Difference between revisions of "Bioinformatics Introduction Structure"
m |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 13: | Line 13: | ||
{{Vspace}} | {{Vspace}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{Vspace}} | {{Vspace}} | ||
| Line 517: | Line 509: | ||
| − | We have spent a significant amount of time to prepare data for the analysis and in practice it usually seems to turn out that way, that the preparation of data occupies the greatest part of our efforts. The actual computational analysis is generally quite fast. And, unfortunately, the '''interpretation of results''' is often somewhat neglected. Don't be that way. Data does not explain itself. The | + | We have spent a significant amount of time to prepare data for the analysis and in practice it usually seems to turn out that way, that the preparation of data occupies the greatest part of our efforts. The actual computational analysis is generally quite fast. And, unfortunately, the '''interpretation of results''' is often somewhat neglected. Don't be that way. Data does not explain itself. The interpretation of your computational results is the most important part. |
We will look at our homology model with two different questions: | We will look at our homology model with two different questions: | ||
| Line 524: | Line 516: | ||
* Can we tell which residues are conserved for functional reasons, rather than for structural reasons? | * Can we tell which residues are conserved for functional reasons, rather than for structural reasons? | ||
| + | {{Vspace}} | ||
| − | + | === The PDB file === | |
| − | |||
| − | |||
| − | |||
{{task|1= | {{task|1= | ||
| Line 540: | Line 530: | ||
}} | }} | ||
| + | As you have seen above, SwissModel numbers the first residue "1" and does not keep the numbering of the template. We should renumber the model so we can compare the model and the template with the same residue numbers. Carefully doing this by hand should not even take you a whole hour. You just need to edit all the residue numbers in a text editor to give the first residue the number "4" and increment numbers from there. | ||
| − | + | Or we could do this with an '''R''' script. | |
| − | |||
| − | |||
{{task|1= | {{task|1= | ||
| − | + | * Renumber your '''model''' PDB file to start with residue number 4 with the code in section <code>PART SEVEN: RENUMBERING THE MODEL</code> of the <code>Structure.R</code> script. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
===First visualization=== | ===First visualization=== | ||
| − | |||
| − | |||
Since a homology model inherits its structural details from the '''template''', your model of the YFO sequence should look very similar to the original 1BM8 structure. | Since a homology model inherits its structural details from the '''template''', your model of the YFO sequence should look very similar to the original 1BM8 structure. | ||
| Line 612: | Line 550: | ||
# In the '''Favourites''' → '''Model Panel''' window you can switch between the two molecules. | # In the '''Favourites''' → '''Model Panel''' window you can switch between the two molecules. | ||
# Hide the ribbon and choose '''backbone only → full'''. You will note that the backbone of the two structures is virtually identical. | # Hide the ribbon and choose '''backbone only → full'''. You will note that the backbone of the two structures is virtually identical. | ||
| − | # Next, choose '''Actions → Atoms/Bonds → show''' to | + | # Next, choose '''Actions → Atoms/Bonds → show''' to display the two molecules in a stick style and note how the sidechains have been modeled. Note especially how sidechain coordinates have been guessed, where the template had shorter sidechains than the target. It may be more clear if you hide H-atoms: '''Select → Chemistry → Element → H''' and '''Actions → Atoms/Bonds → hide''' |
# Display only residue 50 to 74 to focus on the putative helix-turn-helix domain. You can drag your mouse in the '''Favourites → Sequence''', window to select the range then '''Select → Invert (selected model)''' and '''Actions → Atoms/Bonds → hide'''. Or you can use Chimera's commandline: <code>~display</code> to undisplay everything, <code>show #:50-74</code> to show this residue range for all models. | # Display only residue 50 to 74 to focus on the putative helix-turn-helix domain. You can drag your mouse in the '''Favourites → Sequence''', window to select the range then '''Select → Invert (selected model)''' and '''Actions → Atoms/Bonds → hide'''. Or you can use Chimera's commandline: <code>~display</code> to undisplay everything, <code>show #:50-74</code> to show this residue range for all models. | ||
| − | # Study the result: a model of the HTH subdomain of | + | # Study the result: a model of the HTH subdomain of Mbp1_SPIPU. |
}} | }} | ||
| − | + | {{Vspace}} | |
| − | ==Coloring the model by energy == | + | ===Coloring the model by energy === |
SwissModel calculates energies for each residue of the model with a molecular mechanics forcefield. The SwissModel modeling summary page contains a plot of these energies as a function of sequence number like. The values - between 0.0 and 1.0 - are stored in the PDB file's B-factor field. | SwissModel calculates energies for each residue of the model with a molecular mechanics forcefield. The SwissModel modeling summary page contains a plot of these energies as a function of sequence number like. The values - between 0.0 and 1.0 - are stored in the PDB file's B-factor field. | ||
| Line 631: | Line 569: | ||
}} | }} | ||
| − | + | {{Vspace}} | |
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
==Modelling DNA binding== | ==Modelling DNA binding== | ||
| Line 645: | Line 579: | ||
Since there is currently no software available that would reliably model such a complex from first principles<ref>''Rosetta'' may get the structure approximately right, ''Autodock'' may get the complex approximately right, but the coordinate changes involved in induced fit makes the result unreliable - and we have no good way to validate whether the predicted complex is correct. </ref>, we will base a model of a bound complex on homology modelling as well. This means we need to find a similar structure for which the position of bound DNA is known, then superimpose that structure with our model. This places the DNA molecule into the spatial context of the model we are studying. It so happens that early in 2015 an APSES domain structure with bound DNA was published. You probably noticed it as a result of the PDB BLAST search: [http://www.rcsb.org/pdb/explore/explore.do?structureId=4UX5 '''4UX5'''], from the ''Magnaporthe oryzae'' Mbp1 orhologue PCG2<ref>{{#pmid: 25550425}}</ref>. | Since there is currently no software available that would reliably model such a complex from first principles<ref>''Rosetta'' may get the structure approximately right, ''Autodock'' may get the complex approximately right, but the coordinate changes involved in induced fit makes the result unreliable - and we have no good way to validate whether the predicted complex is correct. </ref>, we will base a model of a bound complex on homology modelling as well. This means we need to find a similar structure for which the position of bound DNA is known, then superimpose that structure with our model. This places the DNA molecule into the spatial context of the model we are studying. It so happens that early in 2015 an APSES domain structure with bound DNA was published. You probably noticed it as a result of the PDB BLAST search: [http://www.rcsb.org/pdb/explore/explore.do?structureId=4UX5 '''4UX5'''], from the ''Magnaporthe oryzae'' Mbp1 orhologue PCG2<ref>{{#pmid: 25550425}}</ref>. | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
===A homologous protein/DNA complex structure=== | ===A homologous protein/DNA complex structure=== | ||
| Line 678: | Line 610: | ||
}} | }} | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
===Superimposing your model=== | ===Superimposing your model=== | ||
Both your homology model and the template structure provide valuable information: | Both your homology model and the template structure provide valuable information: | ||
| + | |||
* The template structure shows how conserved the structure is at the protein/DNA interface. You have seen what subtle differences can give rise to a sequence specific complex and a non-specific binding mode. For Mbp1 we know that the APSES domain binds to the same cognate DNA sequence as PCG2. Since your model structure is heavily biased towards the template, evaluating the template in the context of a real protein/DNA complex allows you to judge which binding residues appear to be conserved and possibly modelled in an orientation that is productive for binding. | * The template structure shows how conserved the structure is at the protein/DNA interface. You have seen what subtle differences can give rise to a sequence specific complex and a non-specific binding mode. For Mbp1 we know that the APSES domain binds to the same cognate DNA sequence as PCG2. Since your model structure is heavily biased towards the template, evaluating the template in the context of a real protein/DNA complex allows you to judge which binding residues appear to be conserved and possibly modelled in an orientation that is productive for binding. | ||
| Line 694: | Line 625: | ||
* We need to visualize and evaluate differences in binding between different proteins and for me it works well to colour everything by element, and give the carbon atoms some identifying, distinct colour. This is best achieved through the Chimera command line that you can turn on with the little "computer" icon on the left-hand side of the graphics window. Have a look at the [https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/framecommand.html Chimera Users guide], and choose '''select''' to learn how Chimera's selection syntax works. | * We need to visualize and evaluate differences in binding between different proteins and for me it works well to colour everything by element, and give the carbon atoms some identifying, distinct colour. This is best achieved through the Chimera command line that you can turn on with the little "computer" icon on the left-hand side of the graphics window. Have a look at the [https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/framecommand.html Chimera Users guide], and choose '''select''' to learn how Chimera's selection syntax works. | ||
| − | * Open the Model Panel to check which protein has which Chimera-internal model number. Then you can use the following selection syntax. Instead of the model numbers, I will type <code>< | + | * Open the Model Panel to check which protein has which Chimera-internal model number. Then you can use the following selection syntax. Instead of the model numbers, I will type <code><SPIPU></code>, <code><4ux5></code>, and <code><1BM8></code> - you will certainly know by now that these are placeholder labels and you need to replace them with the numbers <code>0</code>, <code>1</code>, and <code>2</code> instead. |
:* To colour the DNA carbon atoms white, type:<br /> | :* To colour the DNA carbon atoms white, type:<br /> | ||
| Line 705: | Line 636: | ||
::<code>~display #<4ux5>:.B</code> <small>Note: this is the tilde character, not a hyphen or minus sign.</small> | ::<code>~display #<4ux5>:.B</code> <small>Note: this is the tilde character, not a hyphen or minus sign.</small> | ||
| − | :* To colour the | + | :* To colour the SPIPU model carbon atoms a pale reddish color, type:<br /> |
| − | ::<code>color #b06268 #< | + | ::<code>color #b06268 #<SPIPU> & C</code> |
:* To colour the 1BM8 structure carbon atoms a pale greenish color, type:<br /> | :* To colour the 1BM8 structure carbon atoms a pale greenish color, type:<br /> | ||
| Line 714: | Line 645: | ||
** Select all models in the Model Panel and click on '''match'''. | ** Select all models in the Model Panel and click on '''match'''. | ||
** Set 4ux5 Chain A as the Reference chain. | ** Set 4ux5 Chain A as the Reference chain. | ||
| − | ** Select | + | ** Select SPIPU as a '''Chain to match''', select the button for specific reference and specific match, and click '''Apply'''. |
** Repeat this with 1BM8 as the match chain. | ** Repeat this with 1BM8 as the match chain. | ||
| − | * Easy. Now enlarge the binding site. Remember that 4ux5 and 1bm8 are independently determined crystal structures, wheres | + | * Easy. Now enlarge the binding site. Remember that 4ux5 and 1bm8 are independently determined crystal structures, wheres SPIPU was modelled on 1bm8 and is expected to be '''very''' similar to it. To give you some guidance what you should focus on, select 4ux5 residue 84 CA atom and display it as '''Ball & Stick'''. You can also repeat the '''Action''' "Set Pivot in case the pivot has shifted. |
* Study the scene. This is where stereo- vision will help '''a lot'''. | * Study the scene. This is where stereo- vision will help '''a lot'''. | ||
| Line 726: | Line 657: | ||
* Now turn the display of 4ux5 chain B back on and turn chain A off instead. Then superimpose the 1BM8 template and your model on Chain B. | * Now turn the display of 4ux5 chain B back on and turn chain A off instead. Then superimpose the 1BM8 template and your model on Chain B. | ||
| − | |||
* Again, focus on the binding region. What do you think of that? What would you have expected? Do you see a difference? What does this all mean? | * Again, focus on the binding region. What do you think of that? What would you have expected? Do you see a difference? What does this all mean? | ||
| Line 733: | Line 663: | ||
}} | }} | ||
| + | {{Vspace}} | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
== Links and resources == | == Links and resources == | ||
| − | |||
| − | |||
| − | |||
| − | |||
=== The DNA binding site === | === The DNA binding site === | ||
| Line 769: | Line 693: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 960: | Line 707: | ||
{{#pmid: 10747782}} | {{#pmid: 10747782}} | ||
| − | + | :* [http://www.wwpdb.org/documentation/format23/v2.3.html '''PDB file format'''] (see the Coordinate Section if you are unsure about chain identifiers) | |
| − | + | :* [http://en.wikipedia.org/wiki/Structural_alignment Wikipedia on '''Structural Superposition'''] <small>(although the article is called "Structural Alignment")</small> | |
| − | |||
| − | |||
Latest revision as of 03:25, 8 January 2017
Structure
| Data | Sequence | Structure | Phylogeny | Function |
Contents
The Structure Unit
This Unit is part of a brief introduction to bioinformatics. The material is more or less interleaved with the Structure.R Project File which is part of the RStudio project associated with this material. Refer to the course/workshop page for installation instructions.
In the previous units we have discovered homologues of APSES domain containing proteins in all fungal species. This makes the domain an ancient protein family that had already duplicated to several paralogues at the time when the cenancestor of all fungi lived, more than 600,000,000 years ago, in the Vendian period of the Proterozoic era of Precambrian times.
In this unit we will explore the domain's 3-dimensional structure, learn how to compute features from structures, and how to map features computed elsewhere onto structures.
Molecular graphics: UCSF Chimera
To view molecular structures, we need a tool to visualize the three dimensional relationships of atoms. A molecular viewer is a program that takes 3D structure data and allows you to display and explore it. For a number of reasons, I use the UCSF Chimera viewer for this course:
- Chimera is free and open;
- It creates very appealing graphics;
- It is under ongoing development and is well maintained;
- It provides an array of useful utilities for structure analysis; and,
- besides an intuitive, menu driven interface, Chimera can be scripted via its command line, or even programmed via its in-built python interpreter.
Task:
- Access the Chimera homepage and navigate to the Download section.
- Find the the newest version for your platform in the table and click on the file to download it.
- Follow the instructions to install Chimera.
Let's explore Chimera functions first with a simple small molecule:
Modeling small molecules
"Small" molecules are solvent, ligands, substrates, products, prosthetic groups, drugs - in short, essentially everything that is not made by DNA-, RNA-polymerases or the ribosome. Whereas the biopolymers are still front and centre in our quest to understand molecular biology, small molecules are crucial for our quest to interact with the inventory of the cell, create useful products, or advance medicine.
A number of public repositories make small-molecule information available, such as PubChem at the NCBI, the ligand collection at the PDB, the ChEBI database at the European Bioinformatics Institute, the Canadian DrugBank, or the NCI database browser at the US National Cancer Institute. One general way to export topology information from these services is to use SMILES strings—a shorthand notation for the composition and topology of chemical compounds.
Task:
- Access PubChem.
- Enter "caffeine" as a search term in the Compound tab. A number of matches to this keyword search are returned.
- Click on the top hit - 1,3,7-Trimethylxanthine, the Caffeine molecule. Note that the page contains among other items:
- A 2D structural sketch;
- An idealized 3D structural conformer, for which you can download coordinates in several formats;
- The IUPAC name:
1,3,7-trimethylpurine-2,6-dione; - The CAS identifier
58-08-2which is a unique identifier and can be used as a cross-reference ID; - The SMILES strings
CN1C=NC2=C1C(=O)N(C(=O)N2C)C; - ... and much more.
That's great, but let's sketch our own version of caffeine. Several versions of Peter Ertl's Java Molecular Editor (JME) are offered online, PubChem offers this functionality via its Sketcher tool.
Task:
- Return to the PubChem homepage.
- Follow the link to Structure search (in the right hand menu).
- Click on the 3D conformer tab and on the Launch button to launch the molecular editor in its own window.
- Sketch the structure of caffeine. I find the editor quite intuitive but clicking on the Help button will give you a quick, structured overview. Make sure you define your double-bonds correctly.
- Export the SMILES string of your compound to your computer.
Translating SMILES to structure
Chimera can translate SMILES strings to coordinates[1].
Task:
- Open Chimera.
- Select Tools → Structure Editing → Build Structure.
- In the Build Structure window, select the SMILES string button, paste the string from your file, and click Apply.
- The caffeine molecule will be generated and visualized in the graphics window. This is a "stick" representation.
- You can rotate it with your mouse, <command> drag to scale, <shift> drag to translate.
- Use the Actions → Atoms/Bonds → ball & stick or sphere menu items to change appearance.
- Use the Actions → Color → by element menu to change colors.
- Change the display back to stick and use Actions → Surface → show to add a solvent accessible surface. Choosing this command triggers the calculation of the surface, which is then available as an individually selectable object. However, with default parameters the surface appears a bit rough for this small molecule.
- Change the parameters of this solvent accessible surface:
- Select the surface with <control><click> (<control><left mouse button> on windows). A green contour line appears around selected items – it surrounds the surface in this case.
- Open the selection inspector by clicking on the tiny green icon in the lower-right corner of the window (It has a magnifying glass symbol which means "inspect" for Chimera, not "search").
- Select Inspect ...MSMS surface and change the Vertex density value to 50.0 - hit return.
- By default, the surface inherits the colour of the atoms it envelopes. To change the colour of the surface, use the Actions → Color → all options menu. Click the surfaces button to indicate that the color choice should be applied to the surface object (note what else you can apply color to...), then choose cornflower blue.
- Use the Actions → Surface → transparency → 50% menu to see atoms and bonds that are covered by the surface.
- To begin working with molecules in "true" 3D, choose Tools → Viewing Controls → Camera and select camera mode → wall-eye stereo. Also, use the Effects tab of the Viewing window, and check shadows off.
- Your structure should look about like what you see below. Save your session with the File → Save Session dialogue so you can easily recreate the scene.
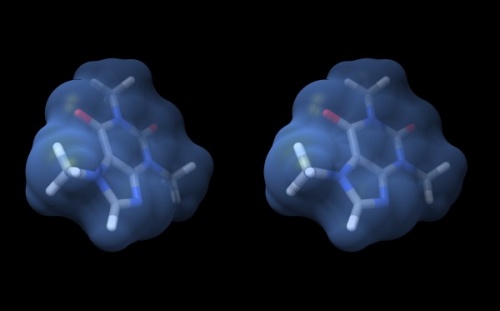
Wall-eye stereo view of the caffeine structure, surrounded by a transparent molecular surface. The image for the left eye is on the left side. For instructions on stereo-viewing, see the next section.
Stereo vision
A simple molecular scene like the caffeine molecule is a great way to practice viewing structures in stereo. This is a learnable skill, but it takes practice.
Task:
Access the Stereo Vision tutorial and practice viewing molecular structures in stereo.
Practice at least ...
- two times daily,
- for 3-5 minutes each session,
Keep up your practice. It is a wonderful skill that will greatly support your understanding of structural molecular biology. Practice with different molecules and try out different colours and renderings.
Note: do not go through your practice sessions mechanically. If you are not making any progress with stereo vision, contact me so I can help you on the right track.
Protein structure features
In this series of tasks we will showcase some of the globally applied tools that help us study molecular structure. Our first task is to find a structure for the sequence we are interested in, to work with.
Structure search
The search options in the PDB structure database are as sophisticated as those at the NCBI. At its simplest, we will try a simple keyword search to get us started.
Task:
- Visit the RCSB PDB website at http://www.rcsb.org/
- Briefly orient yourself regarding the database contents and its information offerings and services.
- Enter
Mbp1into the search field.
Keyword searches are notorious for being imprecise. In our case we retrieve maltose binding proteins, a homologous transcription factor from Magnaporthe oryzae (4UX5), and three Saccharomyces cerevisiae Mbp1 transcription factor structures (1L3G, 1BM8, and 1MB1) – only these contain (partial) sequence of the protein we are interested in. These three are APSES domain structures: an NMR ensemble and two crystal structures of different resolution.
- Click on the
1BM8entry[2] and explore the information and services linked from that page.
Next we will load this molecule in Chimera, work with the sequence interface, use it to select specific parts of a molecule, and colour specific regions (or residues) of a molecule separately.
Task:
- Open Chimera.
- One of the three yeast Mbp1 fragment structures has the PDB ID
1BM8. Load it in Chimera (simply enter the ID into the appropriate field of the File → Fetch by ID... window). - Display the protein in Presets → Interactive 1 mode and familiarize yourself with its topology of helices and strands.
- Open the sequence tool: Tools → Sequence → Sequence. You will see the sequence for each chain - here there is only one chain. By default, coloured rectangles overlay the secondary structure elements of the sequence.
- Hover the mouse over some residues and note that the sequence number and chain is shown at the bottom of the window.
- Click/drag one residue to select it. (Simply a click wont work, you need to drag a little bit for the selection to catch on.) Note that the residue gets a green overlay in the sequence window, as it also gets selected with a green border in the graphics window.
- In the bottom of the sequence window, there are instructions how to select (multiple) regions. Try this: colour the protein white (Select → Select All; Actions → Color → light gray). Clear the selection. Now select all the helical regions (pale yellow boxes) by click/dragging and using the shift key. Color them red. Then select all the strands by clicking into any of the pale green boxes and color them green.
Next, display the DNA binding subdomain.
- In the bottom of the sequence window, there are instructions how to select (multiple) regions. Clear the selection by <control> clicking into an empty spot of the viewer. Now select the region that encompasses the residues that have been reported to form the DNA binding subdomain, residues 50 to 74:
KRTRILEKEVLKETHEKVQGGFGKYQ(Taylor 2000). Show the side chains of these residues by clicking on the little green inspector icon on the viewer window, inspecting Atom and choosing displayed: true, and inspecting Bond and setting the stick radius to 0.4. - Undisplay the Hydrogen atoms by selecting the element H in the Chemistry option of the Selection Menu, and use the Action menu to hide them. Then use the effects pane of the Depiction menu to add a contour.
- Finally, give the scene a gradient grey background grey via the Actions → Color → all options... menu.
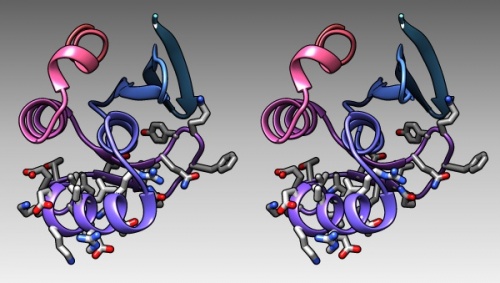
The DNA binding region of Mbp1 according to NMR measurements of DNA contact by Taylor et al. (2000). The backbone of 1BM8 is shown with a colour ramp from blue (N-terminus) to red (C-terminus). The side chains of the region 50-74 are shown coloured by element.
- Finally, generate a stereo-view that shows the molecule well, in which the domain is coloured dark grey, and the DNA binding domain residues (as defined above) are coloured with a colour ramp (Tools → Depiction → Rainbow)[3]
- Show the first and last residue's CA atom[4] as a sphere and colour the first one blue (to mark the N-terminus) and the last one red. E.g.:
- Select → Atom specifier →
:4@CA - Actions → Ribbon → hide
- Actions → Atoms/bonds → show
- Actions → Atoms/bonds → sphere
- Actions → Color → cornflower blue
- Then click on the selection inspector (the green button with the magnifying glass at the lower right of the graphics window) and set the sphere radius to 1.0Å.
- Select → Atom specifier →
A Ramachandran plot
Task:
- Choose Presets → Interactive 2 (all atoms) for a detailed view of the 1BM8 structure.
- Choose Favorites → Model Panel
- Look for the Option Ramachandran plot... in the choices on the right.
- Click the button and study the result. The dots in thisRamachandran Plot represent the phi-psi angle combinations for residue backbones. We see that they are well distributed, this is a high-resolution structure essentially without outliers. Clicking on a dot selects a residue in the structure viewer (selected residues have a green contour), conversely, already selected residues appear as red dots in the Ramachandran plot.
- Choose File → Fetch by ID and fetch
1L3G, an NMR structure of the Mbp1 APSES domain. Chimera loads the 19 models that comprise this structure dataset. - In the Favorites → Model Panel, select 1BM8 and click on hide.
- Then select 1LG3 and click group/ungroup to be able to address the models individually. Select any of the models individually and click again on Ramachandran plot. You will see that the points are much more dispersed, and there are a number of outliers that have comparatively high-energy conformations.
B-factors
Task:
- Choose Favorites → Model Panel, click/drag over the 1LG3 models and click close to remove them again.
- To explore B-Factors in the 1BM8 model, click show to view it again.
- Choose Tools → Structure Analysis → Render byAttribute.
- Select Attributes of atoms, Model 1BM8 and Attribute: bfactor. A histogram appears with sliders that allow you to render the distribution of values found in the structure for this attribute.
- Let's colour the atoms by B-Factor. Click on the colours tab. A standard colouring scheme is blue - white - red, but you can move the sliders, add new thresholds, and colour them individually by clicking on the colour patch to create your own colour spectrum, e.g. from black via red to white, in a black-body spectrum. Click Apply.
- Choose Actions → Atoms/Bonds → stick to give the bonds more volume. You will find that the core of the protein has low temperature factors, and the surface has a number of highly mobile sidechains and loops.
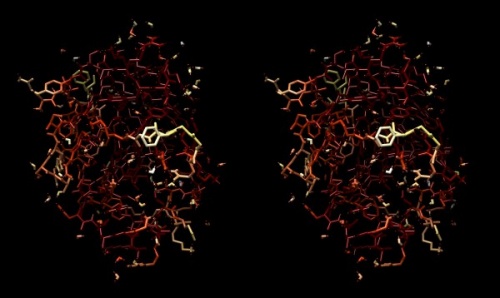
Structure of the yeast transcription factor Mbp1 DNA binding domain (1BM8) coloured by B-factor (thermal factor). The protein bonds are shown in a "stick" model, coloured with a spectrum that emulates black-body radiation. Note that the interior of the protein is less mobile, some of the surface loops are highly mobile (or statically disordered, X-ray structures can't distinguish that) and the discretely bound water molecules that are visible in this high-resolution structure are generally more mobile than the residues they bind to.
Electrostatics
Task:
- To visualize the electrostatic potential of the protein, mapped on the surface, first select Presets → Interactive 2... and Actions → Color → cyan for a vividly contrasting color.
- To apply potential coloring to a surface, we need to calculate a solvent accessible surface first: select Actions → Surface → show.
- A simple electrostatic potential calculation just assumes Coulomb charges. A more accurate calculation of full Poisson-Boltzmann potentials is also available. Select Tools → Surface/Binding Analysis → Coulombic Surface Coloring.
- Make sure the surface object is selected in the form (it should be selected by default since there is only one surface), keep the default parameters and click Apply.
- Use Actions → Surface → Transparency → 30% to make the protein backbone somewhat visible. Select Actions → Atoms/Bonds → hide to turn off the detailed view of coordinates, then Actions → Ribbon → show to display the fold of the domain.
- Open the Tools → Viewing Controls → Lighting window → and set Intensity from two-point to ambient. This reduces shadowing and reflections on the surface and thus emphasizes the color values - here our focus is not on shape, but on property.
- Use the Effects tab to turn shadows off and depth-cueing and silhouettes on. This recreates visual cues of depth which compensate for the loss of shape information by using a flat lighting model.
- Back at the sequence window, use the mouse to select the "DNA recognition domain" (residues 50-74). Then select Actions → Color → all options ... to open the color menu window. Find the control buttons for "Coloring applies to:" and select ribbons only. This is important, otherwise you will overwrite the coloring of your surface. Then choose "red" as the color for the selection.
- Study the resulting structure carefully: what do you expect regarding the electrostic potential of the surface of a DNA binding molecule? What do you find? If there is anything remarkable - how does it relate to the annotated "DNA binding" region? How do you interpret this relationship? Note down your answers, I may ask you to hand them in for credit at a later time in the course.
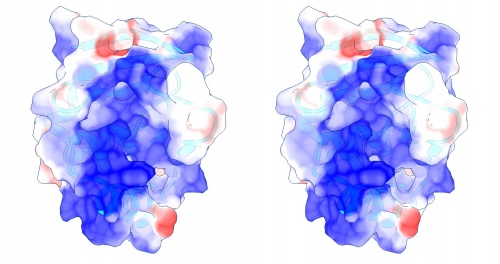
Coulomb (electrostatic) potential mapped to the solvent accessible surface of the yeast transcription factor Mbp1 DNA binding domain (1BM8). The protein backbone is visible through the transparent surface as a cartoon model, note the helix at the bottom of the structure. This helix has been suggested to play a role in forming the domain's DNA binding site and the positive (blue) electrostatic potential of the region is consistent with binding the negatively charged phosphate backbone of DNA. The other side of the domain has a negative (red) charge excess, which balances the molecule's electric charge overall, but also guides the protein-ligand interaction and supports faster on-rates.
Hydrogen bonds
Task:
- Hydrogen bonds encode the basic folding patterns of the protein. To visualize H-bonds select Presets → Publication 1... and Actions → Color → by element.
- Turn the surface and ribbons off with Actions → Surface → hide and Actions → Robbon → hide. Use Actions → Atoms/Bonds → show.
- Use Tools → Structure Analysis → FindHBond and Apply default parameters.
- To emphasize the role of H-bonds in determining the architecture of the protein, select Select → Structure → backbone → full and then Select → Invert (all models). Now Actions → Atoms/bonds → hide will show only the backbone with its H-bonds.
Compute with structures
To practice actual computations with structures we'll use the Grant lab's bio3d package in R.
Task:
- Open an RStudio session, load the project file from the File → Recent projects ... menu.
- Bring code and data resources up to date:
- pull the most recent version of the project from GitHub
- type
init()to load the most recent files and functions. load()the latest version ofmyDB.
- Study and work through the code in the
Structure.Rscript:PART ONE: INTRODUCTION TO bio3d;PART TWO: A RAMACHANDRAN PLOT;PART THREE: H-BOND LENGTH DISTRIBUTIONS; andPART FOUR: MAPPING CALCULATED VALUES TO STRUCTURE.
- There are a number of questions in the code, don't gloss over them but try to answer them for yourself.
Homology Modeling
In order to understand how specific residues in the sequence contribute to the putative function of the protein, and why and how they are conserved throughout evolution, we would need to study an explicit molecular model of an APSES domain protein, bound to its cognate DNA sequence. Explanations of a protein's observed properties and functions can't rely on the general fact that it binds DNA, we need to consider details in terms of specific residues and their spatial arrangement. In particular, it would be interesting to correlate the conservation patterns of key residues with their potential to make specific DNA binding interactions. Unfortunately, the experimental evidence we have considered in Assignment 2 (Taylor et al., 2000) is not sufficient to unambiguously define the details of how a DNA double helix might be bound. Moreover, at least two distinct modes of DNA binding are known for proteins of the winged-helix superfamily, of which the APSES domain is a member.
In this section you will (1) construct a molecular model of the APSES domain from the MBP1_SPIPU sequence.
For the following, please remember the following terminology:
- Target
- The protein that you are planning to model.
- Template
- The protein whose structure you are using as a guide to build the model.
- Model
- The structure that results from the modelling process. It has the Target sequence and is similar to the Template structure.
A brief overview article on the construction and use of homology models is linked to the resource section at the bottom of this page. That section also contains links to other sites and resources you might find useful or interesting.
A Point Mutation
To illustrate how homology modelling works in principle, let's consider changing the sequence of a single amino acid, based on a structural template.
Such minimal changes to structure models can be done directly in Chimera. Let us consider the residue A 42 of the 1BM8 structure. It is oriented towards the core of the protein, but most other Mbp1 orthologs have a larger amino acid in this position, V, or even I.
Task:
- Open
1BM8in Chimera, hide the ribbons and show all atoms as a stick model. - Color the protein white.
- Open the sequence window and select
A 42. Color it red. Choose Actions → Set pivot. Then study how nicely the alanine side chain fits into the cavity formed by its surrounding residues. - To emphasize this better, hide the solvent molecules and select only the protein atoms. Display them as a sphere model to better appreciate the packing, i.e. the Van der Waals contacts we discussed in class. Use the Favorites → Side view panel to move the clipping plane and see a section through the protein. Study the packing, in particular, note that the additional methyl groups of a valine or isoleucine would not have enough space in the structure. Then restore the clipping planes so you can see the whole molecule.
- Lets simplify the view: choose Actions → Atoms/Bonds → backbone only → chain trace. Then select
A 42again in the sequence window and choose Actions → Atoms/Bonds → show. - Add the surrounding residues: choose Select → Zone.... In the window, see that the box is checked that selects all atoms at a distance of less then 5Å to the current selection, and check the lower box to select the whole residue of any atom that matches the distance cutoff criterion. Click OK and choose Actions → Atoms/Bonds → show.
- Select
A 42again: left-click (control click) on any atom of the alanine to select the atom, then up-arrow to select the entire residue. Now let's mutate this residue to isoleucine. - Choose Tools → Structure Editing → Rotamers and select
ILEas the rotamer type. Click OK, a window will pop up that shows you the possible rotamers for isoleucine together with their database-derived probabilities; you can select them in the window and cycle through them with your arrow keys. But note that the probabilities are very different - and thus show you high-energy and low-energy rotamers to choose from. Therefore, unless you have compelling reasons to do otherwise, try to find the highest-probability rotamer that may fit. This is where your stereo viewing practice becomes important, if not essential. It is really, really hard to do this reasonably in a 2D image! It becomes quite obvious in 3D. Btw: I find such "quantitative" work - where the real distances are important - easier in orthographic than in perspective view (cf. the Camera panel). - I find that the first rotamer is actually not such a bad fit. The
CDatom comes close to the sidechains ofI 25andL 96. But we can assume that these are somewhat mobile and can accommodate a denser packing, because - as you can easily verify in your MSA - it is NOT the case that sequences that haveI 42, have a smaller residue in position25and/or96. So let's accept the most frequentILErotamer by selecting it in the rotamer window and clicking OK (while existing side chain(s): replace is selected). - Done.
If you want to go over this in more detail, check the video tutorial on YouTube published by the NIAID bioinformatics group here. I would also encourage you to go over Part 2 of the video tutorial that discusses how to check for and resolve (by energy minimization) steric clashes. But note that it is not clear whether energy minimization will make your structure more correct in the sense of a smaller overall RMSD with the real, mutated protein.
What we have done here with one residue is exactly the way homology modeling works with entire sequences. The homology modelling program simply changes all amino acids to the residues of the target sequence, based on the template structure. Let's now build a homology model for MBP1_SPIPU.
Preparation
- We need to define our Target sequence;
- find a suitable structural Template; and
- build a Model.
Target sequence
We have encountered the PDB 1BM8 structure before, the APSES domain of saccharomyces cerevisiae Mbp1. This is a useful template to model the DNA binding domain of your RBM match. But what exactly is the aligned region of the APSES domain? We could use several approaches to define the APSES domain:
- we could use the biostrings package to calculate a pairwise sequence alignment with the
1BM8sequence, like we did previously for the full-length sequences. This would give us the domain boundaries. - we could calculate a multiple sequence alignment, while including the
1BM8sequence. This would also allow us to infer domain boundaries, actually in all sequences in our database at once. But we have found previously that such multiple sequence alignments are quite sensitive to un-alignable regions of which we have quite a few in the full length sequences. We do need an MSA, but we do need to restrict the length of the sequences we align to a reasonable region. - we could access the domain annotations at CDD or at the SMART Database, but both have interfaces that are difficult to use computationally, and have other issues: NCBI does not recognize APSES domains, only the smaller KilA-N domain, and SMART sometimes does not find APSES domains in our sequences.
- the most straightforward approach of course is to use the annotation that we already have produced for the APSES domain in MBP1_SPIPU. You should be able to simply take the MBP1_SACCE sequence and the one for S. punctatus from the the multiple sequence aligment you have produced with the
msapackage.
Template choice and template sequence
The SWISS-MODEL server provides several different options for constructing homology models. The easiest option requires only a target sequence as input. In this mode the program will automatically choose suitable templates and create an input alignment. I would argue however that that is not the best way to use such a service: template choice and alignment both may be significantly influenced by biochemical reasoning, and an automated algorithm cannot make the necessary decisions. Should you use a structure of reduced resolution that however has a ligand bound? Should you move an indel from an active site to a loop region even though the sequence similarity score might be less? Questions like that may yield answers that are different from the best choices an automated algorithm could make. But Swiss Model is flexible and allows us to upload an explicit alignment between target and template. Please note: the model you will produce is "easy" - the sequence similarity is high and there are no indels to consider, the automated mode would have done just as well. But the strategy we pursue here is suitable also for much more difficult problems. The automated strategy probably is not.
Template choice is the first step. Often more than one related structure can be found in the PDB. The degree of sequence identity is the most important criterion, but there are many other factors to consider. Please refer to the template choice principles page on this Wiki where I discuss more details and alternatives. To find related structures, you can search the PDB itself through its Advanced Search interface; for example one can search for sequence similarity with a BLAST search, or search for structural similarity by accessing structures according to their CATH or SCOP classification. But the BLAST search is probably the method of choice: after all, the most important measure of the probability of success for homology modelling is sequence similarity.
Defining a template means finding a PDB coordinate set that has sufficient sequence similarity to your target that you can build a model based on that template. To find suitable PDB structures, we will perform a BLAST search at the PDB.
Task:
- Retrieve the aligned Mbp1 S. punctatus RBM APSES domain sequence from the
apsesMatmatrix as explained in
PART FIVE: PREPARE APSES DOMAIN SEQUENCES FOR HOMOLOGY MODELING of the Structure.R script. This is your target sequence.
- Navigate to the PDB.
- Click on Advanced to enter the advanced search interface.
- Open the menu to Choose a Query Type:
- Find the Sequence features section and choose Sequence (BLAST...)
- Paste your target sequence into the Sequence field, select not to mask low-complexity regions and Submit Query. Since the E-value is set rather high by default, you will get a number of low-confidence hits as well as the actual homologs, these have very low E-values.
All hits that are homologs are potentially suitable templates, but some are more suitable than others. Consider how the coordinate sets differ and which features would make each more or less suitable for creating a homology model: you should consider ...
- sequence similarity to your target
- size of expected model (= length of alignment)
- presence or absence of ligands
- experimental method and quality of the data set
Sequence similarity is the most important, but we can have the PDB tabulate the other features concisely for this task.
- There is a menu to create Reports: - select customizable table.
- Select (at least) the following information items:
- Structure Summary
- Experimental Method
- Sequence
- Chain Length
- Ligands
- Ligand Name
- Biological details
- Macromolecule Name
- refinement Details
- Resolution
- R Work
- R free
- click: Create report.
Unfortunately you don't get the E-values into the report, and those should strongly influence your final decision. However in our case the sequences and therefore the E-values of the top three hits are all the same. And there is a new structure from January 2015, with a lower resolution. Some of the sequences have a longer chain-length ... but those are only disordered residues (otherwise these would be better suited templates; regrettably, you'd need to check that in the real world, there is no automatic tool to evaluate disorder and its effects on template choice). In my opinion that leaves pretty much only one unambiguous choice for our template: 1BM8.
- Finally
- Click on the 1BM8 ID to navigate to the structure page for the template and save the FASTA sequence to your computer. This is the template sequence.
Sequence numbering
It is not straightforward at all how to number sequence in such a project. A "natural" numbering starts with the start-codon of the full length protein and goes sequentially from there. However, this does not map exactly to other numbering schemes we have encountered. As you know the first residue of the APSES domain (as defined by CDD) is not Residue 1 of the Mbp1 protein. The first residue of the 1BM8 FASTA file (one of the related PDB structures) is the fourth residue of the Mbp1 protein. The first residue in the structure is GLN 3, therefore Q is the first residue in a FASTA sequence derived from the coordinate section of the PDB file (the ATOM records. In the 1MB1 structure, the original N-terminal amino acids are present in the molecule, therefore they are present in the FASTA file which starts with MSNQIY..., but they are disordered in the structure and no coordinates are present for M and S. A sequence derived explicitly from the coordinates is therefore different from the reported FASTA sequence, which is really bad because that is what the modelling program has to work with ... and so on. It can get complicated. You need to remember: a sequence number is not absolute, but assigned in a particular context and you need to be careful how to do this.
Fortunately, the numbering for the residues in the coordinate section of our target structure corresponds not to its FASTA sequence, but to the numbering of the gene. Otherwise we would need to renumber the sequence (e.g. by using the bio3D R package). If we would not do this, the sequence numbers in the model might not correspond to the sequence numbers of our target.
The input alignment
The sequence alignment between target and template is the single most important factor that determines the quality of your model. No comparative modelilng process will repair an incorrect alignment; it is useful to consider a homology model rather like a three-dimensional map of a sequence alignment rather than a structure in its own right. In a homology modelling project, typically the largest amount of time should be spent on preparing the best possible alignment. Even though automated servers like the SwissModel server will align sequences and select template structures for you, it would be unwise to use these just because they are convenient. You should take advantage of the much more sophisticated alignment methods available. Analysis of wrong models can't be expected to produce right results.
The best possible alignment is usually constructed from a multiple sequence alignment that includes at least the target and template sequence and other related sequences as well. The additional sequences are an important aid in identifying the correct placement of insertions and deletions. Your alignment should have been carefully reviewed by you and wherever required, manually adjusted to move insertions or deletions between target and template out of the secondary structure elements of the template structure.
In most of the Mbp1 orthologues, we do not observe indels in the APSES domain regions. Evolutionary pressure on the APSES domains has selected against indels in the more than 600 million years these sequences have evolved independently in their respective species.
Task:
- Confirm the MSA for APSES domain sequences in Mbp1 orthologues with the section
PART SIX: APSES DOMAIN SEQUENCES OF MBP1 ORTHOLOGUESof theStructure.Rscript.
Of course, this one-to-one correspondence is the simplest possible case but the principle of working from a good MSA to recover the best-informed alignment between target and template sequence applies in all cases.
Homology model
The alignment defines the residue by residue relationship between target and template sequence. All we need to do now is to change every residue of the template to the target sequence
SwissModel
Access the Swissmodel server at http://swissmodel.expasy.org and click on the Start Modelling button. Under the Supported Inputs, choose Target-Template Alignment.
Task:
- Paste the aligned sequences of the S. punctatus target and the
1BM8template into the form field. SwissModel will analyse the sequences and ask you to identify target and template. TheMBP1_SPIPUsequence is your target. The1BM8sequence is the template.
- Click Validate Target Template Alignment and check that the returned alignment is correct. All non-identical residues are shown in light-grey.
- Click Build Model to start the modelling process. This will take about a minute or so.
- The resulting page returns information about the resulting model and its quality. You can rotate the model in the window on the right with the mouse. Regions that have a reddish hue have lower quality scores, i.e. they were harder to model or could not be modelled well with good geometry. Hovering the mouse over parts of the structure highlights the respective region of the sequence alignment.
- Mouse over the Model 01 dropdown menu (under the icon of the template structure), and choose the PDB file. Note that the B-factor column of the coordinate section contains the QMEAN scores (between 0 and 1) that the server has calculated. Higher is better. Save the PDB file on your computer.
- Open the SwissModel documentation in a new tab. Read about the modelling process. there are a number of important technical details that help to understand what the computed coordinates of your model mean, you should pay special attention to the GQME and QMEAN quality scores.
- Also save:
- The output page as pdf (for reference)
- The modeling report (as pdf)
Model interpretation
We have spent a significant amount of time to prepare data for the analysis and in practice it usually seems to turn out that way, that the preparation of data occupies the greatest part of our efforts. The actual computational analysis is generally quite fast. And, unfortunately, the interpretation of results is often somewhat neglected. Don't be that way. Data does not explain itself. The interpretation of your computational results is the most important part.
We will look at our homology model with two different questions:
- Can we define the DNA binding residues?
- Can we tell which residues are conserved for functional reasons, rather than for structural reasons?
The PDB file
Task:
Open your model coordinates in a text-editor (make sure you view the PDB file in a fixed-width font (like "courier") so all the columns line up correctly) and consider the following questions:
- What is the residue number of the first residue in the model? What should it be, based on the alignment? If the putative DNA binding region was reported to be residues 50-74 in the Mbp1 protein, which residues of your model correspond to that region?
That's not easy to tell. But it should be.
As you have seen above, SwissModel numbers the first residue "1" and does not keep the numbering of the template. We should renumber the model so we can compare the model and the template with the same residue numbers. Carefully doing this by hand should not even take you a whole hour. You just need to edit all the residue numbers in a text editor to give the first residue the number "4" and increment numbers from there.
Or we could do this with an R script.
Task:
- Renumber your model PDB file to start with residue number 4 with the code in section
PART SEVEN: RENUMBERING THE MODELof theStructure.Rscript.
First visualization
Since a homology model inherits its structural details from the template, your model of the YFO sequence should look very similar to the original 1BM8 structure.
Task:
- Start Chimera and load the model coordinates that you have just renumbered.
- From the PDB, also load the template structure. (Use File → Fetch by ID ...)
- In the Favourites → Model Panel window you can switch between the two molecules.
- Hide the ribbon and choose backbone only → full. You will note that the backbone of the two structures is virtually identical.
- Next, choose Actions → Atoms/Bonds → show to display the two molecules in a stick style and note how the sidechains have been modeled. Note especially how sidechain coordinates have been guessed, where the template had shorter sidechains than the target. It may be more clear if you hide H-atoms: Select → Chemistry → Element → H and Actions → Atoms/Bonds → hide
- Display only residue 50 to 74 to focus on the putative helix-turn-helix domain. You can drag your mouse in the Favourites → Sequence, window to select the range then Select → Invert (selected model) and Actions → Atoms/Bonds → hide. Or you can use Chimera's commandline:
~displayto undisplay everything,show #:50-74to show this residue range for all models. - Study the result: a model of the HTH subdomain of Mbp1_SPIPU.
Coloring the model by energy
SwissModel calculates energies for each residue of the model with a molecular mechanics forcefield. The SwissModel modeling summary page contains a plot of these energies as a function of sequence number like. The values - between 0.0 and 1.0 - are stored in the PDB file's B-factor field.
Task:
- Back in Chimera, use the model panel to close the 1BM8 structure. Select all and show Atoms, bonds to view the entire model structure.
- Choose Tools → Depiction → Render by attribute and select attributes of atoms, Attribute: bfactor, check color atoms and click OK.
- Study the result: It seems that residues in the core of the protein have better energies (higher values) than residues at the surface. Why could that be the case?
Modelling DNA binding
One of the really interesting questions we can discuss with reference to our homology model is how sequence variation might result in changed DNA recognition sites, and then lead to changed cognate DNA binding sequences. In order to address this, we would need to generate a plausible structural model for how DNA is bound to APSES domains.
Since there is currently no software available that would reliably model such a complex from first principles[5], we will base a model of a bound complex on homology modelling as well. This means we need to find a similar structure for which the position of bound DNA is known, then superimpose that structure with our model. This places the DNA molecule into the spatial context of the model we are studying. It so happens that early in 2015 an APSES domain structure with bound DNA was published. You probably noticed it as a result of the PDB BLAST search: 4UX5, from the Magnaporthe oryzae Mbp1 orhologue PCG2[6].
A homologous protein/DNA complex structure
Task:
- The PCG2 / DNA complex
- Open Chimera and load the
4UX5structure. Spend some time exploring it. There are two domains of identical sequence bound to one DNA molecule. The first question I would have is whether the two molecules bind to the same DNA motif - the CGCG core of the "MCB-box", and whether the observed protein:DNA interfaces are actually with the cognate sequence, or whether one (or both) proteins are non-specific complexes. The conditions under which proteins crystallize can be harsh, and physiological function under these conditions is not guaranteed.[7] Indeed, Liu et al. (2015) report that at low concentrations a 1:1 complex is formed and the 2:1 Protein:DNA complex only forms at high concentrations. Figure 3. of their paper shows that the detailed contacts between protein and DNA are in fact not identical.
- Without taking this question too far, let's get a quick view of the comparison by duplicating one domain of the structure and superimposing it on the other. The authors feel that chain
Arepresents the tighter, more specific mode of interaction; so we will duplicate chainBand superpose the copy onA.
- In Chimera, open the Favorites → Model Panel and use the copy/combine button to create a copy of the
4UX5model. Call ittest. - Select chain B of the
testmodel, then use Select → Invert (selected models) to apply the selection to everything in thetestmodel except chain B. - Use Actions → Atoms/Bonds → delete to remove everything but Chain B.
- Select and colour the chain red.
- Back on the Model Panel, select both models and use the match... dialogue to open a MatchMaker dialogue window. Choose the radio button two match two specific chains and select
4UX5chain A as the Reference chain,testchain B as the Chain to match. Click Apply.
You will see that the superimposed structures are very similar, that the main difference is in the orientation of the disordered C-terminus, but also that there is a structural difference between the two structures around Gly 84 which inserts into the minor groove of the double helix.
- Select one of the residues of that loop in chain A by <control>-clicking on it and use Action → Set pivot to set the centre of rotation to that residue: this makes it easier to visualize the binding situation when you make the molecules larger.
- Select residues 81 to 87 and the corresponding (sequence
VQGGYGKY) and in both chains turn their ribbon display off and display this range as "sticks". - Select nucleic acid in the structure submenu and turn ribbons and nucleotide objects off to display the DNA as sticks as well. Colour the DNA by element.
- Study the situation. Focus on Gly 84.A, especially the interaction of its carbonyl oxygen, which hydrogen bonds to the N2 atom of G8.D chain. Gln 89.A hydrogen bonds to the N2 atom of G8.C chain. Gly 84 and Gln 82 thus recognize a G:C C:G pair. In the B chain, Gly 84.B does not contact the DNA well, since it contacts residues of chain A, especially Gln 82.A. The carbonyl atom of Gly 84.B hydrogen bonds to Gln 89.B. and therefore Gln89.B is not available to contact nucleotide bases. What do you think[8]? It seems to me that a crucial interaction for the cognate sequence is contributed by Guanine 8,
- Finally, use the Model Panel to select
testand close it.
Superimposing your model
Both your homology model and the template structure provide valuable information:
- The template structure shows how conserved the structure is at the protein/DNA interface. You have seen what subtle differences can give rise to a sequence specific complex and a non-specific binding mode. For Mbp1 we know that the APSES domain binds to the same cognate DNA sequence as PCG2. Since your model structure is heavily biased towards the template, evaluating the template in the context of a real protein/DNA complex allows you to judge which binding residues appear to be conserved and possibly modelled in an orientation that is productive for binding.
- The model structure maps sequence variation into that context: are the crucial residues for sequence specific binding conserved?
Task:
- Start by loading your model and the 1BM8 structure into your chimera session. Select all, turn all ribbons off, and set all atoms to stick representation. Then select H atoms by element and hide them.
- We need to visualize and evaluate differences in binding between different proteins and for me it works well to colour everything by element, and give the carbon atoms some identifying, distinct colour. This is best achieved through the Chimera command line that you can turn on with the little "computer" icon on the left-hand side of the graphics window. Have a look at the Chimera Users guide, and choose select to learn how Chimera's selection syntax works.
- Open the Model Panel to check which protein has which Chimera-internal model number. Then you can use the following selection syntax. Instead of the model numbers, I will type
<SPIPU>,<4ux5>, and<1BM8>- you will certainly know by now that these are placeholder labels and you need to replace them with the numbers0,1, and2instead.
- To colour the DNA carbon atoms white, type:
color white #<4ux5>:.C,.D & C
- To colour the DNA carbon atoms white, type:
- To colour the 4ux5 A chain carbon atoms grey, type:
color #878795 #<4ux5>:.A & CNote: the color values after the first hash are rgb triplets in the hexadecimal numbering systems - exactly like in R.
- To colour the 4ux5 A chain carbon atoms grey, type:
- To undisplay the 4ux5 B chain, type:
~display #<4ux5>:.BNote: this is the tilde character, not a hyphen or minus sign.
- To undisplay the 4ux5 B chain, type:
- To colour the SPIPU model carbon atoms a pale reddish color, type:
color #b06268 #<SPIPU> & C
- To colour the SPIPU model carbon atoms a pale reddish color, type:
- To colour the 1BM8 structure carbon atoms a pale greenish color, type:
color #92b098 #<1BM8> & C
- To colour the 1BM8 structure carbon atoms a pale greenish color, type:
- Ready? Let's superimpose the chains.
- Select all models in the Model Panel and click on match.
- Set 4ux5 Chain A as the Reference chain.
- Select SPIPU as a Chain to match, select the button for specific reference and specific match, and click Apply.
- Repeat this with 1BM8 as the match chain.
- Easy. Now enlarge the binding site. Remember that 4ux5 and 1bm8 are independently determined crystal structures, wheres SPIPU was modelled on 1bm8 and is expected to be very similar to it. To give you some guidance what you should focus on, select 4ux5 residue 84 CA atom and display it as Ball & Stick. You can also repeat the Action "Set Pivot in case the pivot has shifted.
- Study the scene. This is where stereo- vision will help a lot.
- What do you think? Is this what you expected? Can you explain what you see? Was the modelling process succesful?
- Now turn the display of 4ux5 chain B back on and turn chain A off instead. Then superimpose the 1BM8 template and your model on Chain B.
- Again, focus on the binding region. What do you think of that? What would you have expected? Do you see a difference? What does this all mean?
Links and resources
The DNA binding site
Now, that you know how YFO Mbp1 aligns with yeast Mbp1, you can evaluate functional conservation in these homologous proteins. You probably already downloaded the two Biochemistry papers by Taylor et al. (2000) and by Deleeuw et al. (2008) that we encountered in Assignment 2. These discuss the residues involved in DNA binding[9]. In particular the residues between 50-74 have been proposed to comprise the DNA recognition domain.
Task:
- Using the APSES domain alignment you have just constructed, find the YFO Mbp1 residues that correspond to the range 50-74 in yeast.
- Note whether the sequences are especially highly conserved in this region.
- Using Chimera, look at the region. Use the sequence window to make sure that the sequence numbering between the paper and the PDB file are the same (they are often not identical!). Then select the residues - the proposed recognition domain - and color them differently for emphasis. Study this in stereo to get a sense of the spatial relationships. Check where the conserved residues are.
- A good representation is stick - but other representations that include sidechains will also serve well.
- Calculate a solvent accessible surface of the protein in a separate representation and make it transparent.
- You could combine three representations: (1) the backbone (in ribbon view), (2) the sidechains of residues that presumably contact DNA, distinctly colored, and (3) a transparent surface of the entire protein. This image should show whether residues annotated as DNA binding form a contiguous binding interface.
DNA binding interfaces are expected to comprise a number of positively charged amino acids, that might form salt-bridges with the phosphate backbone.
Task:
- Study and consider whether this is the case here and which residues might be included.
Links and resources
- Molecular Graphics Software Links– a collection of links at the PDB.
| Taylor et al. (2000) Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry 39:3943-54. (pmid: 10747782) |
- PDB file format (see the Coordinate Section if you are unsure about chain identifiers)
- Wikipedia on Structural Superposition (although the article is called "Structural Alignment")
Footnotes and references
- ↑ There also exist several online servers that translate SMILES strings to idealized structures, see e.g. the online SMILES translation service at the NCI.
- ↑
Xu et al. (1997) Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis. Structure 5:349-58. (pmid: 9083114) - ↑ The Rainbow tool can only create color ramps for an entire molecule. In order to achieve this effect: color the molecule with a color ramp, then select the target domain, then invert the selection and color the new selection dark grey.
- ↑ See here for details of the specification syntax.
- ↑ Rosetta may get the structure approximately right, Autodock may get the complex approximately right, but the coordinate changes involved in induced fit makes the result unreliable - and we have no good way to validate whether the predicted complex is correct.
- ↑
Liu et al. (2015) Structural basis of DNA recognition by PCG2 reveals a novel DNA binding mode for winged helix-turn-helix domains. Nucleic Acids Res 43:1231-40. (pmid: 25550425) - ↑ This particular crystal structure however was crystallized from a Tris-buffer with 50mM NaCl at pH 8.0 - comparatively gentle conditions actually.
- ↑ Besides the coordinate difference between the chains, if indeed chain B would be representative of a DNA "scanning" conformation, perhaps one should expect that the local DNA structure that chain B binds to is structurally closer to canonical B-DNA than the DNA binding interface of chain A...
- ↑ (Taylor et al. (2000) Biochemistry 39: 3943-3954 and Deleeuw et al. (2008) Biochemistry. 47:6378-6385)
Ask, if things don't work for you!
- If anything about this page is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- Review Netiquette for the course mailing list.
- Read How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example.
| Data | Sequence | Structure | Phylogeny | Function |