred times with partial data and see which branches and groupings are robust and which depend on the details of the data. <small>(If you are interested, have a look [[BIO_bootstrapping_with_PHYLIP| '''here''']] for the procedure for running a bootstrap analysis on the data set you are working with, but this may require a day or so of computing time on your computer.)</small> In this assignment, we will simply acknowledge that bifurcations that are very close to each other have not been "resolved" and be appropriately cautious in our inferences. In phylogenetic analysis, not all lines a program draws are equally trustworthy. Don't take the trees as a given fact just because a program suggests this. Look at the evidence, include independent information where available, use your reasoning, and analyse the results critically. As you will see, there are some facts that we know for certain: we know which species the genes come from, and we can (usually) make good assumptions about the relationship of the species themselves - the history of speciation events that underlies all evolution of genes. This is extremely helpful information for our work. | ||
| + | |||
| + | |||
| + | If you would like to review concepts of trees, clades, LCAs, OTUs and the like, I have linked an excellent and very understandable introduction-level article on phylogenetic analysis here and to the resource section at the bottom of this page. | ||
| + | |||
| + | {{#pmid: 12801728}} | ||
| + | |||
| + | {{vspace}} | ||
| + | '''R''' packages that may be useful include the following: | ||
| + | * [https://cran.r-project.org/web/views/Phylogenetics.html '''R''' task view Phylogenetics] - this task-view gives an excellent, curated overview of the important '''R'''-packages in the domain. | ||
| + | * [https://cran.r-project.org/web/packages/ape/index.html package '''ape'''] - general purpose phylogenetic analysis, but (as far as I can tell ape only supports analysis with DNA sequences). | ||
| + | * [https://cran.r-project.org/web/packages/ips/index.html package '''ips'''] - wrapper for MrBayes, Beast, RAxML "heavy-duty" phylogenetic analysis packages. | ||
| + | * [https://cran.r-project.org/web/packages/Rphylip/index.html package '''Rphylip'''] - Wrapper for Phylip, the most versatile set of phylogenetic inference tools. | ||
| − | < | + | <!-- Column 2 end --> |
| − | + | </div> | |
| − | + | </div> | |
</div> | </div> | ||
| − | |||
| − | |||
| − | + | {{vspace}} | |
| + | |||
| + | ==Preparing input alignments== | ||
| + | {{vspace}} | ||
| + | You have previously collected homologous sequences and their annotations. We will use these as input for phylogenetic analysis. But let's discuss first how such an input file should be constructed. | ||
| + | |||
| + | {{vspace}} | ||
| + | |||
| + | ===Principles=== | ||
| + | <div class="colmask doublepage"> | ||
| + | <div class="colleft"> | ||
| + | <div class="col1"> | ||
| + | <!-- Column 1 start --> | ||
| + | In order to use molecular sequences for the construction of phylogenetic trees, you have to build a multiple alignment first. This is important: phylogenetic analysis does not build alignments, nor does it revise alignments, it analyses them '''after''' the alignment has been computed. A precondition for the analysis to be meaningful is that all rows of sequences have to contain the exact same number of characters and to hold '''aligned characters in corresponding positions (i.e. columns)'''. The program's inferences are made on a column-wise basis and if your columns contain data from unrelated positions, the inferences are going to be questionable. Clearly, in order for tree-estimation to work, one must not include fragments of sequence which have evolved under a different evolutionary model as all others, e.g. after domain fusion, or after accommodating large stretches of indels. Thus it is appropriate to edit the sequences and pare them down to a most characteristic subset of amino acids. The goal is not to be as comprehensive as possible, but to input those columns of aligned residues that will best represent the true phylogenetic relationships between the sequences. | ||
| + | |||
| + | |||
| + | The result of the tree construction is a decision about the most likely evolutionary relationships. Fundamentally, tree-construction programs decide which sequences had common ancestors. | ||
| + | |||
| + | |||
| + | '''Distance based''' phylogeny programs start by using sequence comparisons to estimate evolutionary distances: | ||
| + | |||
| + | * they apply a model of evolution such as a mutation data matrix, to calculate a score for each pair of sequences, | ||
| + | * this score is stored in a "distance matrix" ... | ||
| + | * ... and used to estimate a tree that groups sequences with close relationships together. (e.g. by using an NJ, Neigbor Joining, algorithm). | ||
| + | |||
| + | They are fast, can work on large numbers of sequences, but are less accurate if genes evolve at different rates. | ||
| + | |||
| + | <!-- Column 1 end --> | ||
| + | </div> | ||
| + | <div class="col2"> | ||
| + | <!-- Column 2 start --> | ||
| + | |||
| + | '''Parsimony based''' phylogeny programs build a tree that minimizes the number of mutation events that are required to get from a common ancestral sequence to all observed sequences. They take all columns into account, not just a single number per sequence pair, as the Distance Methods do. For closely related sequences they work very well, but they construct inaccurate trees when they can't make good estimates for the required number of sequence changes. | ||
| + | |||
| + | |||
| + | '''ML''', or '''Maximum Likelihood''' methods attempt to find the tree for which the observed sequences would be the most likely under a particular evolutionary model. They are based on a rigorous statistical framework and yield the most robust results. But they are also quite compute intensive and a tree of the size that we are building in this assignment is a challenge for the resources of common workstation (runs about an hour on my computer). If the problem is too large, one may split a large problem into smaller, obvious subtrees (e.g. analysing orthologues as a group, only including a few paralogues for comparison) and then merge the smaller trees; this way even very large problems can become tractable. | ||
| − | + | ML methods suffer less from "long-branch attraction" - the phenomenon that weakly similar sequences can be grouped inappropriately close together in a tree due to spuriously shared differences. | |
| − | |||
| − | + | '''Bayesian''' methods don't estimate the tree that gives the highest likelihood for the observed data, but find the most probably tree, given that the data have been observed. If this sounds conceptually similar to you, then you are not wrong. However, the approaches employ very different algorithms. And Bayesian methods need a "prior" on trees before observation. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <!-- Column 2 end --> | |
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | {{vspace}} | ||
| − | + | ===Choosing sequences=== | |
| − | == | + | {{vspace}} |
| − | + | To illustrate the principle we will construct input files by joining APSES domain and Ankyrin domain sequences and for this we will use the Prosite annotations we have collected for the reference set of sequences and your YFO sequences. | |
{{task|1= | {{task|1= | ||
| − | + | ||
| − | + | * Open RStudio. | |
| − | + | * Choose File → Recent Projects → BCH441_2016. | |
| − | + | * Pull the latest version of the project repository from GitHub. | |
| − | + | * type <tt>init()</tt> | |
| − | + | * Open the file <tt>BCH441_A07.R</tt> and work through PART ONE: Choosing sequences. | |
| − | + | ||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| − | + | {{vspace}} | |
| + | |||
| + | ===Adding an Outgroup=== | ||
| + | {{vspace}} | ||
| + | |||
| + | An outgroup is a sequence that is more distantly related to all of the other sequences than any of them are to each other. This allows us to root the tree, because the root - the last common ancestor to all - must be somewhere on the branch that connects the outgroup to the rest. And whenever a molecular clock is assumed, the branching point that connects the outgroup can be assumed to be the oldest divergence event. Having a root that we can compare to the phylogram of species makes the tree interpretation '''much''' more intuitive. In our case, we are facing the problem that our species cover all of the known fungi, thus we can' rightly say that any of them are more distant to the rest. We have to look outside the fungi. The problem is, outside of the fungi there are no proteins with APSES domains<!--, and certainly none that have APSES as well as ankyrin domains in the same gene-->. We can take the ''E. coli'' KilA-N domain sequence - a known, distant homologue to the APSES domain instead, even though it only aligns to a part of the APSES domains<!-- , and we can get an ankyrin region from e.g. a plant. Both outgroup domains then will have the property that they are more distant individually to any of the fungal sequences, even though they don't appear in the same protein -->. | ||
| + | |||
| + | Here is the KilA-N domain sequence in the E. coli Kil-A protein: | ||
| + | |||
| + | >WP_000200358.1 hypothetical protein [Escherichia coli] | ||
| + | <span style="color: #999999;">MTSFQLSLISRE</span>IDGEIIHLRAKDGYINATSMCRTAGKLLSDYTRLKTTQEFFDELSRDMGIPISELIQS | ||
| + | FKGGRPENQGTWVHPDIAINLAQ<span style="color: #999999;">WLSPKFAVQVSRWVREWMSGERTTAEMPVHLKRYMVNRSRIPHTHFS | ||
| + | ILNELTFNLVAPLEQAGYTLPEKMVPDISQGRVFSQWLRDNRNVEPKTFPTYDHEYPDGRVYPARLYPNE | ||
| + | YLADFKEHFNNIWLPQYAPKYFADRDKKALALIEKIMLPNLDGNEQF</span> | ||
| + | |||
| + | ''E. coli'' KilA-N protein. Residues that do not align with APSES domains are shown in grey. | ||
| + | |||
| + | The assignment '''R''' - code contains code to add it to the group of APSES sequences. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- | ||
| + | And here is an ankyrin repeat region, found by BLAST search in ''Solanum tuberosum'', the potato, and confirmed with ScanProsite. Since the potato is more distant in evolution from any fungus than all fungi are to each other, this sequence is suitable to root our ankyrin domain tree. | ||
| − | + | >NP_001275294 ankyrin repeat containing protein [Solanum tuberosum] | |
| + | <span style="color: #999999;">MAPDATDALAVREKVNKFLKAACSGDIELFKKLAKQLDDGKGLAGTVADVKDGNKRGALIFAARESKIEL | ||
| + | CKYLVEELKVDVNEKDDEGETPLLHAAREGHTATVQYLIEQGADPAIP</span>SASGATALHHAAGNGHVELVKL | ||
| + | LLSKGVDVDLQSEAGTPLMWAAGFGQEKVVKVLLEHHANVHAQTKDENNVCPLVSAVATDSLPCVELLAK | ||
| + | AGADVNVRTGDATPLLIAAHNGSAGVINCLLQAGADPNAAEEDGTKPIQVAAASGSREAVEALLPVTERI | ||
| + | QSV<span style="color: #999999;">PEWSVDGVIEFVQSEYKREQERAEAGRKANKSREPIIPKRDLPEVSPEAKKRAADAKARGDEAFKRN | ||
| + | DFATAIDAYTQAIDFDPTDGTLFSNRSLCWLRLGQAERALSDARACRELRPDWAKGCYREGAALRLLQRF | ||
| + | EEAANAFYEGVQINPINMELVTAFREAVEAGRKVHATNKFNSPSSLS</span> | ||
| + | ''S. tuberosum'' "ankyrin repeat and KH domain-containing protein 1-like" protein. Ankyrin repeat region shown in black. | ||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | < | + | <source lang="R"> |
| − | |||
| + | # Let's add our outgroups to the feature sequence tables: | ||
| + | |||
| + | # APSES domain feature from E. coli | ||
| + | apsOutGroupSeq <- paste( | ||
| + | "IDGEIIHLRAKDGYINATSMCRTAGKLLSDYTRLKTTQEFFDELSRDMGI", | ||
| + | "PISELIQSFKGGRPENQGTWVHPDIAINLAQ", | ||
| + | sep = "") | ||
| + | apsOutGroupHead <- ">apses domain from E. coli KilA-N" | ||
| + | apsOutGroupName <- "APS_OUTGRP" | ||
| + | |||
| + | # ankyrin region feature from S. tuberosum | ||
| + | ankOutGroupSeq <- paste( | ||
| + | "PEWSVDGVIEFVQSEYKREQERAEAGRKANKSREPIIPKRDLPEVSPEAK", | ||
| + | "KRAADAKARGDEAFKRNDFATAIDAYTQAIDFDPTDGTLFSNRSLCWLRL", | ||
| + | "GQAERALSDARACRELRPDWAKGCYREGAALRLLQRFEEAANAFYEGVQI", | ||
| + | "NPINMELVTAFREAVEAGRKVHATNKFNSPSSLS", | ||
| + | sep = "") | ||
| + | ankOutGroupHead <- ">ankyrin repeat region from S. tuberosum" | ||
| + | ankOutGroupName <- "ANK_OUTGRP" | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | # add the synthetic proteins to the feature compilations | |
| + | APSES <- rbind(APSES, data.frame(names = apsOutGroupName, | ||
| + | head = apsOutGroupHead, | ||
| + | seq = apsOutGroupSeq, | ||
| + | stringsAsFactors = FALSE)) | ||
| − | + | ANKYRIN <- rbind(ANKYRIN, data.frame(names = ankOutGroupName, | |
| + | head = ankOutGroupHead, | ||
| + | seq = ankOutGroupSeq, | ||
| + | stringsAsFactors = FALSE)) | ||
| − | + | # Remove hyphens, concatenate APSES and ANK_REP_REGION | |
| − | + | # sequences and use names for rownames. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | apsSeq <- character() | |
| + | ankSeq <- character() | ||
| + | for (i in 1:nrow(APSES)) { | ||
| + | aps <- gsub("-", "", APSES$seq[i]) | ||
| + | ank <- gsub("-", "", ANKYRIN$seq[i]) | ||
| + | if (nchar(aps) > 0) { | ||
| + | apsSeq <- c(apsSeq, aps) | ||
| + | names(apsSeq)[length(apsSeq)] <- APSES$names[i] | ||
| + | } | ||
| + | if (nchar(ank) > 0) { | ||
| + | ankSeq <- c(ankSeq, ank) | ||
| + | names(ankSeq)[length(ankSeq)] <- ANKYRIN$names[i] | ||
| + | } | ||
| + | } | ||
| + | head(apsSeq) | ||
| + | head(ankSeq) | ||
| − | + | --> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | {{task|1= | ||
| − | + | *Continue with the R-code: PART TWO: Multiple sequence alignment | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | }} | ||
| − | + | {{Vspace}} | |
| − | + | ===Reviewing and Editing alignments=== | |
| − | + | {{vspace}} | |
| − | |||
| − | + | <div class="colmask doublepage"> | |
| + | <div class="colleft"> | ||
| + | <div class="col1"> | ||
| + | <!-- Column 1 start --> | ||
| + | As discussed in the lecture, it is usually necessary to edit a multiple sequence alignment to make it suitable for phylogenetic inference. Here are the principles: | ||
| + | <div class="emphasis-box"> | ||
| + | '''All characters in a column should be related by homology.''' | ||
</div> | </div> | ||
| + | |||
| + | This implies the following rules of thumb: | ||
| + | |||
| + | *Remove all stretches of residues in which the ''alignment'' appears ambiguous (not just highly variable, but ambiguous regarding the aligned positions). | ||
| + | *Remove all frayed N- and C- termini, especially regions in which not all sequences that are being compared appear homologous and that may stem from unrelated domains. You want to only retain the APSES domains. All the extra residues from the YFO sequence can be deleted. | ||
| + | *Remove all gapped regions that appear to be alignment artefacts due to inappropriate input sequences. | ||
| + | *Remove all but approximately one column from gapped regions '''in those cases where the presence of several related insertions suggest that the indel is real, and not just an alignment artefact.''' (Some researchers simply remove all gapped regions). | ||
| + | *Remove sections N- and C- terminal of gaps where the alignment appears questionable. | ||
| + | *If the sequences fit on a single line you will save yourself potential trouble with block-wise vs. interleaved input. If you do run out of memory try removing columns of sequence. Or remove species that you are less interested in from the alignment. | ||
| + | *Move your outgroup sequence to the first line of your alignment, since this is where PHYLIP will look for it by default. | ||
| + | |||
| + | |||
| + | <!-- Column 1 end --> | ||
| + | </div> | ||
| + | <div class="col2"> | ||
| + | <!-- Column 2 start --> | ||
| + | Indels are even more of a problem than usual. Strictly speaking, the similarity score of an '''alignment''' program as well as the distance score of a '''phylogeny''' program are not calculated for an ordered sequence, but for a sum of independent values, one for each aligned columns of characters. The order of the columns does not change the score. However in an optimal sequence alignment with gaps, this is no longer strictly true since a one-character gap creation has a different penalty score than a one-character gap extension! Most '''alignment''' programs use a model with a constant gap insertion penalty and a linear gap extension penalty. This is not rigorously justified from biology, but parametrized (or you could say "tweaked") to correspond to our observations. However, most '''phylogeny''' programs do not work in this way. They strictly operate on columns of characters and treat a gap character just like a residue with the one letter code "-". Thus gap insertion- and extension- characters get the same score. For short indels, this '''underestimates''' the distance between pairs of sequences, since any evolutionary model should reflect the fact that gaps are much less likely than point mutations. If the gap is very long though, all events are counted individually as many single substitutions (rather than one lengthy one) and this '''overestimates''' the distance. And it gets worse: long stretches of gaps can make sequences appear similar in a way that is not justified, just because they are identical in the "-" character. It is therefore common and acceptable to edit gaps in the alignment and delete all but a few columns of gapped sequence, or to remove such columns altogether. | ||
| + | |||
| + | |||
| + | <!-- Column 2 end --> | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| + | {{Vspace}} | ||
| + | ---- | ||
| + | {{Vspace}} | ||
| + | |||
| + | |||
| + | [[Image:EditingGuide.jpg|frame|none|(Possible) steps in editing a multiple sequence alignment towards a PHYLIP input file. '''a''': raw alignment (CLUSTAL format); '''b''': sequences assembled into single lines; '''c''': columns to be deleted highlighted in red - 1, 3 and 4: large gaps; 2: uncertain alignment and 5: frayed C-terminus: both would put non-homologous characters into the same column; '''d''': input data for PHYLIP: names for sequences must not be longer than 10 characters, the first line must contain the number of sequences and the sequence length. PHYLIP is very picky about incorrectly formatted input, read the [http://evolution.genetics.washington.edu/phylip/doc/sequence.html PHYLIP sequence format guide]. Fortunately Rphylip does the formatting step for you.]] | ||
| + | |||
| + | |||
| + | There is more to learn about this important step of working with aligned sequences, here is an overview of the literature on various algorithms and tools that are available. <!-- Read at least the abstracts. --> | ||
| + | |||
| + | {{#pmid: 17654362}} | ||
| + | {{#pmid: 19505945}} | ||
| + | {{#pmid: 19770262}} | ||
| + | {{#pmid: 20497997}} | ||
| + | {{#pmid: 23193120}} | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ====Sequence masking with R==== | ||
| + | {{Vspace}} | ||
| + | |||
| + | As you saw while inspecting the multiple sequence alignment, there are regions that are poorly suited for phylogenetic analysis | ||
| + | due to the large numbers of gaps. | ||
| + | |||
| + | A good approach to edit the alignment is to import your sequences | ||
| + | into Jalview and remove uncertain columns by hand. | ||
| + | |||
| + | But for this assignment, let's write code for a simple masking heuristic. | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | {{task|1= | ||
| + | |||
| + | * Head back to the '''RStudio project''' and work through <tt>PART THREE: reviewing and editing alignments</tt> | ||
| + | |||
| + | }} | ||
| + | |||
| + | |||
| + | {{Vspace}} | ||
| − | + | ==Calculating trees== | |
| + | {{vspace}} | ||
| − | + | In this section we perform the actual phylogenetic calculation. | |
| + | {{vspace}} | ||
| + | {{task|1= | ||
| − | + | * Download the PHYLIP suite of programs from the [http://evolution.genetics.washington.edu/phylip.html Phylip homepage] and install it on your computer. | |
| − | + | * Return to the '''RStudio project''' and work through <tt>PART FOUR: Calculating trees</tt>. | |
| − | + | }} | |
| − | + | {{Vspace}} | |
| + | <!-- Bootstrapping ... | ||
| + | * run seqboot | ||
| + | * rename outfile to infile | ||
| + | * rerun proml, use option M for multiple datasets with speedy option (use "jumble" of 1) | ||
| + | * rename outtree to intree | ||
| + | * run consense | ||
| + | * Use option R to define trees as rooted | ||
| − | + | Should run at least overnight. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
--> | --> | ||
| + | ==Analysing your tree== | ||
| + | {{vspace}} | ||
| + | In order to analyse your tree, you need a species tree as reference. This really is an absolute prerequisite to make your expectations about the observed tree explicit. Fortunately we have all species nicely documented in our database. | ||
| + | {{vspace}} | ||
| + | |||
| + | ===The reference species tree=== | ||
| + | {{vspace}} | ||
{{task|1= | {{task|1= | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * Navigate to the [http://www.ncbi.nlm.nih.gov/taxonomy '''NCBI Taxonomy page'''] | |
| + | |||
| + | * Execute the following '''R''' command to create an Entrez command that will retrieve all taxonomy records for the species in your database: | ||
| + | <source lang="R"> | ||
| + | cat(paste(paste(c(myDB$taxonomy$ID, "83333"), "[taxid]", sep=""), collapse=" OR ")) | ||
| + | </source> | ||
| + | |||
| + | * Copy the Entrez command, and enter it into the search field of the NCBI taxonomy page. Click on '''Search'''. The resulting page should have twelve species listed - ten "reference" fungi, ''E. coli'' (as the outgroup), and YFO. Make sure YFO is included! If it's not there, you did something wrong that needs to be fixed. | ||
| + | |||
| + | * Click on the '''Summary''' options near the top-left of the page, and select '''Common Tree'''. This places all the species into the universal tree of life and identifies their relationships. | ||
| + | |||
| + | * At the top, there is an option to '''Save as''' ... and the option to select a format to save the tree in. Select '''Phylip Tree''' as the format and click the '''Save as''' button. The file <code>phyliptree.phy</code> will be downloaded to your computer into your default download directory. Move it to the directory you have defined as <code>PROJECTDIR</code>. | ||
| + | |||
| + | *Open the file in a text-editor. This is a tree, specified in the so-called {{WP|Newick_format|'''"Newick format"'''}}. The topology of the tree is defined through the brackets, and the branch-lengths are all the same: this is a cladogram, not a phylogram. The tree contains the long names for the species/strains and for our purposes we really need the "biCodes" instead. I can't think of a very elegant way to make that change programmatically, so just go ahead and replace the species names (not the taxonomic ranks though) with their biCode in your text editor. Remove all the single quotes, and replace any remaining blanks in names with an underscore. Take care however not to delete any colons or parentheses. Save the file. | ||
| + | |||
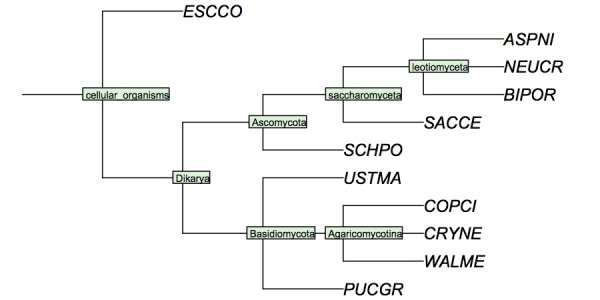
| + | My version looks like this - '''Your version must have YFO somewhere in the tree.'''. | ||
| + | |||
| + | ( | ||
| + | 'ESCCO':4, | ||
| + | ( | ||
| + | ( | ||
| + | 'PUCGR':4, | ||
| + | 'USTMA':4, | ||
| + | ( | ||
| + | 'WALME':4, | ||
| + | 'COPCI':4, | ||
| + | 'CRYNE':4 | ||
| + | )Agaricomycotina:4 | ||
| + | )Basidiomycota:4, | ||
| + | ( | ||
| + | ( | ||
| + | ( | ||
| + | 'ASPNI':4, | ||
| + | 'BIPOR':4, | ||
| + | 'NEUCR':4 | ||
| + | )leotiomyceta:4, | ||
| + | 'SACCE':4 | ||
| + | )saccharomyceta:4, | ||
| + | 'SCHPO':4 | ||
| + | )Ascomycota:4 | ||
| + | )Dikarya:4 | ||
| + | )'cellular organisms':4; | ||
| − | + | *Now read the tree in '''R''' and plot it. | |
| − | |||
| − | |||
| − | |||
| − | + | <source lang="R"> | |
| − | # | + | # Download the EDITED phyliptree.phy |
| − | + | orgTree <- read.tree("phyliptree.phy") | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | # Plot the tree in a new window | |
| + | dev.new(width=6, height=3) | ||
| + | plot(orgTree, cex=1.0, root.edge=TRUE, no.margin=TRUE) | ||
| + | nodelabels(text=orgTree$node.label, cex=0.6, adj=0.2, bg="#D4F2DA") | ||
| − | + | </source> | |
}} | }} | ||
| + | {{vspace}} | ||
| + | |||
| + | I have constructed a cladogram for many of the species we are analysing, based on data published for 1551 fungal ribosomal sequences. The six reference species are included. Such reference trees from rRNA data are a standard method of phylogenetic analysis, supported by the assumption that rRNA sequences are monophyletic and have evolved under comparable selective pressure in all species. | ||
| + | |||
| + | {{vspace}} | ||
| + | <div class="reference-box"> | ||
| + | [[Image:FungiCladogram.jpg|600px|none]] | ||
| − | |||
| + | <small>'''Cladogram of the "reference" fungi''' studied in the assignments. This cladogram is based on a tree returned by the NCBI Common Tree. It is thus a digest of cladistic relationships, not a representation of a specific molecular phylogeny.</small> | ||
| + | </div> | ||
| + | Alternatively, you can look up your species in the latest version of the species tree for the fungi and add it to the tree by hand while resolving the trifurcations. See: | ||
| + | {{#pmid: 22114356}} | ||
| − | + | {{vspace}} | |
| − | + | {{task|1= | |
| − | + | * Return to the RStudio project and continue with the script to its end. Note the deliverable at the end: to print out your trees and bring them to class. | |
| − | + | }} | |
<!-- | <!-- | ||
| − | |||
| − | |||
| + | #Copy the tree-string from the R console. | ||
| + | #Visualize the tree online: navigate to the [http://www.trex.uqam.ca/index.php?action=newick&project=trex Trex-online Newick tree viewer]. Visualize the tree as a phylogram. Explore the options. | ||
| − | + | # A particularly useful viewer is actually Jalview - although this may be more apparent with the larger alignment of '''all''' sequences we'll produce later. | |
| + | ##Open Jalview and load your alignment of all APSES domain proteins. | ||
| + | ##Save the Newick-formatted tree. | ||
| + | ##In the alignment window, choose '''File → Load associated Tree''' and load your tree file. You can click into the tree-window to show which clades branch off at what level - it should be obvious that you can identify three major subclades (plus the outgroup). This view is particularly informative, since you can associate the clades of the tree with the actual sequences in the alignment, and get a good sense what sequence features the tree is based on. | ||
| + | ##Try the '''Calculate → Sort → By Tree Order''' option to sort the sequences by their position in the tree. Also note that you can flip the tree around a node by double-clicking on it. This is especially useful: try to rearrange the tree so that the subdivisions into clades are apparent. Clicking into the window "cuts" the tree and colours your sequences according to the clades in which they are found. This is useful to understand what particular sequences contributed to which part of the phylogenetic inference. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ANALYSIS | |
| − | + | * First, the APS and ANK trees should have the same topology, since they are only different parts of the same protein (unless that protein has swapped its domains with another one during evolution). Clearly, that is not the case. The ''basidiomycota'' are reasonably consistent, although their internal ordering is poorly resolved, particularly in the APS tree. The ''ascomycota'' show two major differences, but they are actually consistent between the APS and the ANK tree: SACCE is less similar to all than we would expect from the species tree. And NEUCR is more similar to the ''basidiomycotal'' proteins. | |
| − | : | + | |
| − | + | * Consider the scale bars: ANK domains have evolved at about twice the rate of the APS domains. This alone should tell us to be cautious with our interpretations since this shows there are different degrees of selective pressure on different parts of the protein. Moreover the <u>relative rates</u> differ as well. NEUCR's APSES domain has evolved much faster by comparison to other proteins than its ankyrin domain. Has its biological function changed? | |
| − | |||
| − | + | * Secondly, both gene trees should follow the species tree. Again, there are differences. But if we exclude SACCE and NEUCR, the remainder actually turns out relatively consistent. | |
| + | In any case: this is what the data tells us. The big picture is mostly conserved, but there are differences in the details. However: now we know what degree of accuracy we can expect from the analysis. | ||
| − | + | {{vspace}} | |
| + | ==The mixed gene tree== | ||
| + | {{vspace}} | ||
| − | + | You have now practiced how to calculate, manipulate, plot, annotate and compare trees. | |
| + | {{task|1= | ||
| − | + | * Now use Rproml to calculate a mixed gene tree based on '''all'' APSES domains. You saved it as <code>APSES.mfa</code>. For the fifty or so domains, each run will take about an hour. Thus run as many <code>random.addition</code> cycles as reasonable during a study break, or overnight. Thus the command will be something like: | |
| − | |||
| − | + | <source lang="R"> | |
| + | allApsIn <- read.protein("APSES.mfa") | ||
| + | fullApsTree <- Rproml(allApsIn, path=PROMLPATH, random.addition=3) | ||
| − | + | #... and don't forget: | |
| + | save(fullApsTree, file="fullApsTree.rda") | ||
| + | </source> | ||
| + | }} | ||
| − | + | {{vspace}} | |
| − | {{ | + | ===Analysis=== |
| − | + | {{vspace}} | |
| + | Here are two principles that will help you make sense of the tree. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | A: '''A gene that is present in an ancestral species is inherited in all descendant species'''. The gene has to be observed in all OTUs, unless its has been lost (which is a rare event). | ||
| − | + | B: '''Paralogous genes in an ancestral species should give rise to monophyletic subtrees for each of the paralogues, in all descendants'''; this means: if the MRCA of a branch has e.g. three genes, we would expect three copies of that branch below this node, one for each of the three genes. Each of these subtrees should recapitulate the reference phylogenetic tree of the species, up to the branchpoint of their MRCA. The precise relationships may not be readily apparent, due to the noise and limited resolution we saw above, but the gene ought to be '''somewhere''' in the tree and you can often assume that it is closest to where it ought to be if the topology was correct. In this way you try to reconcile your expectations with your observations - preferably with as small a number of changes as possible. | |
| − | |||
| − | |||
| − | |||
| − | |||
| + | With these two simple principles (draw them out on a piece of paper if they do not seem obvious to you), you can probably pry your tree apart quite nicely. A few colored pencils and a printout of the tree will help. I would start by identifying all of the Mbp1 RBMs in the tree. | ||
| − | + | Here is a bit of code that you can use to colour the labels of the Mbp1 RBMs: | |
| − | |||
| + | <source lang="R"> | ||
| − | + | # You have previously defined the names for Mbp1 RBMs in | |
| − | + | # the vector apsMbp1Names. You can use these to check | |
| + | # which of the tree tipLabels are in that vector and | ||
| + | # then color them red in the plot. | ||
| − | + | # You'll need to replace <TREE> with whatever you called | |
| − | + | # your full tree with all APSES domain proteins. | |
| − | |||
| − | |||
| − | |||
| + | #First, have a look at the tip labels in your tree: | ||
| + | <TREE>$tip.label | ||
| − | + | # We'll create a vector of black colours of the same length | |
| + | # as the tip label vector: | ||
| + | tipColors = rep("#000000", Ntip(<TREE>)) | ||
| − | + | # ... then we replace each one for which the label is | |
| − | + | # in apsMbp1Names with "#BB0000" (red) | |
| − | + | tipColors[<TREE>$tip.label %in% apsMbp1Names] <- "#BB0000" | |
| − | |||
| − | |||
| − | |||
| + | #inspect: | ||
| + | tipColors | ||
| − | + | # ... and then we plot: | |
| + | plot(<TREE>, tip.color=tipColors, | ||
| + | cex=0.7, root.edge=TRUE, no.margin=TRUE) | ||
| − | |||
| + | </source> | ||
| − | + | {{vspace}} | |
| − | === | + | ===The APSES domains of the MRCA=== |
| + | {{vspace}} | ||
| − | + | Note: A common confusion about cenancestral genes (MRCA = Most Recent Common Ancestor) arises from the fact that by far not all expected genes are present in the OTUs. Some will have been lost, some will have been incorrectly annotated in their genome (frameshifts!) and not been found with PSI-BLAST, some may have diverged beyond recognizability. In general you have to ask: '''given the species represented in a subclade, what is the last common ancestor of that branch'''? The expectation is that '''all''' descendants of that ancestor should be represented in that branch '''unless''' one of the above reasons why a gene might be absent would apply. Eg. if a branch contains species from ''Basidiomycota'' '''and''' ''Ascomycota'', this means that its MRCA was the ancestor of all fungi. | |
| − | |||
{{task|1= | {{task|1= | ||
| − | |||
| − | |||
| − | * | + | * Consider the APSES domain proteins of the fungal cenancestor. What evidence do you see in the tree that identifies them. Note that the hallmark of a clade that originated in the cenancestor is that it contains species from '''all''' subsequent major branches of the species tree. How many of these proteins are there? What arer the names of their SACCE descendants? |
| + | |||
| + | }} | ||
| + | |||
| + | {{vspace}} | ||
| + | |||
| + | ===The APSES domains of YFO=== | ||
| + | {{vspace}} | ||
| − | + | You have identified the APSES domain genes of the fungal cenancestor above. Accordingly, this defines the number of APSES protein genes the ancestor to YFO had. Identify the sequence of duplications and/or gene loss in your organism through which YFO has ended up with the APSES domains it possesses today. | |
| − | + | {{task|1= | |
| + | |||
| + | # Print the tree to a single sheet of paper. | ||
| + | # Mark the clades for the genes of the cenancestor. | ||
| + | # Label all subsequent branchpoints that affect the gene tree for YFO with either '''"D"''' (for duplication) or '''"S"''' (for speciation). Remember that specific speciation events can appear more than once in a tree. Identify such events. | ||
| + | # '''Bring this sheet with you to the quiz on Tuesday. Your annotated printout will be worth half of the phylogeny quiz marks.''' | ||
| − | |||
| − | |||
}} | }} | ||
| − | + | {{vspace}} | |
| − | + | ==Bonus: when did it happen?== | |
| − | + | {{vspace}} | |
| + | |||
| + | A very cool resource is [http://www.timetree.org/ '''Timetree'''] - a tool that allows you to estimate divergence times between species. For example, the speciation event that separated the main branches of the fungi - i.e. the time when the fungal cenacestor lived - is given by the divergence time of ''Schizosaccharomyces pombe'' and ''Saccharomyces cerevisiaea'': 761,000,000 years ago. For comparison, these two fungi are therefore approximately as related to each other as '''you''' are ... | ||
| + | |||
| + | A) to the rabbit?<br> | ||
| + | B) to the opossum?<br> | ||
| + | C) to the chicken?<br> | ||
| + | D) to the rainbow trout?<br> | ||
| + | E) to the warty sea squirt?<br> | ||
| + | F) to the bumblebee?<br> | ||
| + | G) to the earthworm?<br> | ||
| + | H) to the fly agaric?<br> | ||
| + | |||
| + | Check it out - the question will be on the quiz. | ||
| + | |||
| + | {{vspace}} | ||
| + | |||
| + | |||
| + | ==Identifying Orthologs== | ||
| + | {{vspace}} | ||
| + | |||
| + | In the last assignment we discovered homologs to ''S. cerevisiae'' Mbp1 in YFO. Some of these will be orthologs to Mbp1, some will be paralogs. Some will have similar function, some will not. We discussed previously that genes that evolve under continuously similar evolutionary pressure should be most similar in sequence, and should have the most similar "function". | ||
| + | |||
| + | In this assignment we will define the YFO gene that is the most similar ortholog to ''S. cerevisiae'' Mbp1, and perform a multiple sequence alignment with it. | ||
| + | |||
| + | Let us briefly review the basic concepts. | ||
| + | |||
| + | <div style="padding: 2px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> | ||
| − | |||
<br> | <br> | ||
| + | ;All related genes are homologs. | ||
| + | </div> | ||
| − | |||
| − | |||
| − | + | Two central definitions about the mutual relationships between related genes go back to Walter Fitch who stated them in the 1970s: | |
| − | + | <div style="padding: 2px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> | |
| + | |||
| + | <br> | ||
| + | ;Orthologs have diverged after speciation. | ||
| − | < | + | ;Paralogs have diverged after duplication. |
| + | </div> | ||
| − | + | | |
| − | |||
| − | + | [[Image:OrthologParalog.jpg|frame|none|'''Hypothetical evolutionary tree.''' A single gene evolves through two speciation events and one duplication event. A duplication occurs during the evolution from reptilian to synapsid. It is easy to see how this pair of genes (paralogs) in the ancestral synapsid gives rise to two pairs of genes in pig and elephant, respectively. All ''circle'' genes are mutually orthologs, they form a "cluster of orthologs". All genes within one species are mutual paralogs–they are so called ''in-paralogs''. The ''circle'' gene in pig and the ''triangle'' gene in the elephant are so-called ''out-paralogs''. Somewhat counterintuitively, the ''triangle'' gene in the pig and the ''circle'' gene in the raven are also orthologs - but this has to be, since the last common ancestor diverged by '''speciation'''. | |
| − | |||
| − | + | The "phylogram" on the right symbolizes the amount of evolutionary change as proportional to height difference to the "root". It is easy to see how a bidirectional BLAST search will only find pairs of most similar orthologs. If applied to a group of species, bidirectional BLAST searches will find clusters of orthologs only (except if genes were lost, or there are anomalies in the evolutionary rate.)]] | |
| − | |||
| − | |||
| − | + | ==Defining orthologs== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | To be reasonably certain about orthology relationships, we would need to construct and analyze detailed evolutionary trees. This is computationally expensive and the results are not always unambiguous either, as we will see in a later assignment. But a number of different strategies are available that use precomputed results to define orthologs. These are especially useful for large, cross genome surveys. They are less useful for detailed analysis of individual genes. Pay the sites a visit and try a search. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ;Orthologs by eggNOG | |
| + | :The [http://eggnog.embl.de/ '''eggNOG'''] (evolutionary genealogy of genes: Non-supervised Orthologous Groups) database contains orthologous groups of genes at the EMBL. It seems to be continuously updtaed, the search functionality is reasonable and the results for yeast Mbp1 show many genes from several fungi. Importantly, there is only one gene annotated for each species. Alignments and trees are also available, as are database downloads for algorithmic analysis. | ||
| + | <div class="mw-collapsible mw-collapsed" data-expandtext="more..." data-collapsetext="less" style="width:800px"> | ||
| + | | ||
| + | <div class="mw-collapsible-content"> | ||
| − | # ===== | + | {{#pmid: 24297252}} |
| − | # | + | |
| − | + | </div> | |
| + | </div> | ||
| + | |||
| + | |||
| + | ;Orthologs at OrthoDB | ||
| + | :[http://www.orthodb.org/ '''OrthoDB'''] includes a large number of species, among them all of our protein-sequenced fungi. However the search function (by keyword) retrieves many paralogs together with the orthologs, for example, the yeast Soc2 and Phd1 proteins are found in the same orthologous group these two are clearly paralogs. | ||
| + | <div class="mw-collapsible mw-collapsed" data-expandtext="more..." data-collapsetext="less" style="width:800px"> | ||
| + | | ||
| + | <div class="mw-collapsible-content"> | ||
| + | |||
| + | {{#pmid: 23180791}} | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | ;Orthologs at OMA | ||
| + | [http://omabrowser.org/ '''OMA'''] (the Orthologous Matrix) maintained at the Swiss Federal Institute of Technology contains a large number of orthologs from sequenced genomes. Searching with <code>MBP1_YEAST</code> (this is the Swissprot ID) as a "Group" search finds the correct gene in EREGO, KLULA, CANGL and SACCE. But searching with the sequence of the ''Ustilago maydis'' ortholog does not find the yeast protein, but the orthologs in YARLI, SCHPO, LACCBI, CRYNE and USTMA. Apparently the orthologous group has been split into several subgroups across the fungi. However as a whole the database is carefully constructed and available for download and API access; a large and useful resource. | ||
| + | <div class="mw-collapsible mw-collapsed" data-expandtext="more..." data-collapsetext="less" style="width:800px"> | ||
| + | | ||
| + | <div class="mw-collapsible-content"> | ||
| − | # | + | {{#pmid: 21113020}} |
| − | + | ||
| − | + | ... see also the related articles, much innovative and carefully done work on automated orthologue definition by the Dessimoz group. | |
| + | </div> | ||
| + | </div> | ||
| − | |||
| − | |||
| − | + | ;Orthologs by syntenic gene order conservation | |
| + | :We will revisit this when we explore the UCSC genome browser. | ||
| − | |||
| − | |||
| − | |||
| + | ;Orthologs by RBM | ||
| + | :Defining it yourself. RBM (or: Reciprocal Best Match) is easy to compute and half of the work you have already done in [[BIO_Assignment_Week_3|Assignment 3]]. Get the ID for the gene which you have identified and annotated as the best BLAST match for Mbp1 in YFO and confirm that this gene has Mbp1 as the most significant hit in the yeast proteome. <small>The results are unambiguous, but there may be residual doubt whether these two best-matching sequences are actually the most similar orthologs.</small> | ||
| + | |||
| + | {{task|1= | ||
| + | # Navigate to the BLAST homepage. | ||
| + | # Paste the YFO RefSeq sequence identifier into the search field. (You don't have to search with sequences–you can search directly with an NCBI identifier '''IF''' you want to search with the full-length sequence.) | ||
| + | # Set the database to refseq, and restrict the species to ''Saccharomyces cerevisiae''. | ||
| + | # Run BLAST. | ||
| + | # Keep the window open for the next task. | ||
| − | + | The top hit should be yeast Mbp1 (NP_010227). E mail me your sequence identifiers if it is not. | |
| − | + | If it is, you have confirmed the '''RBM''' or '''BBM''' criterion (Reciprocal Best Match or Bidirectional Best Hit, respectively). | |
| − | |||
| − | |||
| + | <small>Technically, this is not perfectly true since you have searched with the APSES domain in one direction, with the full-length sequence in the other. For this task I wanted you to try the ''search-with-accession-number''. Therefore the procedural laxness, I hope it is permissible. In fact, performing the reverse search with the YFO APSES domain should actually be more stringent, i.e. if you find the right gene with the longer sequence, you are even more likely to find the right gene with the shorter one.</small> | ||
| + | }} | ||
| − | |||
| − | |||
| − | |||
| − | + | ;Orthology by annotation | |
| + | :The NCBI precomputes BLAST results and makes them available at the RefSeq database entry for your protein. | ||
| − | # | + | {{task|1= |
| − | # | + | # In your BLAST result page, click on the RefSeq link for your query to navigate to the RefSeq database entry for your protein. |
| + | # Follow the '''Blink''' link in the right-hand column under '''Related information'''. | ||
| + | # Restrict the view RefSeq under the "Display options" and to Fungi. | ||
| − | + | You should see a number of genes with low E-values and high coverage in other fungi - however this search is problematic since the full length gene across the database finds mostly Ankyrin domains. | |
}} | }} | ||
| + | |||
| + | |||
| + | You will find that '''all''' of these approaches yield '''some''' of the orthologs. But none finds them all. The take home message is: precomputed results are good for large-scale survey-type investigations, where you can't humanly process the information by hand. But for more detailed questions, careful manual searches are still indsipensable. | ||
| + | |||
| + | <div class="mw-collapsible mw-collapsed" data-expandtext="Expand for crowdsourcing" data-collapsetext="Collapse"> | ||
| + | ;Orthology by crowdsourcing | ||
| + | :Luckily a crowd of willing hands has prepared the necessary sequences for you: in the section below you will find a link to the annotated and verified Mbp1 orthologs from last year's course :-) | ||
| + | |||
| + | <div class="mw-collapsible-content"> | ||
| + | We could call this annotation by many hands {{WP|Crowdsourcing|"crowdsourcing"}} - handing out small parcels of work to many workers, who would typically allocate only a small share of their time, but here the strength is in numbers and especially projects that organize via the Internet can tally up very impressive manpower, for free, or as {{WP|Microwork}}. These developments have some interest for bioinformatics: many of our more difficult tasks can not be easily built into an algorithm, language related tasks such as text-mining, or pattern matching tasks come to mind. Allocating this to a large number of human contributors may be a viable alternative to computation. A marketplace where this kind of work is already a reality is {{WP|Amazon Mechanical Turk|Amazon's "Mechanical Turk" Marketplace}}: programmers–"requesters"– use an open interface to post tasks for payment, "providers" from all over the world can engage in these. Tasks may include matching of pictures, or evaluating the aesthetics of competing designs. A quirky example I came across recently was when information designer David McCandless had 200 "Mechanical Turks" draw a small picture of their soul for his collection. | ||
| + | |||
| + | The name {{WP|The Turk|"Mechanical Turk"}} by the way relates to a famous ruse, when a Hungarian inventor and adventurer toured the imperial courts of 18<sup>th</sup> century Europe with an automaton, dressed in turkish robes and turban, that played chess at the grandmaster level against opponents that included Napoleon Bonaparte and Benjamin Franklin. No small mechanical feat in any case, it was only in the 19<sup>th</sup> century that it was revealed that the computational power was actually provided by a concealed human. | ||
| + | |||
| + | Are you up for some "Turking"? Before the next quiz, edit [http://biochemistry.utoronto.ca/steipe/abc/students/index.php/BCH441_2014_Assignment_7_RBM '''the Mbp1 RBM page on the Student Wiki] and include the RBM for Mbp1, for a 10% bonus on the next quiz. | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| | ||
| − | |||
| − | |||
| − | + | ===Coloring a 3D model by conservation=== | |
| + | |||
| + | With the superimposed coordinates, you can begin to get a sense whether either or both binding modes could be appropriate for a protein-DNA complex in your Mbp1 orthologue. But these are geometrical criteria only, and the protein in your species may be flexible enough to adopt a different conformation in a complex, and different again from your model. A more powerful way to analyze such hypothetical complexes is to look at conservation patterns. With VMD, you can import a sequence alignment into the MultiSeq extension and color residies by conservation. The protocol below assumes | ||
| + | |||
| + | *You have prealigned the reference Mbp1 proteins with your species' Mbp1 orthologue; | ||
| + | *You have saved the alignment in a CLUSTAL format. | ||
| + | |||
| + | You can use Jalview or any other MSA server to do so. You can even do this by hand - there should be few if any indels and the correct alignment is easy to see. | ||
{{task|1= | {{task|1= | ||
| − | + | ;Load the Mbp1 APSES alignment into MultiSeq. | |
| − | + | ||
| − | + | :(A) In the MultiSeq Window, navigate to '''File → Import Data...'''; Choose "From Files" and Browse to the location of the alignment you have saved. The File navigation window gives you options which files to enable: choose to Enable <code>ALN</code> files (these are CLUSTAL formatted multiple sequence alignments). | |
| − | + | :(B) Open the alignment file, click on '''Ok''' to import the data, it will take a short while to load. If the data can't be loaded, the file may have the wrong extension: .aln is required. | |
| − | + | :(C) find the Mbp1_SACCE sequence in the list, click on it and move it to the top of the Sequences list with your mouse (the list is not static, you can re-order the sequences in any way you like). | |
| − | |||
| − | |||
| − | |||
| − | + | You will see that the 1MB1 sequence and the APSES domain sequence do not match: at the N-terminus the sequence that corresponds to the PDB structure has extra residues, and in the middle the APSES sequences may have gaps inserted. | |
| − | |||
| − | + | ;Bring the 1MB1 sequence in register with the APSES alignment. | |
| + | :(A)MultiSeq supports typical text-editor selection mechanisms. Clicking on a residue selects it, clicking on a row selects the whole sequence. Dragging with the mouse selects several residues, shift-clicking selects ranges, and option-clicking toggles the selection on or off for individual residues. Using the mouse and/or the shift key as required, select the '''entire first column''' of the sequences you have imported. | ||
| + | :(B) Select '''Edit → Enable Editing... → Gaps only''' to allow changing indels. | ||
| + | :(C) Pressing the spacebar once should insert a gap character before the '''selected column''' in all sequences. Insert as many gaps as you need to align the beginning of sequences with the corresponding residues of 1MB1: <code>S I M ...</code> | ||
| + | :(D) Now insert as many gaps as you need into the structure sequence, to align it completely with the Mbp1_SACCE APSES domain sequence. (Simply select residues in the sequence and use the space bar to insert gaps. (Note: I have noticed a bug that sometimes prevents slider or keyboard input to the MultiSeq window; it fails to regain focus after operations in a different window. I don't know whether this is a Mac related problem or a more general bug in MultiSeq. When this happens I quit VMD and restore the session from a saved state. It is a bit annoying but not mission-critical.) | ||
| + | :(E) When you are done, it may be prudent to save the state of your alignment. Use '''File → Save Session...''' | ||
| − | + | ;Color by similarity | |
| + | :(A) Use the '''View → Coloring → Sequence similarity → BLOSUM30''' option to color the residues in the alignment and structure. This clearly shows you where conserved and variable residues are located and allows to analyze their structural context. | ||
| + | :(B) You can adjust the color scale in the usual way by navigating to '''VMD main → Graphics → Colors...''', choosing the Color Scale tab and adjusting the scale midpoint. | ||
| + | :(C) Navigate to the '''Representations''' window and apply the color scheme to your tube-and-sidechain representation: double-click on the NewCartoon representation to hide it and use '''User''' coloring of your ''Tube'' and ''Licorice'' representations to apply the sequence similarity color gradient that MultiSeq has calculated. | ||
| + | <br><div style="padding: 5px; background: #DDDDEE;"> | ||
| + | * Once you have colored the residues of your model by conservation, create another informative stereo-image and paste it into your assignment. | ||
| + | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| − | + | {{vspace}} | |
| − | |||
| + | --> | ||
| − | + | {{Vspace}} | |
==Links and Resources== | ==Links and Resources== | ||
| + | ;Literature | ||
| + | {{#pmid: 26323765}} | ||
| + | {{#pmid: 22114356}} | ||
| + | {{#pmid: 19190756}} | ||
| − | + | Also: [http://www.nature.com/scitable/topicpage/reading-a-phylogenetic-tree-the-meaning-of-41956 Nature-Scitable (2008): '''Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups'''] | |
| − | {{#pmid: | + | {{#pmid: 12801728}} |
| − | + | :* [http://evolution.genetics.washington.edu/phylip/phylip.html '''PHYLIP''' documentation] | |
| − | {{ | + | {{PDF |
| + | |authors= Tuimala, Jarno | ||
| + | |year= 2006 | ||
| + | |title= A primer to phylogenetic analysis using the PHYLIP package | ||
| + | |journal= | ||
| + | |volume= | ||
| + | |pages= | ||
| + | |URL= http://koti.mbnet.fi/tuimala/oppaat/phylip2.pdf | ||
| + | |doi= | ||
| + | |file= Tuimala_PHYLIP.pdf | ||
| + | |abstract= The purpose of this tutorial is to demonstrate how to use PHYLIP, a collection of phylogenetic analysis software, and some of the options that are available. This tutorial is not intended to be a course in phylogenetics, although some phylogenetic concepts will be discussed briefly. There are other books available which cover the theoretical sides of the phylogenetic analysis, but the actual data analysis work is less well covered. Here we will mostly deal with molecular sequence data analysis in the current PHYLIP version 3.66. | ||
| + | }} | ||
| − | :* [http:// | + | ;Software |
| − | :* [http:// | + | :* [http://evolution.genetics.washington.edu/phylip.html '''PHYLIP''' home page] |
| + | <!-- not currently active | ||
| + | :* [http://itol.embl.de/ '''iTOL''' - Interactive Tree of Life project'''] | ||
| + | --> | ||
| + | ;Sequences | ||
| + | :* [[Reference APSES domains (reference species)|'''reference APSES domains page''']] | ||
| − | |||
| − | |||
| − | |||
<!-- {{#pmid: 19957275}} --> | <!-- {{#pmid: 19957275}} --> | ||
| Line 485: | Line 794: | ||
| | ||
{{#lst:BIO_Assignment_Week_1|assignment_footer}} | {{#lst:BIO_Assignment_Week_1|assignment_footer}} | ||
| + | |||
| + | <table style="width:100%;"><tr> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:left; font-size:80%;">[[BIO_Assignment_Week_6|< Assignment 6]]</td> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:right; font-size:80%;">[[BIO_Assignment_Week_8|Assignment 8 >]]</td> | ||
| + | </tr></table> | ||
Latest revision as of 15:19, 27 August 2017
Assignment for Week 7
Phylogenetic Analysis
| < Assignment 6 | Assignment 8 > |
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Contents
- Nothing in Biology makes sense except in the light of evolution.
- Theodosius Dobzhansky
... but does evolution make sense in the light of biology?
As we have seen in the previous assignments, the Mbp1 transcription factor has homologues in all other fungi, yet there is not always a clear one-to-one mapping between members of a family in distantly related species. It appears that various systems of APSES domain transcription factors have evolved independently. Of course this bears directly on our notion of function - what it means to say that two genes in different organisms have the "same" function. In case two organisms both have an orthologous gene for the same, distinct function, calling these functions "the same" may be warranted. But what if that gene has duplicated in one species, and the two paralogues now perform different, related functions in one organism? Theses two are still orthologues to both their homologues in the other species, but now we expect functionally significant residues to have adapted to the new - and possibly distinct - roles of each paralogue. In order to be able to even ask such questions, we need to make the evolutionary history of gene families explicit. This is the domain of phylogenetic analysis. We can ask questions like: how many paralogues did the cenancestor of a clade possess? Which of these underwent additional duplications in the phylogenesis of the organism I am studying? Did any genes get lost? And - adding additional biological insight to the picture - did the observed duplications lead to the "invention" of new biological systems? When was that? And perhaps even: how did the species benefit from this event?
We will develop this kind of analysis in this assignment. In the previous assignment you have established which gene in your species is the reciprocally most closely related orthologue to yeast Mbp1 (with reciprocal best match) and you have identified the full complement of APSES domain genes in your assigned organism (as a result of your PSI-BLAST search). In this assignment, we will analyse these genes' evolutionary relationship and compare it to the evolutionary relationship of other fungal APSES domains. The goal is to define families of related transcription factors and their evolutionary history. All APSES domain annotations are now available in your protein "database". Now we will attempt to compute the phylogram for these proteins. The goal is to identify orthologues and paralogues.
A number of excellent tools for phylogenetic analysis exist; general purpose packages include the (free) PHYLIP package, the MEGA package and the (commercial) PAUP* package. Of these, only MEGA is still under active development, although PHYLIP still functions perfectly (except for problems with graphical windows under Mac OS 10.6). Specialized tools for tree-building include Treepuzzle or Mr. Bayes. This assignment is constructed around programs that are available in PHYLIP, however you are welcome to use other tools that fulfill a similar purpose if you wish. In this field, researchers consider trees that have been built with ML (maximum likelihood) methods to be more reliable than trees that are built with parsimony methods, or distance methods such as NJ (Neighbor Joining). However ML methods are also much more compute-intensive. Just like with multiple sequence alignments, some algorithms will come closer to guessing the truth and others will not and usually it is hard to tell which is the more trustworthy of two diverging results. The prudent researcher tries out alternatives and forms her own opinion. Specifically, we may usually assume results that converge when computed with different algorithms, to be more reliable than those that depend strongly on a particular algorithm, parameters, or details of input data.
In this assignment, we will take a computational shortcut, (something you should not do in real life). We will skip establishing the reliability of the tree with a bootstrap procedure, i.e. repeat the tree-building a hundred times with partial data and see which branches and groupings are robust and which depend on the details of the data. (If you are interested, have a look here for the procedure for running a bootstrap analysis on the data set you are working with, but this may require a day or so of computing time on your computer.) In this assignment, we will simply acknowledge that bifurcations that are very close to each other have not been "resolved" and be appropriately cautious in our inferences. In phylogenetic analysis, not all lines a program draws are equally trustworthy. Don't take the trees as a given fact just because a program suggests this. Look at the evidence, include independent information where available, use your reasoning, and analyse the results critically. As you will see, there are some facts that we know for certain: we know which species the genes come from, and we can (usually) make good assumptions about the relationship of the species themselves - the history of speciation events that underlies all evolution of genes. This is extremely helpful information for our work.
If you would like to review concepts of trees, clades, LCAs, OTUs and the like, I have linked an excellent and very understandable introduction-level article on phylogenetic analysis here and to the resource section at the bottom of this page.
| Baldauf (2003) Phylogeny for the faint of heart: a tutorial. Trends Genet 19:345-51. (pmid: 12801728) |
R packages that may be useful include the following:
- R task view Phylogenetics - this task-view gives an excellent, curated overview of the important R-packages in the domain.
- package ape - general purpose phylogenetic analysis, but (as far as I can tell ape only supports analysis with DNA sequences).
- package ips - wrapper for MrBayes, Beast, RAxML "heavy-duty" phylogenetic analysis packages.
- package Rphylip - Wrapper for Phylip, the most versatile set of phylogenetic inference tools.
Preparing input alignments
You have previously collected homologous sequences and their annotations. We will use these as input for phylogenetic analysis. But let's discuss first how such an input file should be constructed.
Principles
In order to use molecular sequences for the construction of phylogenetic trees, you have to build a multiple alignment first. This is important: phylogenetic analysis does not build alignments, nor does it revise alignments, it analyses them after the alignment has been computed. A precondition for the analysis to be meaningful is that all rows of sequences have to contain the exact same number of characters and to hold aligned characters in corresponding positions (i.e. columns). The program's inferences are made on a column-wise basis and if your columns contain data from unrelated positions, the inferences are going to be questionable. Clearly, in order for tree-estimation to work, one must not include fragments of sequence which have evolved under a different evolutionary model as all others, e.g. after domain fusion, or after accommodating large stretches of indels. Thus it is appropriate to edit the sequences and pare them down to a most characteristic subset of amino acids. The goal is not to be as comprehensive as possible, but to input those columns of aligned residues that will best represent the true phylogenetic relationships between the sequences.
The result of the tree construction is a decision about the most likely evolutionary relationships. Fundamentally, tree-construction programs decide which sequences had common ancestors.
Distance based phylogeny programs start by using sequence comparisons to estimate evolutionary distances:
- they apply a model of evolution such as a mutation data matrix, to calculate a score for each pair of sequences,
- this score is stored in a "distance matrix" ...
- ... and used to estimate a tree that groups sequences with close relationships together. (e.g. by using an NJ, Neigbor Joining, algorithm).
They are fast, can work on large numbers of sequences, but are less accurate if genes evolve at different rates.
Parsimony based phylogeny programs build a tree that minimizes the number of mutation events that are required to get from a common ancestral sequence to all observed sequences. They take all columns into account, not just a single number per sequence pair, as the Distance Methods do. For closely related sequences they work very well, but they construct inaccurate trees when they can't make good estimates for the required number of sequence changes.
ML, or Maximum Likelihood methods attempt to find the tree for which the observed sequences would be the most likely under a particular evolutionary model. They are based on a rigorous statistical framework and yield the most robust results. But they are also quite compute intensive and a tree of the size that we are building in this assignment is a challenge for the resources of common workstation (runs about an hour on my computer). If the problem is too large, one may split a large problem into smaller, obvious subtrees (e.g. analysing orthologues as a group, only including a few paralogues for comparison) and then merge the smaller trees; this way even very large problems can become tractable.
ML methods suffer less from "long-branch attraction" - the phenomenon that weakly similar sequences can be grouped inappropriately close together in a tree due to spuriously shared differences.
Bayesian methods don't estimate the tree that gives the highest likelihood for the observed data, but find the most probably tree, given that the data have been observed. If this sounds conceptually similar to you, then you are not wrong. However, the approaches employ very different algorithms. And Bayesian methods need a "prior" on trees before observation.
Choosing sequences
To illustrate the principle we will construct input files by joining APSES domain and Ankyrin domain sequences and for this we will use the Prosite annotations we have collected for the reference set of sequences and your YFO sequences.
Task:
- Open RStudio.
- Choose File → Recent Projects → BCH441_2016.
- Pull the latest version of the project repository from GitHub.
- type init()
- Open the file BCH441_A07.R and work through PART ONE: Choosing sequences.
Adding an Outgroup
An outgroup is a sequence that is more distantly related to all of the other sequences than any of them are to each other. This allows us to root the tree, because the root - the last common ancestor to all - must be somewhere on the branch that connects the outgroup to the rest. And whenever a molecular clock is assumed, the branching point that connects the outgroup can be assumed to be the oldest divergence event. Having a root that we can compare to the phylogram of species makes the tree interpretation much more intuitive. In our case, we are facing the problem that our species cover all of the known fungi, thus we can' rightly say that any of them are more distant to the rest. We have to look outside the fungi. The problem is, outside of the fungi there are no proteins with APSES domains. We can take the E. coli KilA-N domain sequence - a known, distant homologue to the APSES domain instead, even though it only aligns to a part of the APSES domains.
Here is the KilA-N domain sequence in the E. coli Kil-A protein:
>WP_000200358.1 hypothetical protein [Escherichia coli] MTSFQLSLISREIDGEIIHLRAKDGYINATSMCRTAGKLLSDYTRLKTTQEFFDELSRDMGIPISELIQS FKGGRPENQGTWVHPDIAINLAQWLSPKFAVQVSRWVREWMSGERTTAEMPVHLKRYMVNRSRIPHTHFS ILNELTFNLVAPLEQAGYTLPEKMVPDISQGRVFSQWLRDNRNVEPKTFPTYDHEYPDGRVYPARLYPNE YLADFKEHFNNIWLPQYAPKYFADRDKKALALIEKIMLPNLDGNEQF
E. coli KilA-N protein. Residues that do not align with APSES domains are shown in grey.
The assignment R - code contains code to add it to the group of APSES sequences.
Task:
- Continue with the R-code: PART TWO: Multiple sequence alignment
Reviewing and Editing alignments
As discussed in the lecture, it is usually necessary to edit a multiple sequence alignment to make it suitable for phylogenetic inference. Here are the principles:
All characters in a column should be related by homology.
This implies the following rules of thumb:
- Remove all stretches of residues in which the alignment appears ambiguous (not just highly variable, but ambiguous regarding the aligned positions).
- Remove all frayed N- and C- termini, especially regions in which not all sequences that are being compared appear homologous and that may stem from unrelated domains. You want to only retain the APSES domains. All the extra residues from the YFO sequence can be deleted.
- Remove all gapped regions that appear to be alignment artefacts due to inappropriate input sequences.
- Remove all but approximately one column from gapped regions in those cases where the presence of several related insertions suggest that the indel is real, and not just an alignment artefact. (Some researchers simply remove all gapped regions).
- Remove sections N- and C- terminal of gaps where the alignment appears questionable.
- If the sequences fit on a single line you will save yourself potential trouble with block-wise vs. interleaved input. If you do run out of memory try removing columns of sequence. Or remove species that you are less interested in from the alignment.
- Move your outgroup sequence to the first line of your alignment, since this is where PHYLIP will look for it by default.
Indels are even more of a problem than usual. Strictly speaking, the similarity score of an alignment program as well as the distance score of a phylogeny program are not calculated for an ordered sequence, but for a sum of independent values, one for each aligned columns of characters. The order of the columns does not change the score. However in an optimal sequence alignment with gaps, this is no longer strictly true since a one-character gap creation has a different penalty score than a one-character gap extension! Most alignment programs use a model with a constant gap insertion penalty and a linear gap extension penalty. This is not rigorously justified from biology, but parametrized (or you could say "tweaked") to correspond to our observations. However, most phylogeny programs do not work in this way. They strictly operate on columns of characters and treat a gap character just like a residue with the one letter code "-". Thus gap insertion- and extension- characters get the same score. For short indels, this underestimates the distance between pairs of sequences, since any evolutionary model should reflect the fact that gaps are much less likely than point mutations. If the gap is very long though, all events are counted individually as many single substitutions (rather than one lengthy one) and this overestimates the distance. And it gets worse: long stretches of gaps can make sequences appear similar in a way that is not justified, just because they are identical in the "-" character. It is therefore common and acceptable to edit gaps in the alignment and delete all but a few columns of gapped sequence, or to remove such columns altogether.
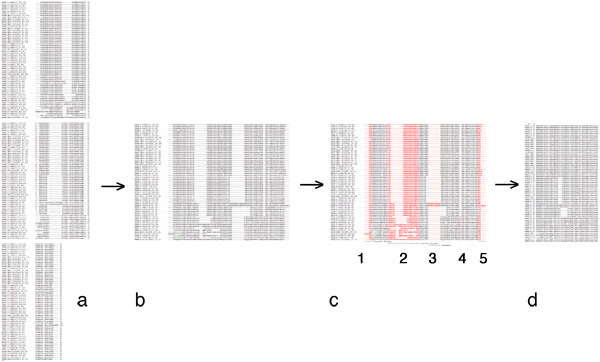
There is more to learn about this important step of working with aligned sequences, here is an overview of the literature on various algorithms and tools that are available.
| Talavera & Castresana (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564-77. (pmid: 17654362) |
| Capella-Gutiérrez et al. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972-3. (pmid: 19505945) |
| Blouin et al. (2009) Reproducing the manual annotation of multiple sequence alignments using a SVM classifier. Bioinformatics 25:3093-8. (pmid: 19770262) |
| Penn et al. (2010) GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res 38:W23-8. (pmid: 20497997) |
| Rajan (2013) A method of alignment masking for refining the phylogenetic signal of multiple sequence alignments. Mol Biol Evol 30:689-712. (pmid: 23193120) |
Sequence masking with R
As you saw while inspecting the multiple sequence alignment, there are regions that are poorly suited for phylogenetic analysis due to the large numbers of gaps.
A good approach to edit the alignment is to import your sequences into Jalview and remove uncertain columns by hand.
But for this assignment, let's write code for a simple masking heuristic.
Task:
- Head back to the RStudio project and work through PART THREE: reviewing and editing alignments
Calculating trees
In this section we perform the actual phylogenetic calculation.
Task:
- Download the PHYLIP suite of programs from the Phylip homepage and install it on your computer.
- Return to the RStudio project and work through PART FOUR: Calculating trees.
Analysing your tree
In order to analyse your tree, you need a species tree as reference. This really is an absolute prerequisite to make your expectations about the observed tree explicit. Fortunately we have all species nicely documented in our database.
The reference species tree
Task:
- Navigate to the NCBI Taxonomy page
- Execute the following R command to create an Entrez command that will retrieve all taxonomy records for the species in your database:
cat(paste(paste(c(myDB$taxonomy$ID, "83333"), "[taxid]", sep=""), collapse=" OR "))- Copy the Entrez command, and enter it into the search field of the NCBI taxonomy page. Click on Search. The resulting page should have twelve species listed - ten "reference" fungi, E. coli (as the outgroup), and YFO. Make sure YFO is included! If it's not there, you did something wrong that needs to be fixed.
- Click on the Summary options near the top-left of the page, and select Common Tree. This places all the species into the universal tree of life and identifies their relationships.
- At the top, there is an option to Save as ... and the option to select a format to save the tree in. Select Phylip Tree as the format and click the Save as button. The file
phyliptree.phywill be downloaded to your computer into your default download directory. Move it to the directory you have defined asPROJECTDIR.
- Open the file in a text-editor. This is a tree, specified in the so-called "Newick format". The topology of the tree is defined through the brackets, and the branch-lengths are all the same: this is a cladogram, not a phylogram. The tree contains the long names for the species/strains and for our purposes we really need the "biCodes" instead. I can't think of a very elegant way to make that change programmatically, so just go ahead and replace the species names (not the taxonomic ranks though) with their biCode in your text editor. Remove all the single quotes, and replace any remaining blanks in names with an underscore. Take care however not to delete any colons or parentheses. Save the file.
My version looks like this - Your version must have YFO somewhere in the tree..
( 'ESCCO':4, ( ( 'PUCGR':4, 'USTMA':4, ( 'WALME':4, 'COPCI':4, 'CRYNE':4 )Agaricomycotina:4 )Basidiomycota:4, ( ( ( 'ASPNI':4, 'BIPOR':4, 'NEUCR':4 )leotiomyceta:4, 'SACCE':4 )saccharomyceta:4, 'SCHPO':4 )Ascomycota:4 )Dikarya:4 )'cellular organisms':4;
- Now read the tree in R and plot it.
# Download the EDITED phyliptree.phy
orgTree <- read.tree("phyliptree.phy")
# Plot the tree in a new window
dev.new(width=6, height=3)
plot(orgTree, cex=1.0, root.edge=TRUE, no.margin=TRUE)
nodelabels(text=orgTree$node.label, cex=0.6, adj=0.2, bg="#D4F2DA")
I have constructed a cladogram for many of the species we are analysing, based on data published for 1551 fungal ribosomal sequences. The six reference species are included. Such reference trees from rRNA data are a standard method of phylogenetic analysis, supported by the assumption that rRNA sequences are monophyletic and have evolved under comparable selective pressure in all species.
Cladogram of the "reference" fungi studied in the assignments. This cladogram is based on a tree returned by the NCBI Common Tree. It is thus a digest of cladistic relationships, not a representation of a specific molecular phylogeny.
Alternatively, you can look up your species in the latest version of the species tree for the fungi and add it to the tree by hand while resolving the trifurcations. See:
| Ebersberger et al. (2012) A consistent phylogenetic backbone for the fungi. Mol Biol Evol 29:1319-34. (pmid: 22114356) |
Task:
- Return to the RStudio project and continue with the script to its end. Note the deliverable at the end: to print out your trees and bring them to class.
Links and Resources
- Literature
| Szöllősi et al. (2015) Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos Trans R Soc Lond., B, Biol Sci 370:20140335. (pmid: 26323765) |
| Ebersberger et al. (2012) A consistent phylogenetic backbone for the fungi. Mol Biol Evol 29:1319-34. (pmid: 22114356) |
| Marcet-Houben & Gabaldón (2009) The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome. PLoS ONE 4:e4357. (pmid: 19190756) |
Also: Nature-Scitable (2008): Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups
| Baldauf (2003) Phylogeny for the faint of heart: a tutorial. Trends Genet 19:345-51. (pmid: 12801728) |
| Tuimala, Jarno (2006) A primer to phylogenetic analysis using the PHYLIP package. |
| (pmid: None) [ Source URL ] Abstract |
- Software
- Sequences
Footnotes and references
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.
| < Assignment 6 | Assignment 8 > |