Difference between revisions of "BIO Assignment Week 5"
m |
|||
| Line 9: | Line 9: | ||
</tr></table> | </tr></table> | ||
| − | {{Template: | + | {{Template:Active}} |
| − | + | <small>Concepts and activities (and reading, if applicable) for this assignment will be topics on the upcoming quiz.</small> | |
| − | Concepts and activities (and reading, if applicable) for this assignment will be topics on | ||
| − | |||
| − | |||
| − | |||
| Line 20: | Line 16: | ||
__TOC__ | __TOC__ | ||
| + | | ||
| + | <div style="padding: 2px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> | ||
| − | + | <br> | |
| + | ;How could the search for ultimate truth have revealed so hideous and visceral-looking an object? | ||
| + | :''[https://en.wikipedia.org/wiki/Max_Perutz Max Perutz] <small>(on his first glimpse of the Hemoglobin structure)</small>'' | ||
| + | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
| + | ==Introduction== | ||
| + | Where is the hidden beauty in structure, and where, the "ultimate truth"? In the previous assignments we have discovered homologues of APSES domain containing proteins in all fungal species. This makes the domain an ancient protein family that had already duplicated to several paralogues at the time when the cenancestor of all fungi lived, more than 600,000,000 years ago, in the [http://www.ucmp.berkeley.edu/fungi/fungifr.html Vendian period] of the Proterozoic era of Precambrian times. | ||
| + | In this assignment we will explore its molecular structure. | ||
| − | |||
| − | |||
| − | + | | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Molecular graphics: UCSF Chimera== | |
| + | To view molecular structures, we need a tool to visualize the three dimensional relationships of atoms. A ''molecular viewer'' is a program that takes 3D structure data and allows you to display and explore it. For a number of reasons, I use the UCSF Chimera viewer for this course: | ||
| + | # Chimera is free and open; | ||
| + | # It creates very appealing graphics; | ||
| + | # It is under ongoing development and is well maintained; | ||
| + | # It provides an array of useful utilities for structure analysis; and, | ||
| + | # besides an intuitive, menu driven interface, Chimera can be scripted via its command line, or even programmed via its in-built python interpreter. | ||
| − | + | {{#lst:UCSF_Chimera|Installation}} | |
| + | Let's explore Chimera functions first with a simple small molecule: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | === Modeling small molecules === | |
| + | "Small" molecules are solvent, ligands, substrates, products, prosthetic groups, drugs - in short, essentially everything that is not made by DNA-, RNA-polymerases or the ribosome. Whereas the biopolymers are still front and centre in our quest to understand molecular biology, small molecules are crucial for our quest to interact with the inventory of the cell, create useful products, or advance medicine. | ||
| − | + | A number of public repositories make small-molecule information available, such as [http://pubchem.ncbi.nlm.nih.gov/ PubChem] at the NCBI, the ligand collection at the [http://pdb.org '''PDB'''], the [http://www.ebi.ac.uk/chebi/ ChEBI] database at the European Bioinformatics Institute, or the [http://cactus.nci.nih.gov/ncidb2.2/ NCI database browser] at the US National Cancer Institute. One general way to export topology information from these services is to use {{WP|SMILES|SMILES strings}}—a shorthand notation for the composition and topology of chemical compounds. | |
| − | |||
| − | |||
| − | |||
| − | A number of public repositories make small molecule information available, such as [http://pubchem.ncbi.nlm.nih.gov/ PubChem] at the NCBI, the ligand collection at the [http://pdb.org '''PDB'''], the [http://www.ebi.ac.uk/chebi/ ChEBI] database at the European Bioinformatics Institute, or the [http://cactus.nci.nih.gov/ncidb2.2/ NCI database browser] at the US National Cancer Institute. One general way to export topology information from these services is to use {{WP|SMILES|SMILES strings}}—a shorthand notation for the composition and topology of chemical compounds. | ||
{{task| | {{task| | ||
| − | # Access | + | # Access [http://pubchem.ncbi.nlm.nih.gov/ PubChem]. |
| − | # Enter "caffeine" as a search term. | + | # Enter "caffeine" as a search term in the '''Compound''' tab. A number of matches to this keyword search are returned. |
| − | # | + | # Click on the [http://pubchem.ncbi.nlm.nih.gov/compound/2519 top hit - the Caffeine molecule]. Note that the page contains among other items: |
| + | ## A 2D structural sketch; | ||
| + | ## An idealized 3D structural conformer, for which you can download coordinates in several formats; | ||
| + | ## The IUPAC name: <code>1,3,7-trimethylpurine-2,6-dione</code>; | ||
| + | ## The CAS identifier <code>58-08-2</code> which is a unique identifier and can be used as a cross-reference ID; | ||
| + | ## The {{WP|SMILES|SMILES strings|SMILES string}} <code>CN1C{{=}}NC2{{=}}C1C({{=}}O)N(C({{=}}O)N2C)C</code>; | ||
| + | ## ... and much more. | ||
}} | }} | ||
| − | + | That's great, but let's sketch our own version of caffeine. Several versions of Peter Ertl's {{WP|JME_editor|Java Molecular Editor (JME)}} are offered online, PubChem offers this functionality via its '''Sketcher''' tool. | |
{{task| | {{task| | ||
| − | # | + | # Return to the [http://pubchem.ncbi.nlm.nih.gov/ PubChem homepage]. |
| − | # Follow the link to ''' | + | # Follow the link to '''Structure search''' (in the right hand menu). |
# Click on the '''3D conformer''' tab and on the '''Launch''' button to launch the molecular editor in its own window. | # Click on the '''3D conformer''' tab and on the '''Launch''' button to launch the molecular editor in its own window. | ||
| − | # Sketch the structure of caffeine. I find the editor quite intuitive but | + | # Sketch the structure of caffeine. I find the editor quite intuitive but clicking on the '''Help''' button will give you a quick, structured overview. Make sure you define your double-bonds correctly. |
| − | + | # '''Export''' the SMILES string of your compound to your project folder. | |
| − | # | ||
}} | }} | ||
| + | |||
=== Translating SMILES to structure === | === Translating SMILES to structure === | ||
| − | + | Chimera can translate SMILES strings to coordinates<ref>There are several online servers that translate SMILES strings to idealized structures, see e.g. the [http://cactus.nci.nih.gov/translate/ online SMILES translation service] at the NCI.</ref>. | |
| − | |||
{{task| | {{task| | ||
| − | # | + | # Open Chimera. |
| − | # | + | # Select '''Tools''' → '''Structure Editing''' → '''Build Structure'''. |
| − | # | + | # In the '''Build Structure''' window, select the '''SMILES string''' button, paste the string from your file, and click '''Apply'''. |
| − | + | # The caffeine molecule will be generated and visualized in the graphics window. This is a "stick" representation. | |
| + | # You can rotate it with your mouse, <command> drag to scale, <shift> drag to translate. | ||
| + | # Use the '''Actions''' → '''Atoms/Bonds''' → '''ball & stick''' or '''sphere''' menu items to change appearance. | ||
| + | # Use the '''Actions''' → '''Color''' → '''by element''' menu to change colors. | ||
| + | # Change the display back to stick and use '''Actions''' → '''Surface''' → '''show''' to add a solvent accessible surface. It is a bit rough for this small molecule... | ||
| + | ## Select the surface with <control><click> (<control><left mouse button> on windows). A green contour line appears around selected items. | ||
| + | ## Open the selection inspector by clicking on the tiny green icon in the lower-right corner of the window (It has a magnifying glass symbol which means "inspect" for Chimera, not "search"). | ||
| + | ## Select Inspect ...'''MSMS surface''' and change the '''Vertex density''' value to 50.0 - hit return. | ||
| + | # Use the '''Actions''' → '''Color''' → '''all options''' menu to change colors: use the '''surfaces''' button to apply a color choice to the surface and choose '''cornflower blue'''. | ||
| + | # Use the '''Actions''' → '''Surface''' → '''transparency''' → '''50%''' menu to see bonds contained in the surface. | ||
| + | # To begin working with molecules in "true" 3D, choose '''Tools''' → '''Viewing Controls''' → '''Camera''' and select '''camera mode''' → '''wall-eye stereo'''. Also, use the '''Effects''' tab of the '''Viewing''' window, and ''check'' '''shadows''' off. | ||
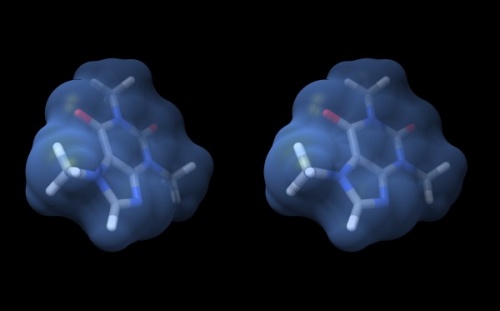
| + | # Your structure should look about like what you see below. Save your session with the '''File''' → '''Save Session''' dialogue so you can easily recreate the scene. | ||
| − | |||
| − | {{ | + | {{stereo|Caffeine_stereo.jpg|'''Wall-eye stereo view''' of the caffeine structure, surrounded by a transparent molecular surface. The image for the left eye is on the left side. For instructions on ''stereo-viewing'', see the next section. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
| − | === | + | === Stereo vision === |
| − | + | A simple molecular scene like the caffeine molecule is a great way to practice viewing structures in stereo. This is a learnable skill, but it takes practice. | |
| − | {{task| | + | {{task| |
| − | + | Access the '''[[Stereo Vision]]''' tutorial and practice viewing molecular structures in stereo. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | Practice at least ... | ||
| + | * two times daily, | ||
| + | * for 3-5 minutes each session, | ||
}} | }} | ||
| − | + | Keep up your practice throughout the course. It is a wonderful skill that will greatly support your understanding of structural molecular biology. Practice with different molecules and try out different colours and renderings. | |
| − | + | '''Note: do not go through your practice sessions mechanically. If you are not making any progress with stereo vision, contact me so we can help you on the right track.''' | |
| − | |||
| − | |||
| − | + | ;TBC | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | == Links and resources == | |
| − | |||
| − | |||
| − | + | * [[UCSF Chimera|'''Chimera page''']] | |
| − | + | * [https://www.cgl.ucsf.edu/chimera/current/docs/UsersGuide/framecontrib.html Chimera Tools Index] | |
| − | + | * [[Stereo Vision|'''Stereo vision tutorial''']] | |
| − | |||
| − | |||
| − | |||
| − | + | *[http://www.rcsb.org/pdb/static.do?p=software/software_links/molecular_graphics.html Molecular Graphics Software Links]– a collection of links at the PDB | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{#pmid: 10679470}} | {{#pmid: 10679470}} | ||
{{#pmid: 15808743}} | {{#pmid: 15808743}} | ||
Revision as of 04:11, 19 October 2015
Assignment for Week 5
Structure Analysis
| < Assignment 4 | Assignment 6 > |
Note! This assignment is currently active. All significant changes will be announced on the mailing list.
Concepts and activities (and reading, if applicable) for this assignment will be topics on the upcoming quiz.
Contents
- How could the search for ultimate truth have revealed so hideous and visceral-looking an object?
- Max Perutz (on his first glimpse of the Hemoglobin structure)
Introduction
Where is the hidden beauty in structure, and where, the "ultimate truth"? In the previous assignments we have discovered homologues of APSES domain containing proteins in all fungal species. This makes the domain an ancient protein family that had already duplicated to several paralogues at the time when the cenancestor of all fungi lived, more than 600,000,000 years ago, in the Vendian period of the Proterozoic era of Precambrian times.
In this assignment we will explore its molecular structure.
Molecular graphics: UCSF Chimera
To view molecular structures, we need a tool to visualize the three dimensional relationships of atoms. A molecular viewer is a program that takes 3D structure data and allows you to display and explore it. For a number of reasons, I use the UCSF Chimera viewer for this course:
- Chimera is free and open;
- It creates very appealing graphics;
- It is under ongoing development and is well maintained;
- It provides an array of useful utilities for structure analysis; and,
- besides an intuitive, menu driven interface, Chimera can be scripted via its command line, or even programmed via its in-built python interpreter.
Task:
- Access the Chimera homepage and navigate to the Download section.
- Find the the newest version for your platform in the table and click on the file to download it.
- Follow the instructions to install Chimera.
Let's explore Chimera functions first with a simple small molecule:
Modeling small molecules
"Small" molecules are solvent, ligands, substrates, products, prosthetic groups, drugs - in short, essentially everything that is not made by DNA-, RNA-polymerases or the ribosome. Whereas the biopolymers are still front and centre in our quest to understand molecular biology, small molecules are crucial for our quest to interact with the inventory of the cell, create useful products, or advance medicine.
A number of public repositories make small-molecule information available, such as PubChem at the NCBI, the ligand collection at the PDB, the ChEBI database at the European Bioinformatics Institute, or the NCI database browser at the US National Cancer Institute. One general way to export topology information from these services is to use SMILES strings—a shorthand notation for the composition and topology of chemical compounds.
Task:
- Access PubChem.
- Enter "caffeine" as a search term in the Compound tab. A number of matches to this keyword search are returned.
- Click on the top hit - the Caffeine molecule. Note that the page contains among other items:
- A 2D structural sketch;
- An idealized 3D structural conformer, for which you can download coordinates in several formats;
- The IUPAC name:
1,3,7-trimethylpurine-2,6-dione; - The CAS identifier
58-08-2which is a unique identifier and can be used as a cross-reference ID; - The SMILES strings
CN1C=NC2=C1C(=O)N(C(=O)N2C)C; - ... and much more.
That's great, but let's sketch our own version of caffeine. Several versions of Peter Ertl's Java Molecular Editor (JME) are offered online, PubChem offers this functionality via its Sketcher tool.
Task:
- Return to the PubChem homepage.
- Follow the link to Structure search (in the right hand menu).
- Click on the 3D conformer tab and on the Launch button to launch the molecular editor in its own window.
- Sketch the structure of caffeine. I find the editor quite intuitive but clicking on the Help button will give you a quick, structured overview. Make sure you define your double-bonds correctly.
- Export the SMILES string of your compound to your project folder.
Translating SMILES to structure
Chimera can translate SMILES strings to coordinates[1].
Task:
- Open Chimera.
- Select Tools → Structure Editing → Build Structure.
- In the Build Structure window, select the SMILES string button, paste the string from your file, and click Apply.
- The caffeine molecule will be generated and visualized in the graphics window. This is a "stick" representation.
- You can rotate it with your mouse, <command> drag to scale, <shift> drag to translate.
- Use the Actions → Atoms/Bonds → ball & stick or sphere menu items to change appearance.
- Use the Actions → Color → by element menu to change colors.
- Change the display back to stick and use Actions → Surface → show to add a solvent accessible surface. It is a bit rough for this small molecule...
- Select the surface with <control><click> (<control><left mouse button> on windows). A green contour line appears around selected items.
- Open the selection inspector by clicking on the tiny green icon in the lower-right corner of the window (It has a magnifying glass symbol which means "inspect" for Chimera, not "search").
- Select Inspect ...MSMS surface and change the Vertex density value to 50.0 - hit return.
- Use the Actions → Color → all options menu to change colors: use the surfaces button to apply a color choice to the surface and choose cornflower blue.
- Use the Actions → Surface → transparency → 50% menu to see bonds contained in the surface.
- To begin working with molecules in "true" 3D, choose Tools → Viewing Controls → Camera and select camera mode → wall-eye stereo. Also, use the Effects tab of the Viewing window, and check shadows off.
- Your structure should look about like what you see below. Save your session with the File → Save Session dialogue so you can easily recreate the scene.
Wall-eye stereo view of the caffeine structure, surrounded by a transparent molecular surface. The image for the left eye is on the left side. For instructions on stereo-viewing, see the next section.
Stereo vision
A simple molecular scene like the caffeine molecule is a great way to practice viewing structures in stereo. This is a learnable skill, but it takes practice.
Task:
Access the Stereo Vision tutorial and practice viewing molecular structures in stereo.
Practice at least ...
- two times daily,
- for 3-5 minutes each session,
Keep up your practice throughout the course. It is a wonderful skill that will greatly support your understanding of structural molecular biology. Practice with different molecules and try out different colours and renderings.
Note: do not go through your practice sessions mechanically. If you are not making any progress with stereo vision, contact me so we can help you on the right track.
- TBC
Links and resources
- Molecular Graphics Software Links– a collection of links at the PDB
| Gajiwala & Burley (2000) Winged helix proteins. Curr Opin Struct Biol 10:110-6. (pmid: 10679470) |
|
[ PubMed ] [ DOI ] The winged helix proteins constitute a subfamily within the large ensemble of helix-turn-helix proteins. Since the discovery of the winged helix/fork head motif in 1993, a large number of topologically related proteins with diverse biological functions have been characterized by X-ray crystallography and solution NMR spectroscopy. Recently, a winged helix transcription factor (RFX1) was shown to bind DNA using unprecedented interactions between one of its eponymous wings and the major groove. This surprising observation suggests that the winged helix proteins can be subdivided into at least two classes with radically different modes of DNA recognition. |
| Aravind et al. (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231-62. (pmid: 15808743) |
|
[ PubMed ] [ DOI ] The helix-turn-helix (HTH) domain is a common denominator in basal and specific transcription factors from the three super-kingdoms of life. At its core, the domain comprises of an open tri-helical bundle, which typically binds DNA with the 3rd helix. Drawing on the wealth of data that has accumulated over two decades since the discovery of the domain, we present an overview of the natural history of the HTH domain from the viewpoint of structural analysis and comparative genomics. In structural terms, the HTH domains have developed several elaborations on the basic 3-helical core, such as the tetra-helical bundle, the winged-helix and the ribbon-helix-helix type configurations. In functional terms, the HTH domains are present in the most prevalent transcription factors of all prokaryotic genomes and some eukaryotic genomes. They have been recruited to a wide range of functions beyond transcription regulation, which include DNA repair and replication, RNA metabolism and protein-protein interactions in diverse signaling contexts. Beyond their basic role in mediating macromolecular interactions, the HTH domains have also been incorporated into the catalytic domains of diverse enzymes. We discuss the general domain architectural themes that have arisen amongst the HTH domains as a result of their recruitment to these diverse functions. We present a natural classification, higher-order relationships and phyletic pattern analysis of all the major families of HTH domains. This reconstruction suggests that there were at least 6-11 different HTH domains in the last universal common ancestor of all life forms, which covered much of the structural diversity and part of the functional versatility of the extant representatives of this domain. In prokaryotes the total number of HTH domains per genome shows a strong power-equation type scaling with the gene number per genome. However, the HTH domains in two-component signaling pathways show a linear scaling with gene number, in contrast to the non-linear scaling of HTH domains in single-component systems and sigma factors. These observations point to distinct evolutionary forces in the emergence of different signaling systems with HTH transcription factors. The archaea and bacteria share a number of ancient families of specific HTH transcription factors. However, they do not share any orthologous HTH proteins in the basal transcription apparatus. This differential relationship of their basal and specific transcriptional machinery poses an apparent conundrum regarding the origins of their transcription apparatus. |
Footnotes and references
- ↑ There are several online servers that translate SMILES strings to idealized structures, see e.g. the online SMILES translation service at the NCI.
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.
| < Assignment 4 | Assignment 6 > |