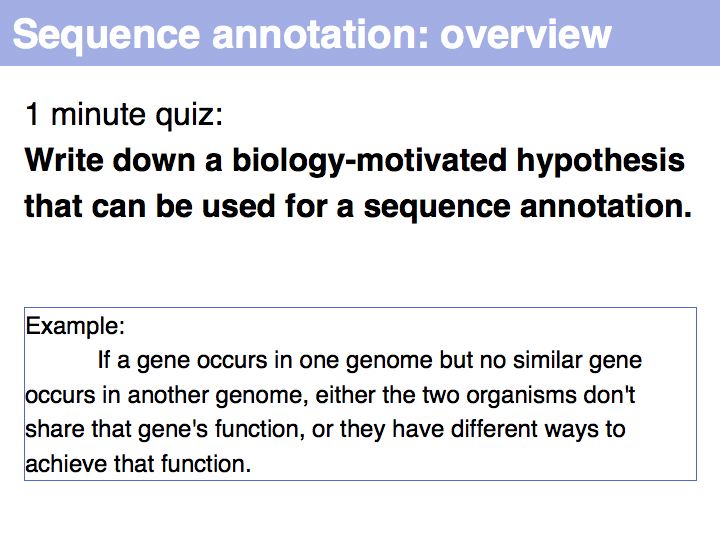
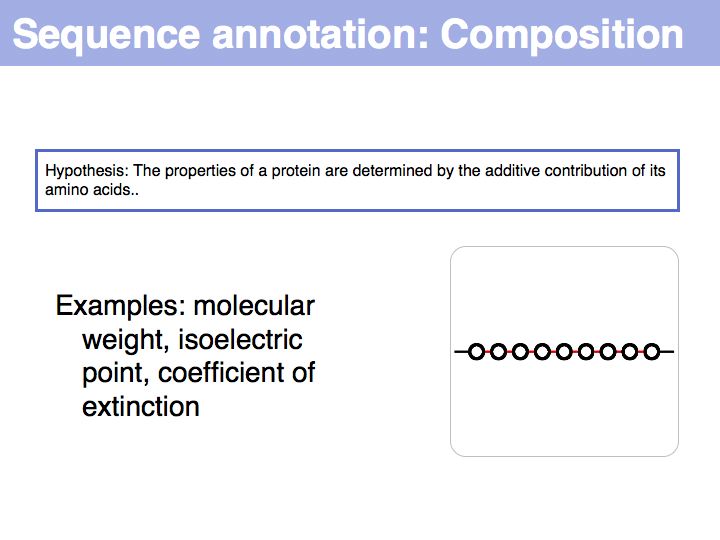
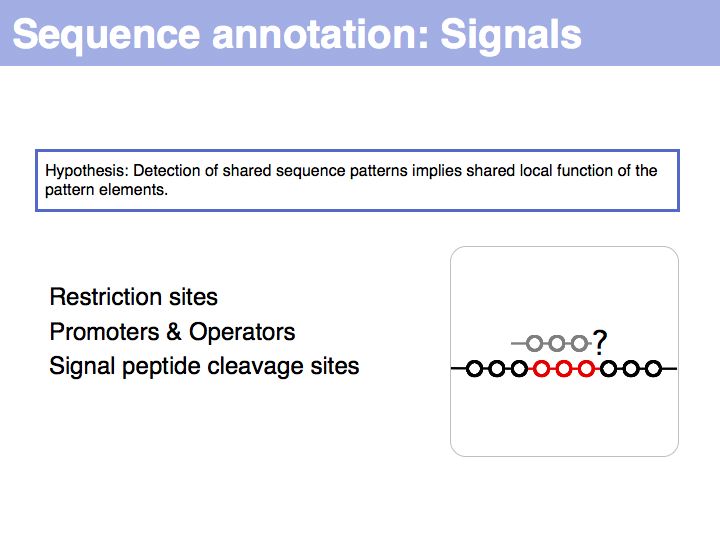
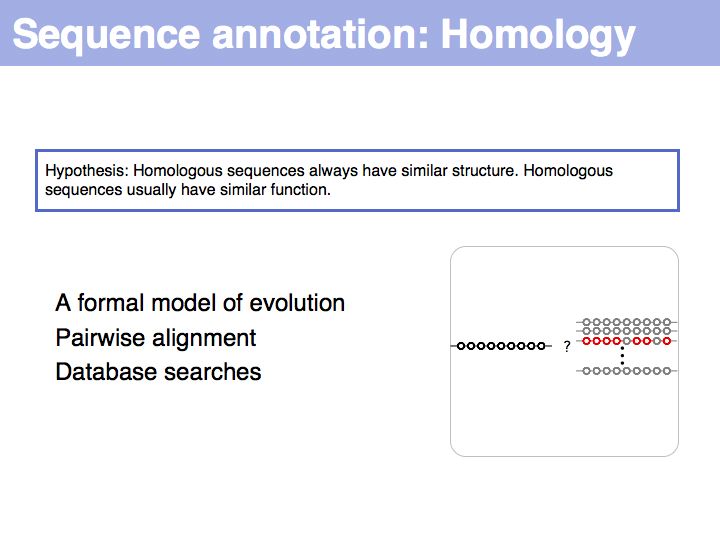
Difference between revisions of "Lecture 03"
Jump to navigation
Jump to search





| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <!-- div style="padding: 5px; background: #FF4560; border:solid 2px #000000;"> | ||
| + | '''Update Warning!''' | ||
| + | This page has not been revised yet for the 2007 Fall term. Some of the slides may be reused, but please consider the page as a whole out of date as long as this warning appears here. | ||
| + | </div --> | ||
| + | | ||
| + | |||
| + | | ||
| + | |||
__NOTOC__ | __NOTOC__ | ||
<small>[[Lecture_02|(Previous lecture)]] ... [[Lecture_04|(Next lecture)]]</small> | <small>[[Lecture_02|(Previous lecture)]] ... [[Lecture_04|(Next lecture)]]</small> | ||
| Line 4: | Line 12: | ||
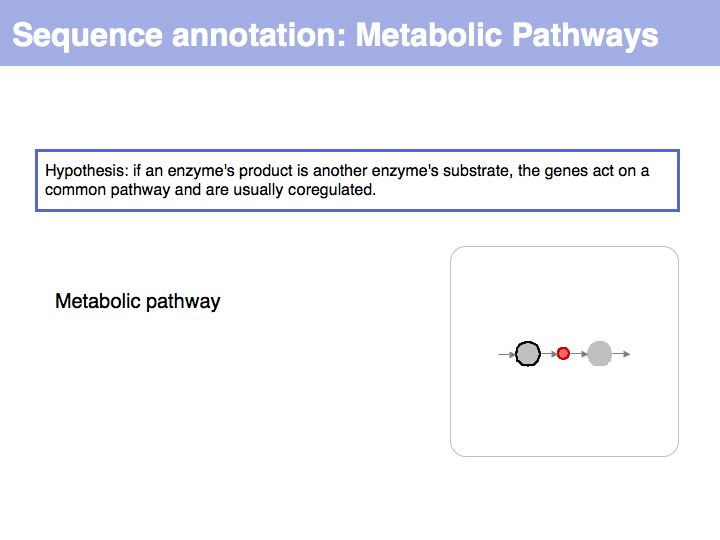
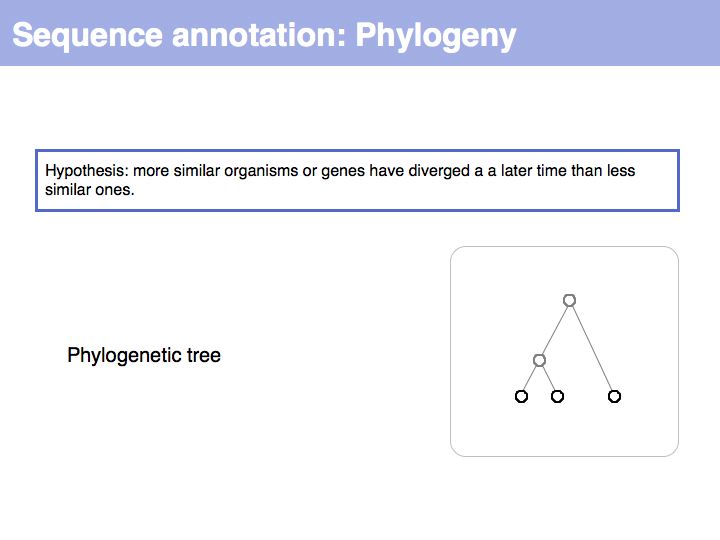
==Sequence Properties== | ==Sequence Properties== | ||
| − | |||
| − | + | ;What you should take home from this part of the course: | |
| − | * | + | *Understand the ideas of analysis by composition and analysis by signal; |
| − | * Exercises | + | *Know what '''deterministic pattern matching''' is; |
| + | *Recognize and understand the term '''regular expression'''; | ||
| + | *Be familiar with common sequence signals in DNA,RNA and proteins; | ||
| + | *Be familiar with the Prosite database and the Prosite scan server; | ||
| + | *Kow where to find EMBOSS tools and how to use them; | ||
| + | *Know about the offerings on the ExPASy tools collection page; | ||
| + | *Work on an understanding how biological facts can be translated into hypotheses and how hypotheses can be translated into computational procedures for analysis. | ||
| + | | ||
| + | |||
| + | ;Links summary: | ||
| + | *[http://www.nature.com/news/ ''Nature'' news] | ||
| + | *[http://sciencenow.sciencemag.org/ ''Science'' news] | ||
| + | *[http://rebase.neb.com/ ReBase] | ||
| + | *[http://wishart.biology.ualberta.ca/PlasMapper/index.html PlasMapper] | ||
| + | *[http://www.expasy.org/prosite prosite] | ||
| + | *[http://www.expasy.org/tools the ExPASy tools collection] | ||
| + | | ||
| + | |||
| + | ;Exercises | ||
| + | |||
| + | *Retrieve and read the Prosite documentation entry for the Leucine Zipper. | ||
| + | *Download entry 1NWQ from the PDB, visualize the Leucine Zipper with VMD and study its architecture (stereo vision!). | ||
| + | |||
| + | | ||
| + | | ||
==Lecture Slides== | ==Lecture Slides== | ||
| + | |||
======Slide 001====== | ======Slide 001====== | ||
| − | [[Image:L03_s001.jpg|frame|none|Lecture 03, Slide 001]] | + | [[Image:L03_s001.jpg|frame|none|Lecture 03, Slide 001<br> |
| + | This finding made the news. You should be aware of important new developments: subscribe to read at least the news items from [http://www.nature.com/news/ ''Nature''] and [http://sciencenow.sciencemag.org/ ''Science''], preferably subscribe to and browse their tables of contents too. In this particular new finding, researchers challenge our current concept of "genome": what is a genome, if the same physical DNA molecule can contain coding information for more than one species? Also, this finding further emphasizes the importance of horizontal gene transfer in evolution. | ||
| + | ]] | ||
| + | |||
======Slide 002====== | ======Slide 002====== | ||
| − | [[Image:L03_s002.jpg|frame|none|Lecture 03, Slide 002]] | + | [[Image:L03_s002.jpg|frame|none|Lecture 03, Slide 002<br> |
| + | What properties of a sequence can you analyze to describe what it is or does? | ||
| + | ]] | ||
======Slide 003====== | ======Slide 003====== | ||
| − | [[Image:L03_s003.jpg|frame|none| | + | [[Image:L03_s003.jpg|frame|none|Lecture 03, Slide 003<br> |
| + | ]] | ||
======Slide 004====== | ======Slide 004====== | ||
| − | [[Image:L03_s004.jpg|frame|none|Lecture 03, Slide 004]] | + | [[Image:L03_s004.jpg|frame|none|Lecture 03, Slide 004<br> |
| + | |||
| + | ]] | ||
======Slide 005====== | ======Slide 005====== | ||
| − | [[Image:L03_s005.jpg|frame|none|Lecture 03, Slide 005]] | + | [[Image:L03_s005.jpg|frame|none|Lecture 03, Slide 005<br> |
| + | |||
| + | ]] | ||
======Slide 006====== | ======Slide 006====== | ||
| − | [[Image:L03_s006.jpg|frame|none|Lecture 03, Slide 006]] | + | [[Image:L03_s006.jpg|frame|none|Lecture 03, Slide 006<br> |
| + | |||
| + | ]] | ||
======Slide 007====== | ======Slide 007====== | ||
| − | [[Image:L03_s007.jpg|frame|none|Lecture 03, Slide 007]] | + | [[Image:L03_s007.jpg|frame|none|Lecture 03, Slide 007<br> |
| + | (#: Number of ...) A protein's isoelectric point depends on the pK values of the amino acids; the pK values characterize the propensity fo an amino acid sidechain to dissociate, which in turn depends on how energetically favourable dissociation is. For example: since a negatively charged amino acid will be stabilized in a positive electrostatic field, such a field will shift a pK value '''down'''. This means the pH value at which the side chain will be 50% ionized is lower, or in other words, in a positive electrostatic field the concentration of protons must be higher to keep a proton associated to the sidechain.<br> | ||
| + | <br> | ||
| + | Compositional properties of nucleic acids include hybridization temperature and helix structure. | ||
| + | ]] | ||
| + | |||
======Slide 008====== | ======Slide 008====== | ||
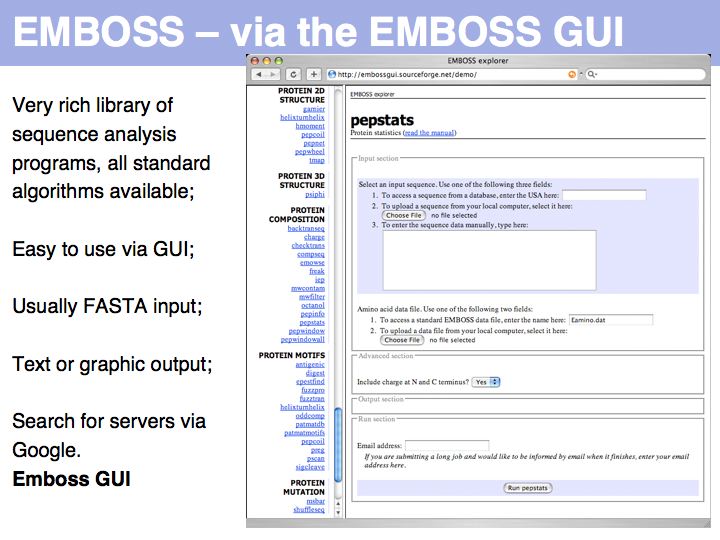
| − | [[Image:L03_s008.jpg|frame|none|Lecture 03, Slide 008]] | + | [[Image:L03_s008.jpg|frame|none|Lecture 03, Slide 008<br> |
| + | Simple tools exist to conveniently calculate compositional properties for peptide sequences. For example the EMBOSS GUI serves [http://emboss.sourceforge.net/ EMBOSS] tools on the Web on [http://www.google.com/search?&rls=en&q=%22EMBOSS+GUI%22 many freely accessible servers] in the world. One of these tools ist the '''pepstats''' routine that was used to create the output above. | ||
| + | ]] | ||
======Slide 009====== | ======Slide 009====== | ||
| − | [[Image:L03_s009.jpg|frame|none|Lecture 03, Slide 009]] | + | [[Image:L03_s009.jpg|frame|none|Lecture 03, Slide 009<br> |
| + | |||
| + | ]] | ||
======Slide 010====== | ======Slide 010====== | ||
| − | [[Image:L03_s010.jpg|frame|none|Lecture 03, Slide 010]] | + | [[Image:L03_s010.jpg|frame|none|Lecture 03, Slide 010<br> |
| + | Comparison of our unknown text (the German translation of "What's in a name ...") with letter frequencies of German, English and French shows a tendency towards the German origins but we can't immediately say that this is statistically significant. It is, after all, a very short sample. It is interesting to consider the outliers W (for reasons of [http://en.wikipedia.org/wiki/Alliteration alliteration]) and I and S (for [http://en.wikipedia.org/wiki/Assonance assonance]). Both poetic devices can be regarded to create constraints on the choice of words that will lead to deviations from expected distributions. | ||
| + | ]] | ||
======Slide 011====== | ======Slide 011====== | ||
| − | [[Image:L03_s011.jpg|frame|none|Lecture 03, Slide 011]] | + | [[Image:L03_s011.jpg|frame|none|Lecture 03, Slide 011<br> |
| + | This has a corollary in the non-random distribution of amino acids across species. Some of this may be due to physicochemical properties of amino acids in a particular ecological niche, but such effects may also be due to chance characteristics of the biochemical machinery of replication and translation. | ||
| + | ]] | ||
======Slide 012====== | ======Slide 012====== | ||
| − | [[Image:L03_s012.jpg|frame|none|Lecture 03, Slide 012]] | + | [[Image:L03_s012.jpg|frame|none|Lecture 03, Slide 012<br> |
| + | Sometimes the excess or depletion of a component may carry important information. In the example above case, the choice of words has been dictated by their letter composition. The poem [http://www.ubu.com/contemp/bok/eunoia_final.html Eunoia] is an example of a univocalic [http://en.wikipedia.org/wiki/Lipogram lipogram], a form of [http://en.wikipedia.org/wiki/Concrete_poetry concrete poetry], in which the author has constrained himself to use only a single vowel in each of the poem's chapters. | ||
| + | ]] | ||
======Slide 013====== | ======Slide 013====== | ||
| − | [[Image:L03_s013.jpg|frame|none|Lecture 03, Slide 013]] | + | [[Image:L03_s013.jpg|frame|none|Lecture 03, Slide 013<br> |
| + | The atypical distribution and clustering of particular amino acids suggests consequences for folding and interactions of the encoded protein. | ||
| + | ]] | ||
======Slide 014====== | ======Slide 014====== | ||
| − | [[Image:L03_s014.jpg|frame|none|Lecture 03, Slide 014]] | + | [[Image:L03_s014.jpg|frame|none|Lecture 03, Slide 014<br> |
| + | In this graph, amino acids have been ordered according to their standard frequencies in proteins (blue) to emphasize deviations in a particular protein (white). The sequence of the [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&id=6325066 single stranded RNA binding protein Nab3p] is remarkable, possessing long tracts of Glutamine and Glutamic acid and an excess of proline. Just looking at these anomalies allows us to infer that large tracts of sequence are likely not structured (and thus fulfill their function as an adaptable, flexible polypeptide) and that large segments are highly negatively charged (and thus presumably have a highly positively charged ligand, such as the (+)charges of exposed, single stranded nucleotide bases). | ||
| + | ]] | ||
======Slide 015====== | ======Slide 015====== | ||
| − | [[Image:L03_s015.jpg|frame|none|Lecture 03, Slide 015]] | + | [[Image:L03_s015.jpg|frame|none|Lecture 03, Slide 015<br> |
| + | |||
| + | ]] | ||
======Slide 016====== | ======Slide 016====== | ||
| − | [[Image:L03_s016.jpg|frame|none|Lecture 03, Slide 016]] | + | [[Image:L03_s016.jpg|frame|none|Lecture 03, Slide 016<br> |
| + | A sequence is fundamentally different from an unordered set, since it places its components into a context. Here is where biology differs from human language: A pattern with a different sequence is a different pattern. Constraints on patterns can be structural or functional. | ||
| + | ]] | ||
======Slide 017====== | ======Slide 017====== | ||
| − | [[Image:L03_s017.jpg|frame|none|Lecture 03, Slide 017]] | + | [[Image:L03_s017.jpg|frame|none|Lecture 03, Slide 017<br> |
| + | |||
| + | ]] | ||
======Slide 018====== | ======Slide 018====== | ||
| − | [[Image:L03_s018.jpg|frame|none|Lecture 03, Slide 018]] | + | [[Image:L03_s018.jpg|frame|none|Lecture 03, Slide 018<br> |
| + | |||
| + | ]] | ||
======Slide 019====== | ======Slide 019====== | ||
| − | [[Image:L03_s019.jpg|frame|none|Lecture 03, Slide 019]] | + | [[Image:L03_s019.jpg|frame|none|Lecture 03, Slide 019<br> |
| + | |||
| + | ]] | ||
======Slide 020====== | ======Slide 020====== | ||
| − | [[Image:L03_s020.jpg|frame|none|Lecture 03, Slide 020]] | + | [[Image:L03_s020.jpg|frame|none|Lecture 03, Slide 020<br> |
| + | Restriction endonucleases are the quintessential pattern recognition molecules. They bind strongly the specific conformation of DNA that is associated with a particular DNA sequence. Even though the structural differences between DNA strands of similar sequence is small, evolutionary pressure has resulted in enzymes that are highly specific for their cognate sequence. An excellent site for endonuclease information is [http://rebase.neb.com/ '''Rebase''']. | ||
| + | ]] | ||
======Slide 021====== | ======Slide 021====== | ||
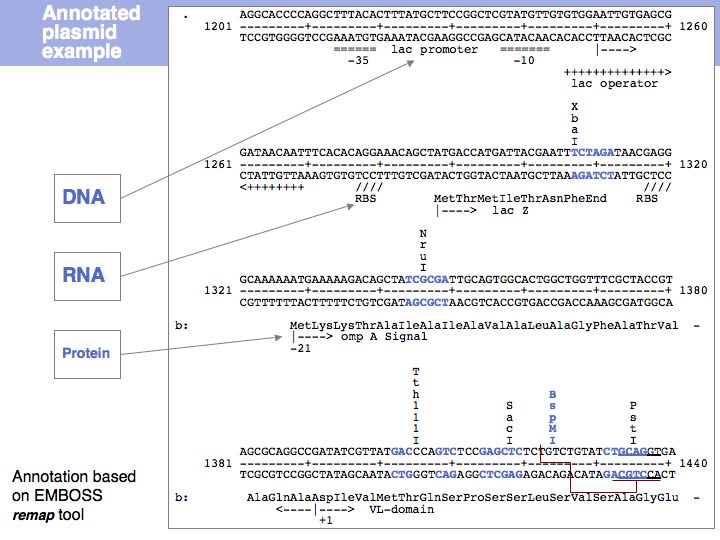
| − | [[Image:L03_s021.jpg|frame|none|Lecture 03, Slide 021]] | + | [[Image:L03_s021.jpg|frame|none|Lecture 03, Slide 021<br> |
| + | Sequence maps collect annotations from various sources - for plasmids in the laboratory either in linear form (based on the '''remap''' tool of the EMBOSS suite) ... | ||
| + | ]] | ||
======Slide 022====== | ======Slide 022====== | ||
| − | [[Image:L03_s022.jpg|frame|none|Lecture 03, Slide 022]] | + | [[Image:L03_s022.jpg|frame|none|Lecture 03, Slide 022<br> |
| + | ... or as a circular map, e.g. from [http://wishart.biology.ualberta.ca/PlasMapper/index.html PlasMapper]. | ||
| + | ]] | ||
======Slide 023====== | ======Slide 023====== | ||
| − | [[Image:L03_s023.jpg|frame|none|Lecture 03, Slide 023]] | + | [[Image:L03_s023.jpg|frame|none|Lecture 03, Slide 023<br> |
| + | |||
| + | ]] | ||
======Slide 024====== | ======Slide 024====== | ||
| − | [[Image:L03_s024.jpg|frame|none|Lecture 03, Slide 024]] | + | [[Image:L03_s024.jpg|frame|none|Lecture 03, Slide 024<br> |
| + | [http://en.wikipedia.org/wiki/Pattern_matching '''Pattern search''' ''(or pattern matching)''] means inspecting an entity and stating whether that entity is an example of a given pattern. Usually the entity is a substring of a ''sequence'', but patterns in ''protein structure'', biological ''networks'' or ''morphogenesis'' can also be computationally defined. '''Pattern discovery''' means finding patterns that have not been defined ''a priori''. | ||
| + | ]] | ||
======Slide 025====== | ======Slide 025====== | ||
| − | [[Image:L03_s025.jpg|frame|none|Lecture 03, Slide 025]] | + | [[Image:L03_s025.jpg|frame|none|Lecture 03, Slide 025<br> |
| + | Deterministic pattern search is a well understood field of computer science. Much more elegant solutions than "brute force" search exist ... | ||
| + | ]] | ||
======Slide 026====== | ======Slide 026====== | ||
| − | [[Image:L03_s026.jpg|frame|none|Lecture 03, Slide 026]] | + | [[Image:L03_s026.jpg|frame|none|Lecture 03, Slide 026<br> |
| + | ... such as the [http://en.wikipedia.org/wiki/Boyer–Moore_string_search_algorithm Boyer-Moore] algorithm. For a step-by-step version see [http://www.cs.utexas.edu/users/moore/best-ideas/string-searching/fstrpos-example.html '''here''']. Defining optimal algorithms and analyzing their resource requirements is the domain of computer science. | ||
| + | ]] | ||
======Slide 027====== | ======Slide 027====== | ||
| − | [[Image:L03_s027.jpg|frame|none|Lecture 03, Slide 027]] | + | [[Image:L03_s027.jpg|frame|none|Lecture 03, Slide 027<br> |
| + | If searches are to be repeated, precomputed '''index trees''' are much faster than examining the entire sequence. Simply look up where a pattern could be. Time (and storage space) invested in constructing the index pays off manyfold for every lookup. | ||
| + | ]] | ||
======Slide 028====== | ======Slide 028====== | ||
| − | [[Image:L03_s028.jpg|frame|none|Lecture 03, Slide 028]] | + | [[Image:L03_s028.jpg|frame|none|Lecture 03, Slide 028<br> |
| + | |||
| + | ]] | ||
======Slide 029====== | ======Slide 029====== | ||
| − | [[Image:L03_s029.jpg|frame|none|Lecture 03, Slide 029]] | + | [[Image:L03_s029.jpg|frame|none|Lecture 03, Slide 029<br> |
| + | To be able search for patterns we need a convention to define them. In particular, we would like to be able to find degenerate patterns: patterns in which we allow a number of alternative choices for particular positions. Such patterns are commonly written as [http://en.wikipedia.org/wiki/Regular_expression '''Regular Expressions'''] (even though some sites, such as the [http://www.expasy.org/prosite '''ProSite database'''] use a custom variant of the concept). | ||
| + | ]] | ||
| + | |||
======Slide 030====== | ======Slide 030====== | ||
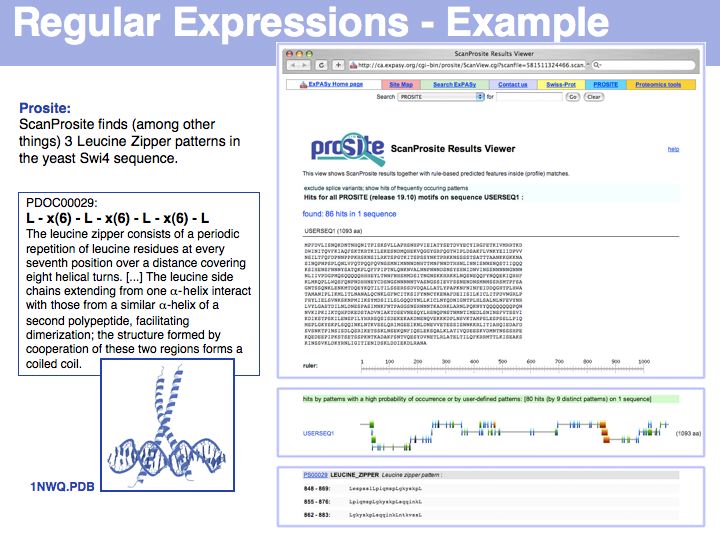
| − | [[Image:L03_s030.jpg|frame|none|Lecture 03, Slide 030]] | + | [[Image:L03_s030.jpg|frame|none|Lecture 03, Slide 030<br> |
| + | Here is an example of regular expression searching: the leucine zipper, a protein dimerization element found frequently in transcription factors is defined by PROSITE as <tt>'''L-x(6)-L-x(6)-L-x(6)-L'''</tt>. | ||
| + | ]] | ||
| + | |||
======Slide 031====== | ======Slide 031====== | ||
| − | [[Image:L03_s031.jpg|frame|none|Lecture 03, Slide 031]] | + | [[Image:L03_s031.jpg|frame|none|Lecture 03, Slide 031<br> |
| + | A crude Perl program to find the Leucine Zipper pattern uses a regular expression at its core. <tt>'''L.{6}){3,}L'''</tt> means: a string matching an "L", followed by 6 occurrences of any character ("."), repeated three or more times, and terminated by a final "L". (The arcane-looking <tt>print</tt> statement is just there to capture the sequence number of the pattern.) | ||
| + | ]] | ||
| + | |||
======Slide 032====== | ======Slide 032====== | ||
| − | [[Image:L03_s032.jpg|frame|none|Lecture 03, Slide 032]] | + | [[Image:L03_s032.jpg|frame|none|Lecture 03, Slide 032<br> |
| + | Having this as a Perl program on the computer makes it trivially easy to adjust the query, for example to allow any of the amino acids V, I, L, or M in the pattern and thus find examples that may be functionally related to the core leucine zipper. | ||
| + | ]] | ||
| + | ======Slide 033====== | ||
| + | [[Image:L03_s033.jpg|frame|none|Lecture 03, Slide 033<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 034====== | ||
| + | [[Image:L03_s034.jpg|frame|none|Lecture 03, Slide 034<br> | ||
| + | see http://www.expasy.org/tools | ||
| + | ]] | ||
| + | ======Slide 035====== | ||
| + | [[Image:L03_s035.jpg|frame|none|Lecture 03, Slide 035<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 036====== | ||
| + | [[Image:L03_s036.jpg|frame|none|Lecture 03, Slide 036<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 037====== | ||
| + | [[Image:L03_s037.jpg|frame|none|Lecture 03, Slide 037<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 038====== | ||
| + | [[Image:L03_s038.jpg|frame|none|Lecture 03, Slide 038<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 039====== | ||
| + | [[Image:L03_s039.jpg|frame|none|Lecture 03, Slide 039<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 040====== | ||
| + | [[Image:L03_s040.jpg|frame|none|Lecture 03, Slide 040<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 041====== | ||
| + | [[Image:L03_s041.jpg|frame|none|Lecture 03, Slide 041<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 042====== | ||
| + | [[Image:L03_s042.jpg|frame|none|Lecture 03, Slide 042<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 043====== | ||
| + | [[Image:L03_s043.jpg|frame|none|Lecture 03, Slide 043<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 044====== | ||
| + | [[Image:L03_s044.jpg|frame|none|Lecture 03, Slide 044<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 045====== | ||
| + | [[Image:L03_s045.jpg|frame|none|Lecture 03, Slide 045<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 046====== | ||
| + | [[Image:L03_s046.jpg|frame|none|Lecture 03, Slide 046<br> | ||
| + | |||
| + | ]] | ||
| + | |||
| + | |||
| + | | ||
| + | ---- | ||
| + | <small>[[Lecture_02|(Previous lecture)]] ... [[Lecture_04|(Next lecture)]]</small> | ||
Latest revision as of 16:21, 17 November 2007
(Previous lecture) ... (Next lecture)
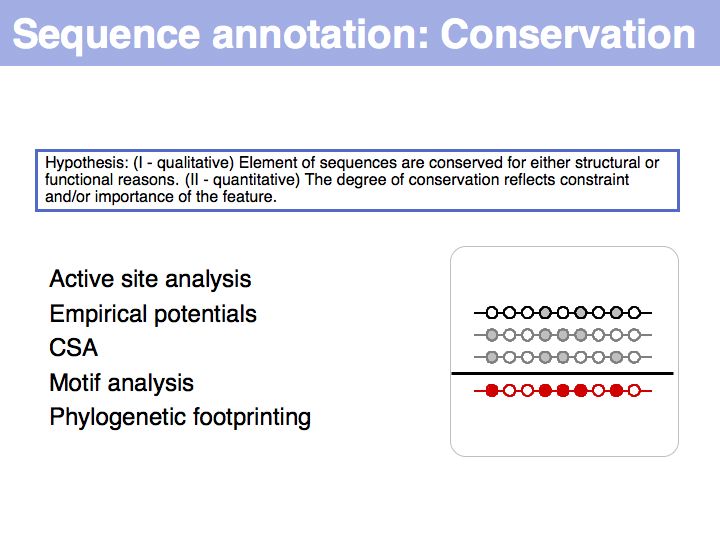
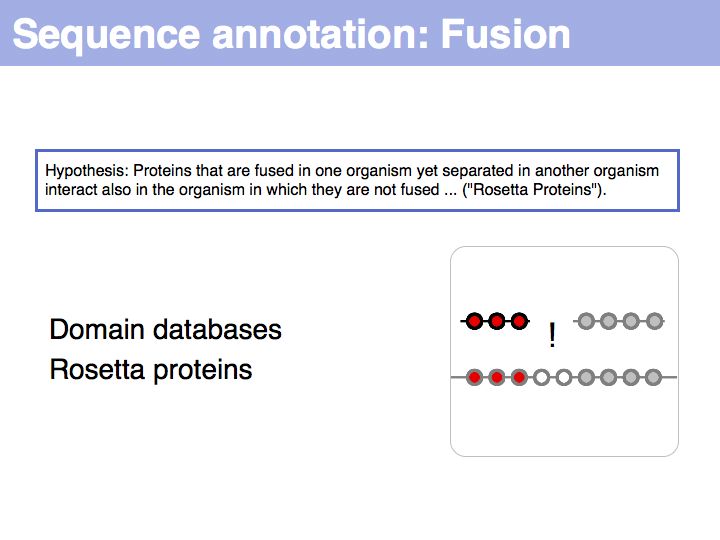
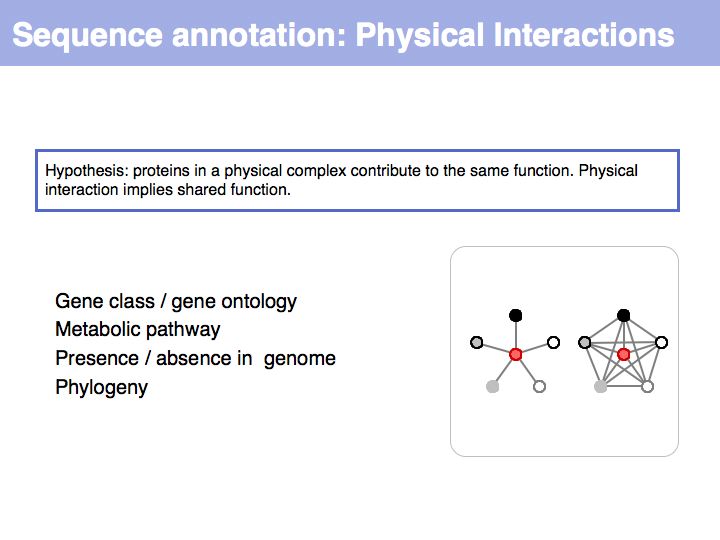
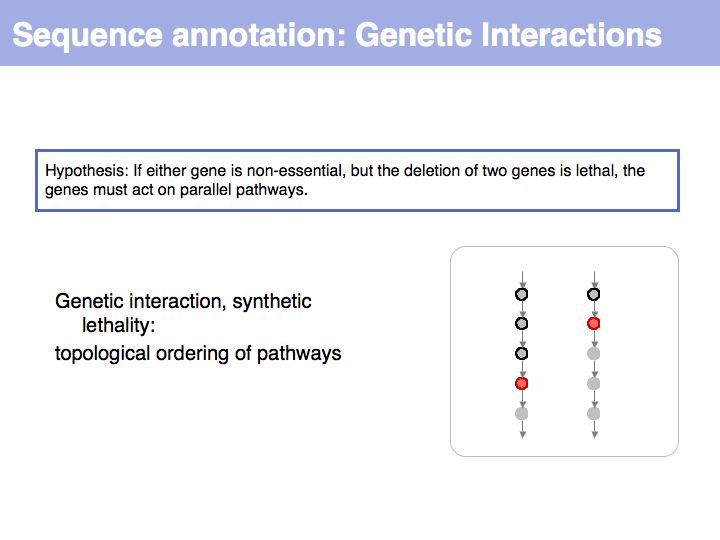
Sequence Properties
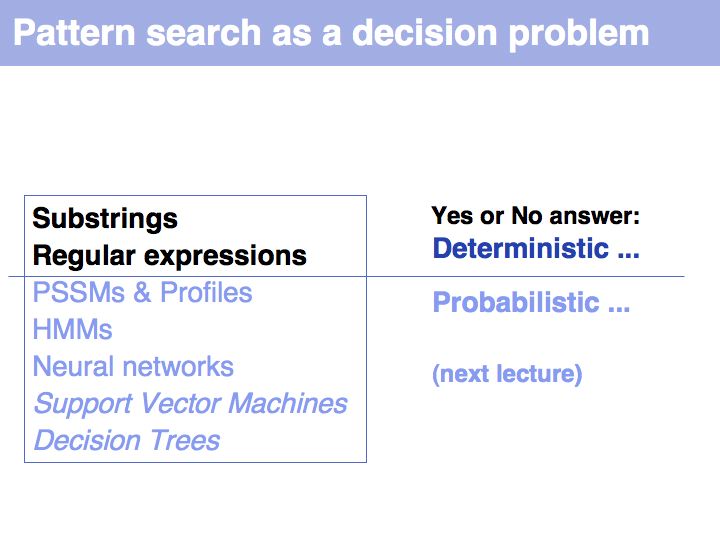
- What you should take home from this part of the course
- Understand the ideas of analysis by composition and analysis by signal;
- Know what deterministic pattern matching is;
- Recognize and understand the term regular expression;
- Be familiar with common sequence signals in DNA,RNA and proteins;
- Be familiar with the Prosite database and the Prosite scan server;
- Kow where to find EMBOSS tools and how to use them;
- Know about the offerings on the ExPASy tools collection page;
- Work on an understanding how biological facts can be translated into hypotheses and how hypotheses can be translated into computational procedures for analysis.
- Links summary
- Exercises
- Retrieve and read the Prosite documentation entry for the Leucine Zipper.
- Download entry 1NWQ from the PDB, visualize the Leucine Zipper with VMD and study its architecture (stereo vision!).
Lecture Slides
Slide 001

Lecture 03, Slide 001
This finding made the news. You should be aware of important new developments: subscribe to read at least the news items from Nature and Science, preferably subscribe to and browse their tables of contents too. In this particular new finding, researchers challenge our current concept of "genome": what is a genome, if the same physical DNA molecule can contain coding information for more than one species? Also, this finding further emphasizes the importance of horizontal gene transfer in evolution.
This finding made the news. You should be aware of important new developments: subscribe to read at least the news items from Nature and Science, preferably subscribe to and browse their tables of contents too. In this particular new finding, researchers challenge our current concept of "genome": what is a genome, if the same physical DNA molecule can contain coding information for more than one species? Also, this finding further emphasizes the importance of horizontal gene transfer in evolution.
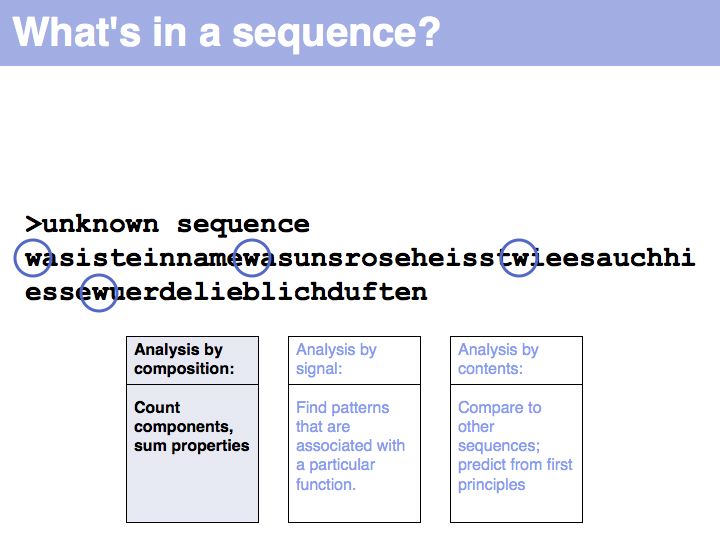
Slide 002
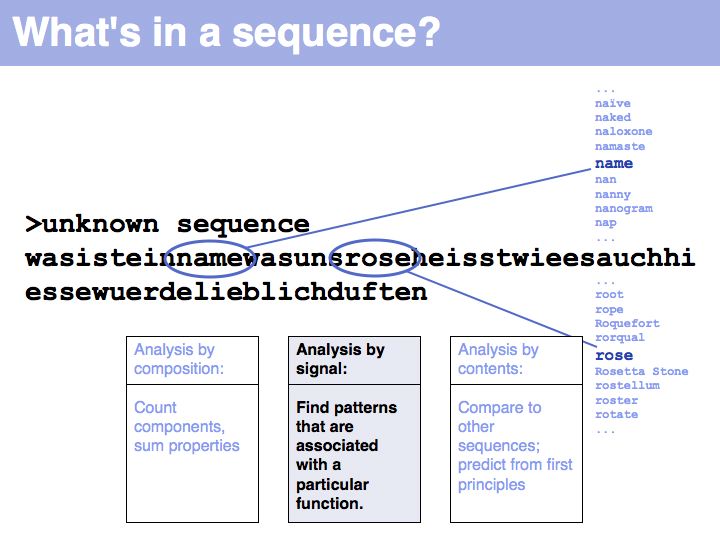
Slide 003
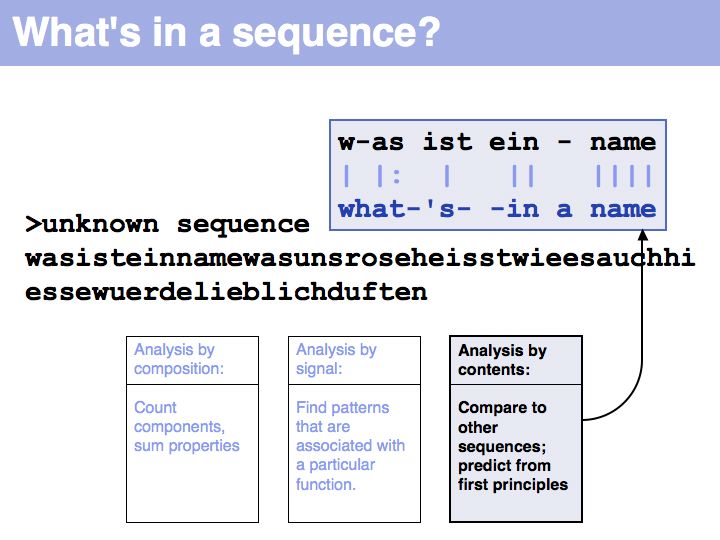
Slide 004
Slide 005
Slide 006
Slide 007

Lecture 03, Slide 007
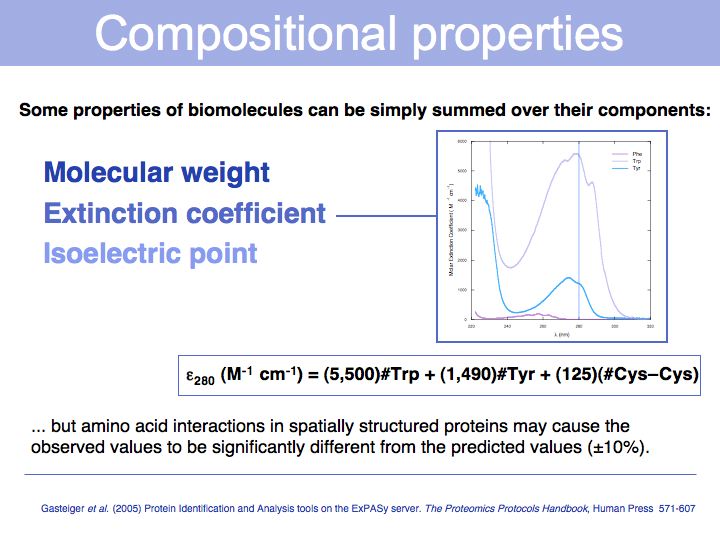
(#: Number of ...) A protein's isoelectric point depends on the pK values of the amino acids; the pK values characterize the propensity fo an amino acid sidechain to dissociate, which in turn depends on how energetically favourable dissociation is. For example: since a negatively charged amino acid will be stabilized in a positive electrostatic field, such a field will shift a pK value down. This means the pH value at which the side chain will be 50% ionized is lower, or in other words, in a positive electrostatic field the concentration of protons must be higher to keep a proton associated to the sidechain.
Compositional properties of nucleic acids include hybridization temperature and helix structure.
(#: Number of ...) A protein's isoelectric point depends on the pK values of the amino acids; the pK values characterize the propensity fo an amino acid sidechain to dissociate, which in turn depends on how energetically favourable dissociation is. For example: since a negatively charged amino acid will be stabilized in a positive electrostatic field, such a field will shift a pK value down. This means the pH value at which the side chain will be 50% ionized is lower, or in other words, in a positive electrostatic field the concentration of protons must be higher to keep a proton associated to the sidechain.
Compositional properties of nucleic acids include hybridization temperature and helix structure.
Slide 008

Lecture 03, Slide 008
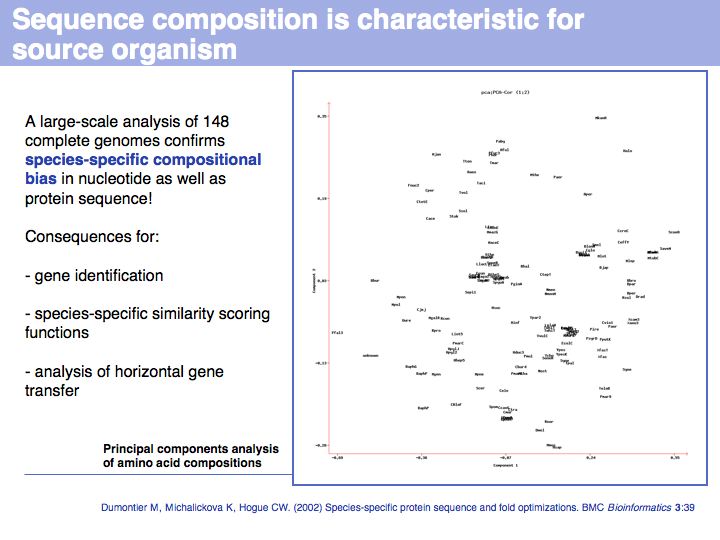
Simple tools exist to conveniently calculate compositional properties for peptide sequences. For example the EMBOSS GUI serves EMBOSS tools on the Web on many freely accessible servers in the world. One of these tools ist the pepstats routine that was used to create the output above.
Simple tools exist to conveniently calculate compositional properties for peptide sequences. For example the EMBOSS GUI serves EMBOSS tools on the Web on many freely accessible servers in the world. One of these tools ist the pepstats routine that was used to create the output above.
Slide 009
Slide 010

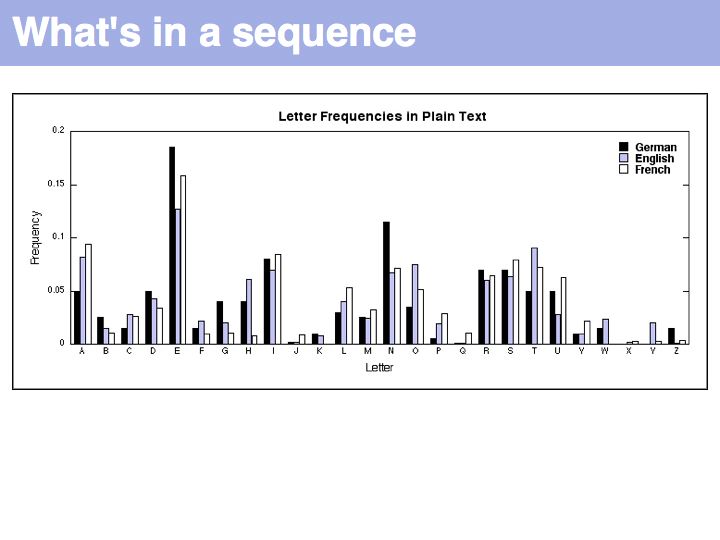
Lecture 03, Slide 010
Comparison of our unknown text (the German translation of "What's in a name ...") with letter frequencies of German, English and French shows a tendency towards the German origins but we can't immediately say that this is statistically significant. It is, after all, a very short sample. It is interesting to consider the outliers W (for reasons of alliteration) and I and S (for assonance). Both poetic devices can be regarded to create constraints on the choice of words that will lead to deviations from expected distributions.
Comparison of our unknown text (the German translation of "What's in a name ...") with letter frequencies of German, English and French shows a tendency towards the German origins but we can't immediately say that this is statistically significant. It is, after all, a very short sample. It is interesting to consider the outliers W (for reasons of alliteration) and I and S (for assonance). Both poetic devices can be regarded to create constraints on the choice of words that will lead to deviations from expected distributions.
Slide 011

Lecture 03, Slide 011
This has a corollary in the non-random distribution of amino acids across species. Some of this may be due to physicochemical properties of amino acids in a particular ecological niche, but such effects may also be due to chance characteristics of the biochemical machinery of replication and translation.
This has a corollary in the non-random distribution of amino acids across species. Some of this may be due to physicochemical properties of amino acids in a particular ecological niche, but such effects may also be due to chance characteristics of the biochemical machinery of replication and translation.
Slide 012

Lecture 03, Slide 012
Sometimes the excess or depletion of a component may carry important information. In the example above case, the choice of words has been dictated by their letter composition. The poem Eunoia is an example of a univocalic lipogram, a form of concrete poetry, in which the author has constrained himself to use only a single vowel in each of the poem's chapters.
Sometimes the excess or depletion of a component may carry important information. In the example above case, the choice of words has been dictated by their letter composition. The poem Eunoia is an example of a univocalic lipogram, a form of concrete poetry, in which the author has constrained himself to use only a single vowel in each of the poem's chapters.
Slide 013
Slide 014

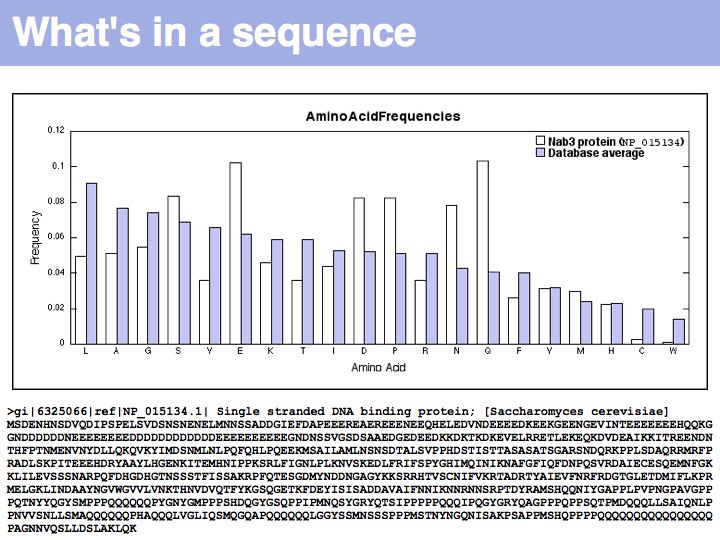
Lecture 03, Slide 014
In this graph, amino acids have been ordered according to their standard frequencies in proteins (blue) to emphasize deviations in a particular protein (white). The sequence of the single stranded RNA binding protein Nab3p is remarkable, possessing long tracts of Glutamine and Glutamic acid and an excess of proline. Just looking at these anomalies allows us to infer that large tracts of sequence are likely not structured (and thus fulfill their function as an adaptable, flexible polypeptide) and that large segments are highly negatively charged (and thus presumably have a highly positively charged ligand, such as the (+)charges of exposed, single stranded nucleotide bases).
In this graph, amino acids have been ordered according to their standard frequencies in proteins (blue) to emphasize deviations in a particular protein (white). The sequence of the single stranded RNA binding protein Nab3p is remarkable, possessing long tracts of Glutamine and Glutamic acid and an excess of proline. Just looking at these anomalies allows us to infer that large tracts of sequence are likely not structured (and thus fulfill their function as an adaptable, flexible polypeptide) and that large segments are highly negatively charged (and thus presumably have a highly positively charged ligand, such as the (+)charges of exposed, single stranded nucleotide bases).
Slide 015
Slide 016
Slide 017
Slide 018
Slide 019
Slide 020

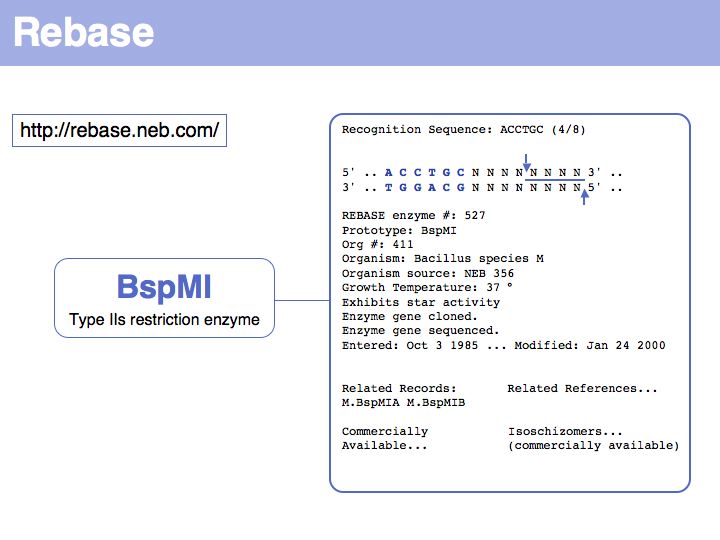
Lecture 03, Slide 020
Restriction endonucleases are the quintessential pattern recognition molecules. They bind strongly the specific conformation of DNA that is associated with a particular DNA sequence. Even though the structural differences between DNA strands of similar sequence is small, evolutionary pressure has resulted in enzymes that are highly specific for their cognate sequence. An excellent site for endonuclease information is Rebase.
Restriction endonucleases are the quintessential pattern recognition molecules. They bind strongly the specific conformation of DNA that is associated with a particular DNA sequence. Even though the structural differences between DNA strands of similar sequence is small, evolutionary pressure has resulted in enzymes that are highly specific for their cognate sequence. An excellent site for endonuclease information is Rebase.
Slide 021
Slide 022

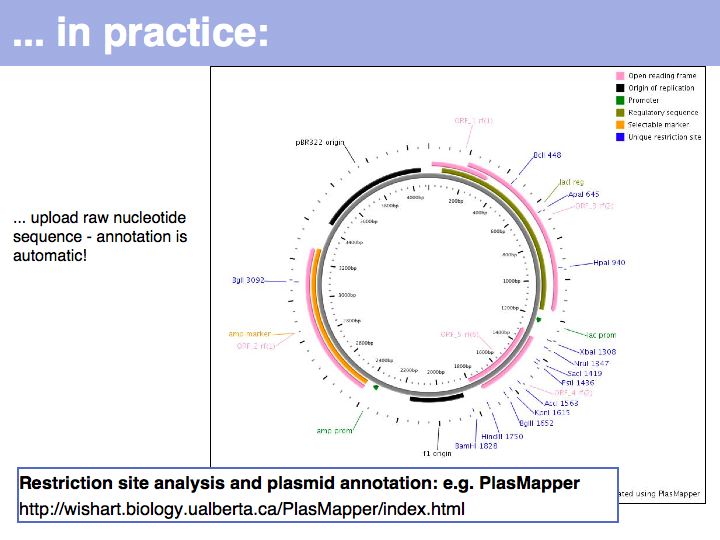
Lecture 03, Slide 022
... or as a circular map, e.g. from PlasMapper.
... or as a circular map, e.g. from PlasMapper.
Slide 023
Slide 024

Lecture 03, Slide 024
Pattern search (or pattern matching) means inspecting an entity and stating whether that entity is an example of a given pattern. Usually the entity is a substring of a sequence, but patterns in protein structure, biological networks or morphogenesis can also be computationally defined. Pattern discovery means finding patterns that have not been defined a priori.
Pattern search (or pattern matching) means inspecting an entity and stating whether that entity is an example of a given pattern. Usually the entity is a substring of a sequence, but patterns in protein structure, biological networks or morphogenesis can also be computationally defined. Pattern discovery means finding patterns that have not been defined a priori.
Slide 025
Slide 026

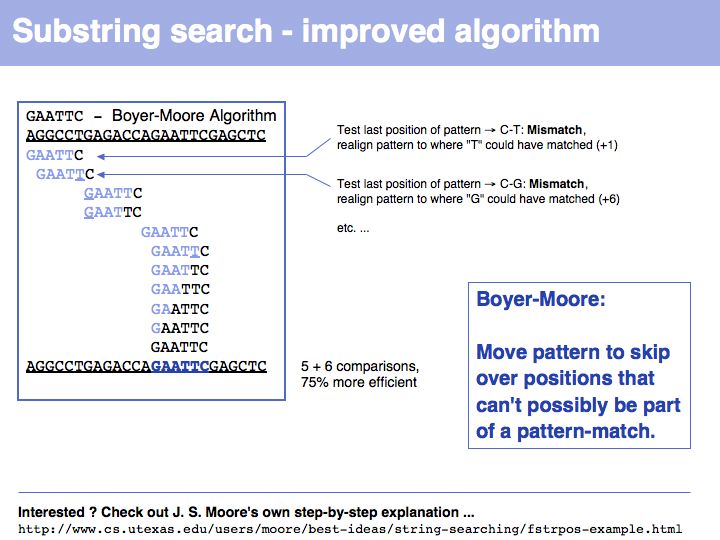
Lecture 03, Slide 026
... such as the Boyer-Moore algorithm. For a step-by-step version see here. Defining optimal algorithms and analyzing their resource requirements is the domain of computer science.
... such as the Boyer-Moore algorithm. For a step-by-step version see here. Defining optimal algorithms and analyzing their resource requirements is the domain of computer science.
Slide 027
Slide 028
Slide 029

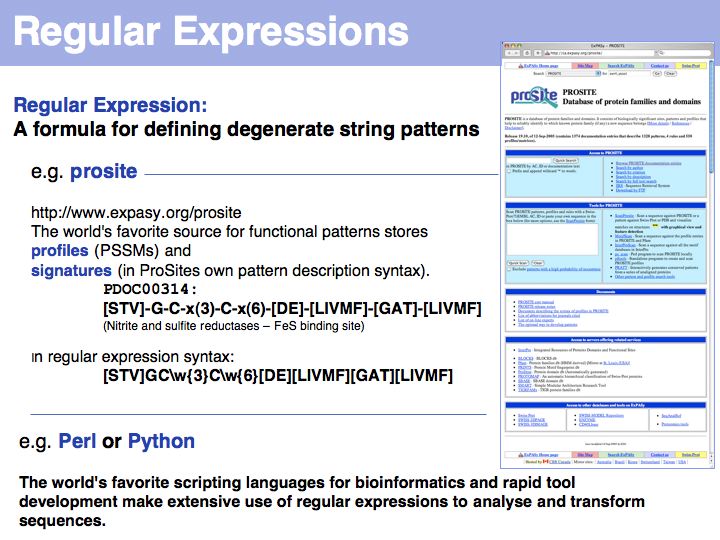
Lecture 03, Slide 029
To be able search for patterns we need a convention to define them. In particular, we would like to be able to find degenerate patterns: patterns in which we allow a number of alternative choices for particular positions. Such patterns are commonly written as Regular Expressions (even though some sites, such as the ProSite database use a custom variant of the concept).
To be able search for patterns we need a convention to define them. In particular, we would like to be able to find degenerate patterns: patterns in which we allow a number of alternative choices for particular positions. Such patterns are commonly written as Regular Expressions (even though some sites, such as the ProSite database use a custom variant of the concept).
Slide 030
Slide 031

Lecture 03, Slide 031
A crude Perl program to find the Leucine Zipper pattern uses a regular expression at its core. L.{6}){3,}L means: a string matching an "L", followed by 6 occurrences of any character ("."), repeated three or more times, and terminated by a final "L". (The arcane-looking print statement is just there to capture the sequence number of the pattern.)
A crude Perl program to find the Leucine Zipper pattern uses a regular expression at its core. L.{6}){3,}L means: a string matching an "L", followed by 6 occurrences of any character ("."), repeated three or more times, and terminated by a final "L". (The arcane-looking print statement is just there to capture the sequence number of the pattern.)
Slide 032
Slide 033
Slide 034

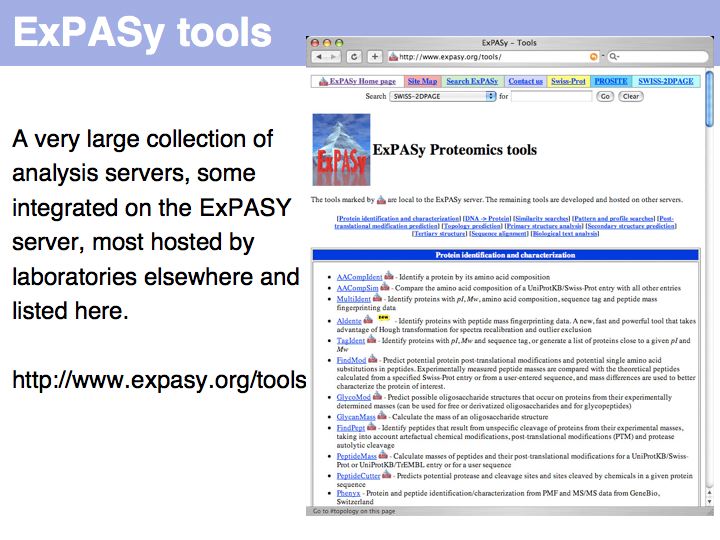
Lecture 03, Slide 034
see http://www.expasy.org/tools
see http://www.expasy.org/tools
Slide 035
Slide 036
Slide 037
Slide 038
Slide 039
Slide 040
Slide 041
Slide 042
Slide 043
Slide 044
Slide 045
Slide 046