Difference between revisions of "Lecture 09"
Jump to navigation
Jump to search


| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
| − | = | + | <!-- div style="padding: 5px; background: #FF4560; border:solid 2px #000000;"> |
| + | '''Update Warning!''' | ||
| + | This page has not been revised yet for the 2008 Fall term. | ||
| + | Some of the slides will probably be reused, but please consider the page as a whole out of date | ||
| + | as long as this warning appears here. Also, the lectures may be taught in a different sequence. | ||
| + | </div --> | ||
| + | | ||
| + | | ||
| + | <small>[[Lecture_08|(Previous lecture)]] ... [[Lecture_10|(Next lecture)]]</small> | ||
| − | |||
| − | + | <div class="toclimit-3">__TOC__</div> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | ======Slide | + | |
| − | [[Image: | + | <br> |
| + | <br> | ||
| + | <div style="padding: 2px; background: #879BFA; border:solid 1px #AAAAAA;"> | ||
| + | ==Protein Structure Databases== | ||
| + | </div><br> | ||
| + | | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ====Objectives for this part of the course==== | ||
| + | </div><br> | ||
| + | * Understand that "sequence" and "structure" are abstractions of biopolymers.<br> | ||
| + | * Understand that "structure" is an idealized concept, applied to an ensemble of dynamic molecules.<br> | ||
| + | * Be aware of principal methods of experimental structure determination and some of their limitations regarding interpretation of data and resulting accuracy.<br> | ||
| + | * Understand that structures may have considerable local and global uncertainties.<br> | ||
| + | * Know that structure abstractions can be stored, retrieved and visualized and become familiar with the principal databases and information sources for that purpose.<br> | ||
| + | * Be familiar with the contents of a PDB formatted file. | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ====Links summary==== | ||
| + | </div><br> | ||
| + | *[http://www.rcsb.org/pdb The '''PDB''']<br> | ||
| + | |||
| + | *[http://ruppweb.dyndns.org/Xray/101index.html see: Bernhard Rupp's introduction to crystal structure]<br> | ||
| + | *[http://www.biochem.ucl.ac.uk/~roman/procheck/procheck.html '''Procheck''']<br> | ||
| + | *[http://www.wwpdb.org/documentation/format23/sect9.html Coordinate section of the PDB format specification (V 2.3)]<br> | ||
| + | *[http://xray.bmc.uu.se/hicup/ the HIC-Up database of hetero compounds in PDB files]<br> | ||
| + | *[http://pqs.ebi.ac.uk/ The PQS service for Probable Quarternary Structures at the EB]<br> | ||
| + | *[http://ndbserver.rutgers.edu/ The Nucleic Acid Structure Database]<br> | ||
| + | *[http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/ '''PDBsum''']<br> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #879BFA; border:solid 1px #AAAAAA;"> | ||
| + | |||
| + | ==Lecture slides== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ==="Sequence" and "structure" are abstractions of biopolymers=== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | ======Slide 004====== | ||
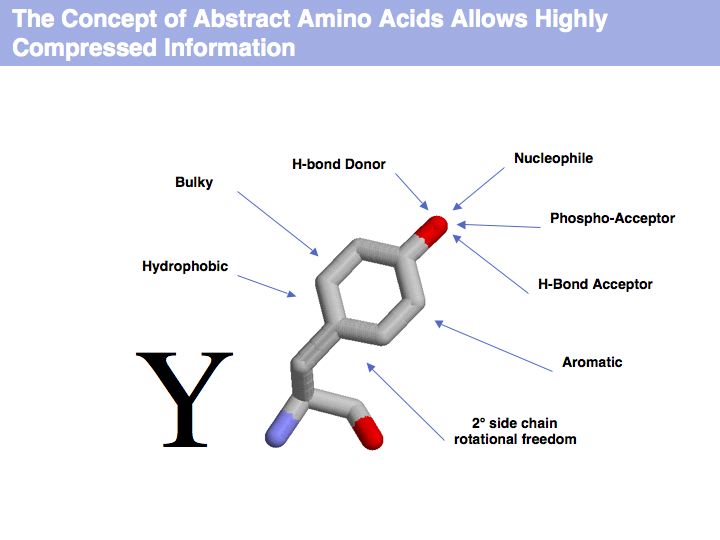
| + | [[Image:09_slide004.jpg|frame|none|Lecture 09, Slide 004<br> | ||
| + | The letter Y represents highly compressed information. | ||
| + | ]] | ||
| + | ======Slide 005====== | ||
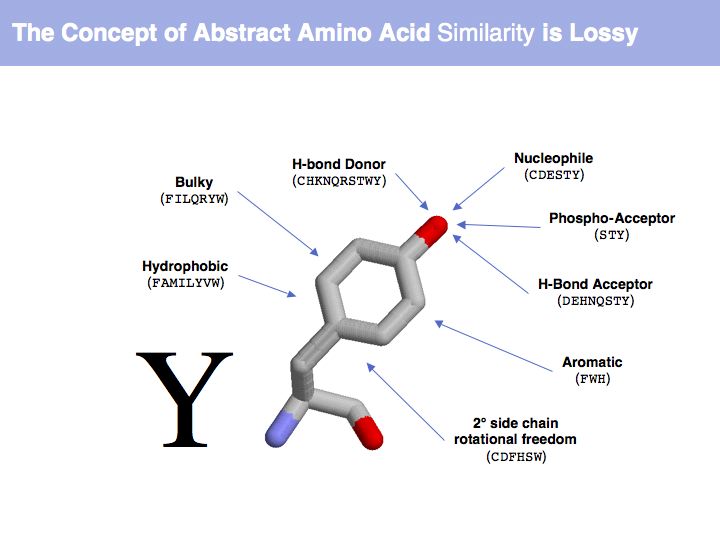
| + | [[Image:09_slide005.jpg|frame|none|Lecture 09, Slide 005<br> | ||
| + | Which amino acid tyrosine is regarded to be similar to depends on which property we are considering. | ||
| + | ]] | ||
| + | ======Slide 006====== | ||
| + | [[Image:09_slide006.jpg|frame|none|Lecture 09, Slide 006<br> | ||
| + | |||
| + | ]] | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ===Structure can be determined experimentally=== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | ======Slide 008====== | ||
| + | [[Image:09_slide008.jpg|frame|none|Lecture 09, Slide 008<br> | ||
| + | |||
| + | ]] | ||
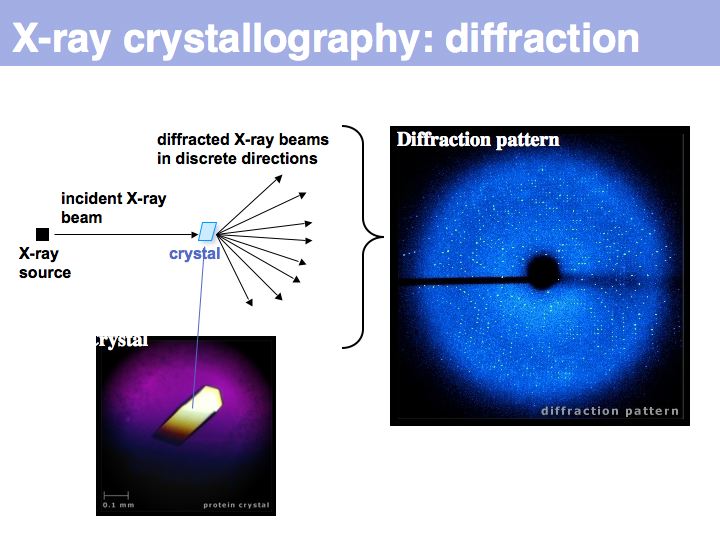
| + | ======Slide 009====== | ||
| + | [[Image:09_slide009.jpg|frame|none|Lecture 09, Slide 009<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 010====== | ||
| + | [[Image:09_slide010.jpg|frame|none|Lecture 09, Slide 010<br> | ||
| + | [http://ruppweb.dyndns.org/Xray/101index.html see: Bernhard Rupp's introduction to crystal structure] | ||
| + | ]] | ||
| + | ======Slide 011====== | ||
| + | [[Image:09_slide011.jpg|frame|none|Lecture 09, Slide 011<br> | ||
| + | The inability to measure the '''phases''' of diffracted photons prevents the reconstruction of the diffracting objects from on set of experimental measurements alone. Additional information must be sought, based on the fact that photons that are '''in phase''' enhance the measured intensities, whereas photons that are '''phase-shifted''' by 180° cancel each other's intnesities. Thus measuring intensity changes if additional diffraction centres are placed into the structure allows us to infer relative phases, and if several relative phases are known, we can triangulate thier absolute values. Experimental error makes this a difficult problem, but undr favourable circumstances, the electron density map will be interpretable; a structurl model can the be built and refined. | ||
| + | ]] | ||
| + | ======Slide 012====== | ||
| + | [[Image:09_slide012.jpg|frame|none|Lecture 09, Slide 012<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 013====== | ||
| + | [[Image:09_slide013.jpg|frame|none|Lecture 09, Slide 013<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 014====== | ||
| + | [[Image:09_slide014.jpg|frame|none|Lecture 09, Slide 014<br> | ||
]] | ]] | ||
| − | ======Slide | + | ======Slide 015====== |
| − | [[Image: | + | [[Image:09_slide015.jpg|frame|none|Lecture 09, Slide 015<br> |
]] | ]] | ||
| − | ======Slide | + | ======Slide 016====== |
| − | [[Image: | + | [[Image:09_slide016.jpg|frame|none|Lecture 09, Slide 016<br> |
| + | *[http://www.biochem.ucl.ac.uk/~roman/procheck/procheck.html '''Procheck'''] | ||
| + | ]] | ||
| + | |||
| + | ======Slide 017====== | ||
| + | [[Image:09_slide017.jpg|frame|none|Lecture 09, Slide 017<br> | ||
]] | ]] | ||
| − | |||
| − | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ===Structure abstractions can be held in databases=== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | ======Slide 019====== | ||
| + | [[Image:09_slide019.jpg|frame|none|Lecture 09, Slide 019<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 020====== | ||
| + | [[Image:09_slide020.jpg|frame|none|Lecture 09, Slide 020<br> | ||
| + | [http://www.rcsb.org/pdb The '''PDB'''] | ||
| + | ]] | ||
| + | ======Slide 021====== | ||
| + | [[Image:09_slide021.jpg|frame|none|Lecture 09, Slide 021<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 022====== | ||
| + | [[Image:09_slide022.jpg|frame|none|Lecture 09, Slide 022<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 023====== | ||
| + | [[Image:09_slide023.jpg|frame|none|Lecture 09, Slide 023<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 024====== | ||
| + | [[Image:09_slide024.jpg|frame|none|Lecture 09, Slide 024<br> | ||
| + | Additional complications arise from "insertion codes". These are leters that allow the insertion of residues in a common numbering scheme of sequences that are compared between several homologes. In principle this is a good idea, since this makes comparison of residues much easier. But strings such as "23A" can no longer be tretaed as "sequence '''numbers'''" - thet are sequence labels and using them correctly can be a challenge. | ||
]] | ]] | ||
| − | ======Slide | + | ======Slide 025====== |
| − | [[Image: | + | [[Image:09_slide025.jpg|frame|none|Lecture 09, Slide 025<br> |
| − | + | Potential pitfalls:<br> | |
| + | * Record type: changes not consistently applied for modifications<br> | ||
| + | * Atom number: rarely used and a nuisance to update when changing.<br> | ||
| + | * Item<br> | ||
| + | * Atom name: careful about columns<br> | ||
| + | * Amino acid type: selenocysteine. Some very old files use TRY for TRP<br> | ||
| + | * Chain<br> | ||
| + | * Alternate location<br> | ||
| + | * Sequence number<br> | ||
| + | * X,Y, and Z are given in Å (10<sup-10</sup> m = 0.1 nm) values in a cartesian (i.e. orthogonal) coordinate system; but origin and orientation is arbitrary!<br> | ||
| + | * Occupancy can describe: special locations, partially bound ligands, unobserved fragments of structure<br> | ||
| + | * B-values, (also called temperature factors) are a measure of the volume of space around into which a <br> | ||
| + | <br> | ||
| + | Read the [http://www.wwpdb.org/documentation/format23/sect9.html Coordinate section of the PDB format specification (V 2.3)] | ||
| + | ]] | ||
| + | ======Slide 026====== | ||
| + | [[Image:09_slide026.jpg|frame|none|Lecture 09, Slide 026<br> | ||
| + | See [http://xray.bmc.uu.se/hicup/ the HIC-Up database of hetero compounds in PDB files]. | ||
]] | ]] | ||
| + | ======Slide 027====== | ||
| + | [[Image:09_slide027.jpg|frame|none|Lecture 09, Slide 027<br> | ||
| − | |||
| − | |||
| − | |||
]] | ]] | ||
| + | ======Slide 028====== | ||
| + | [[Image:09_slide028.jpg|frame|none|Lecture 09, Slide 028<br> | ||
| − | |||
| − | |||
| − | |||
]] | ]] | ||
| + | ======Slide 029====== | ||
| + | [[Image:09_slide029.jpg|frame|none|Lecture 09, Slide 029<br> | ||
| + | [http://pqs.ebi.ac.uk/ The PQS service for Probable Quarternary Structures at the EB] | ||
| + | ]] | ||
| + | ======Slide 030====== | ||
| + | [[Image:09_slide030.jpg|frame|none|Lecture 09, Slide 030<br> | ||
| − | ======Slide | + | ]] |
| − | [[Image: | + | ======Slide 031====== |
| + | [[Image:09_slide031.jpg|frame|none|Lecture 09, Slide 031<br> | ||
| + | [http://ndbserver.rutgers.edu/ The Nucleic Acid Structure Database] | ||
| + | ]] | ||
| + | ======Slide 032====== | ||
| + | [[Image:09_slide032.jpg|frame|none|Lecture 09, Slide 032<br> | ||
| + | [http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/ '''PDBsum'''] is a secondary database thst stores analyss and interpretation information for PDB coordinate sets. | ||
| + | ]] | ||
| + | ======Slide 033====== | ||
| + | [[Image:09_slide033.jpg|frame|none|Lecture 09, Slide 033<br> | ||
]] | ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | ---- | ||
| + | <small>[[Lecture_08|(Previous lecture)]] ... [[Lecture_10|(Next lecture)]]</small> | ||
Latest revision as of 20:53, 9 December 2007
(Previous lecture) ... (Next lecture)
Protein Structure Databases
Objectives for this part of the course
- Understand that "sequence" and "structure" are abstractions of biopolymers.
- Understand that "structure" is an idealized concept, applied to an ensemble of dynamic molecules.
- Be aware of principal methods of experimental structure determination and some of their limitations regarding interpretation of data and resulting accuracy.
- Understand that structures may have considerable local and global uncertainties.
- Know that structure abstractions can be stored, retrieved and visualized and become familiar with the principal databases and information sources for that purpose.
- Be familiar with the contents of a PDB formatted file.
Links summary
- see: Bernhard Rupp's introduction to crystal structure
- Procheck
- Coordinate section of the PDB format specification (V 2.3)
- the HIC-Up database of hetero compounds in PDB files
- The PQS service for Probable Quarternary Structures at the EB
- The Nucleic Acid Structure Database
- PDBsum
Lecture slides
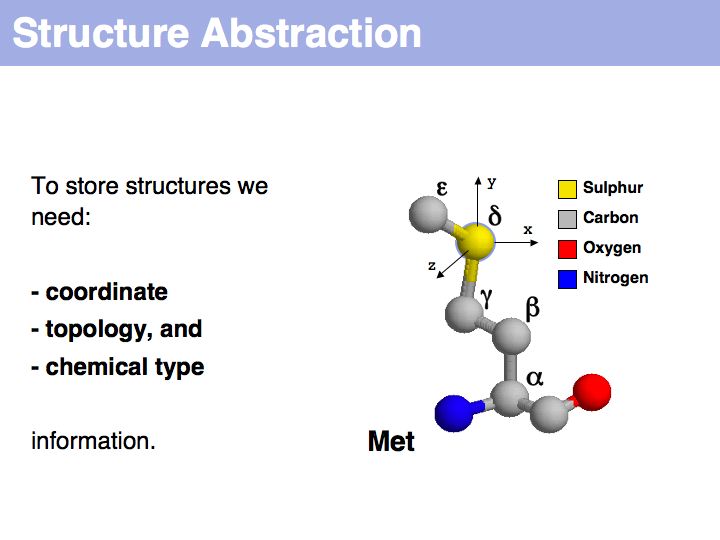
"Sequence" and "structure" are abstractions of biopolymers
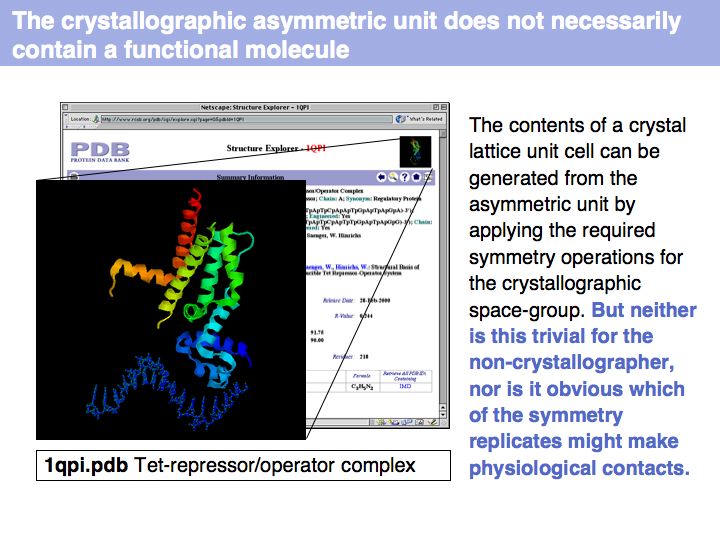
Slide 004
Slide 005
Slide 006
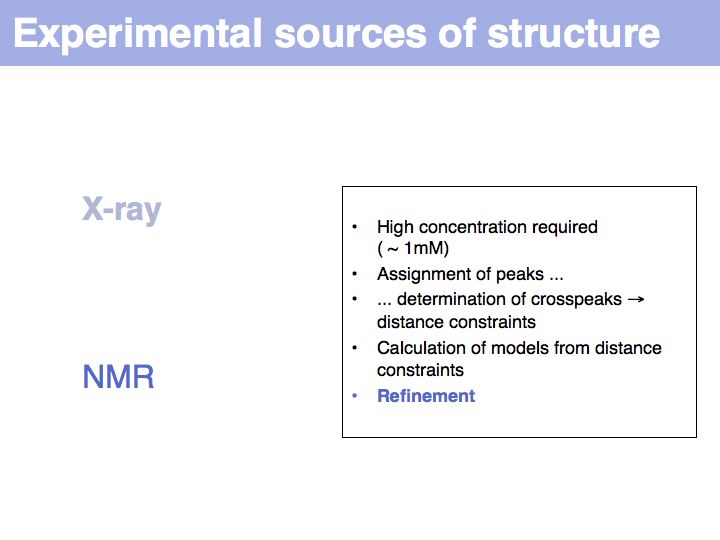
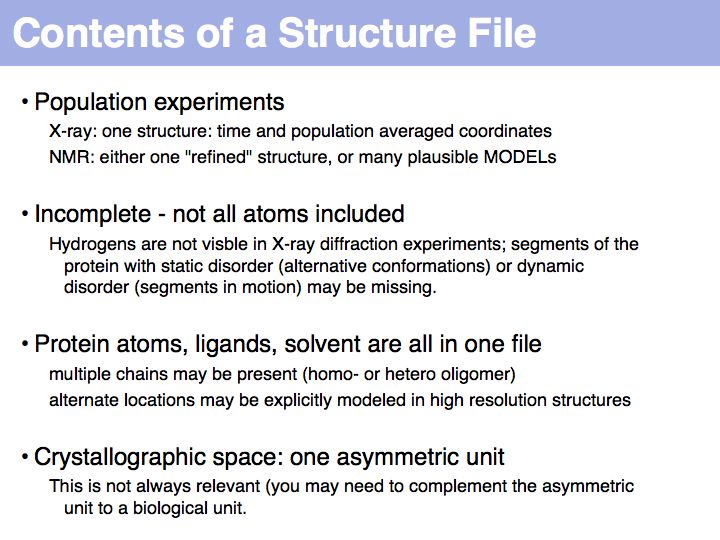
Structure can be determined experimentally
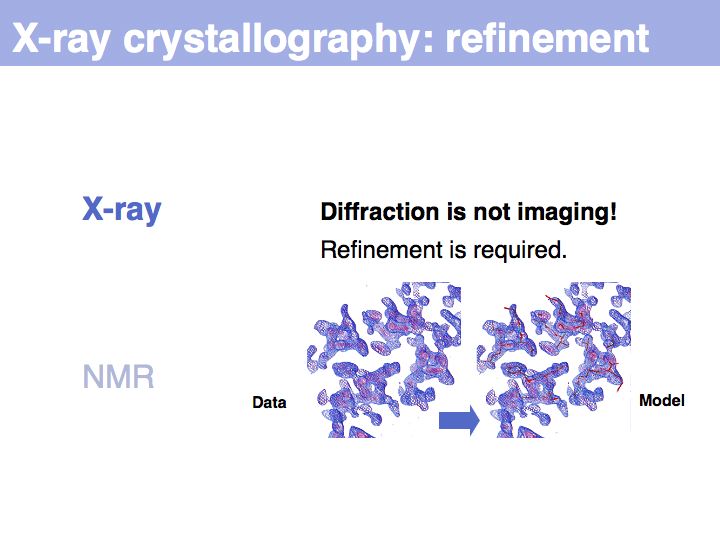
Slide 008
Slide 009
Slide 010

Lecture 09, Slide 010
see: Bernhard Rupp's introduction to crystal structure
see: Bernhard Rupp's introduction to crystal structure
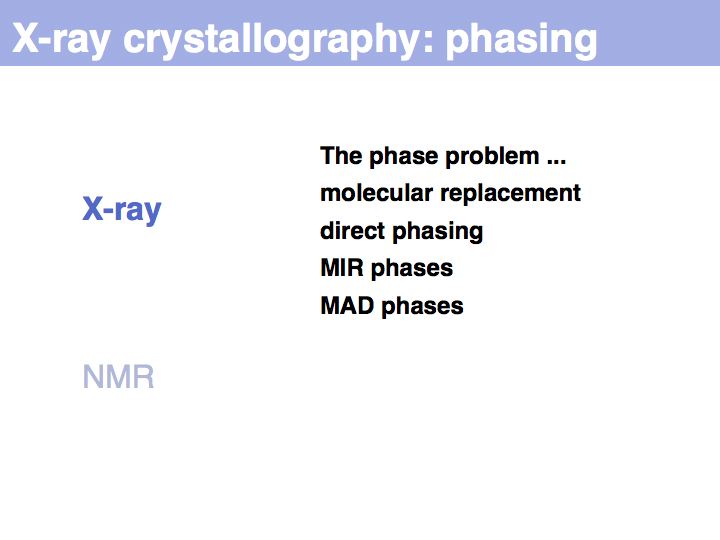
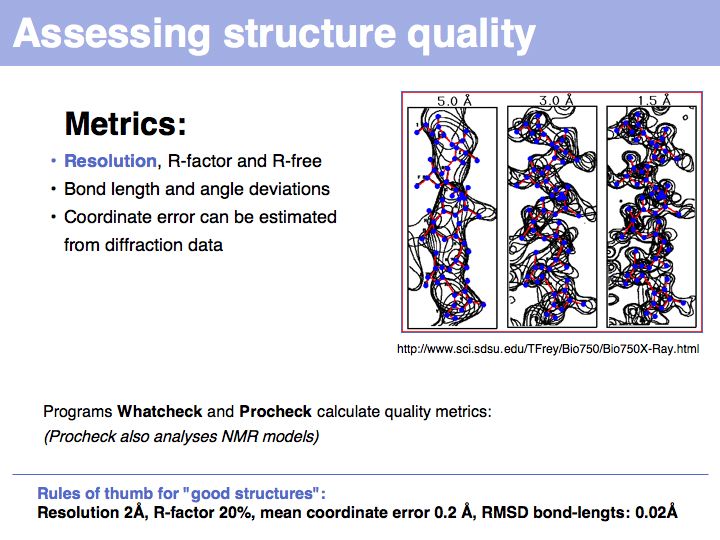
Slide 011

Lecture 09, Slide 011
The inability to measure the phases of diffracted photons prevents the reconstruction of the diffracting objects from on set of experimental measurements alone. Additional information must be sought, based on the fact that photons that are in phase enhance the measured intensities, whereas photons that are phase-shifted by 180° cancel each other's intnesities. Thus measuring intensity changes if additional diffraction centres are placed into the structure allows us to infer relative phases, and if several relative phases are known, we can triangulate thier absolute values. Experimental error makes this a difficult problem, but undr favourable circumstances, the electron density map will be interpretable; a structurl model can the be built and refined.
The inability to measure the phases of diffracted photons prevents the reconstruction of the diffracting objects from on set of experimental measurements alone. Additional information must be sought, based on the fact that photons that are in phase enhance the measured intensities, whereas photons that are phase-shifted by 180° cancel each other's intnesities. Thus measuring intensity changes if additional diffraction centres are placed into the structure allows us to infer relative phases, and if several relative phases are known, we can triangulate thier absolute values. Experimental error makes this a difficult problem, but undr favourable circumstances, the electron density map will be interpretable; a structurl model can the be built and refined.
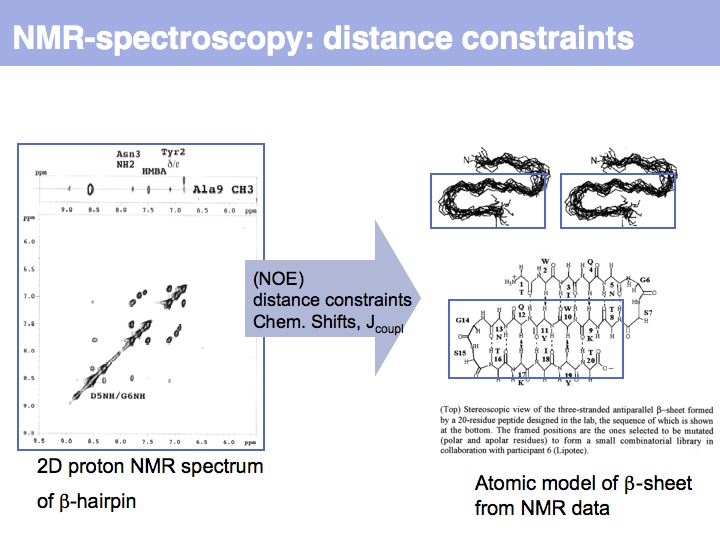
Slide 012
Slide 013
Slide 014
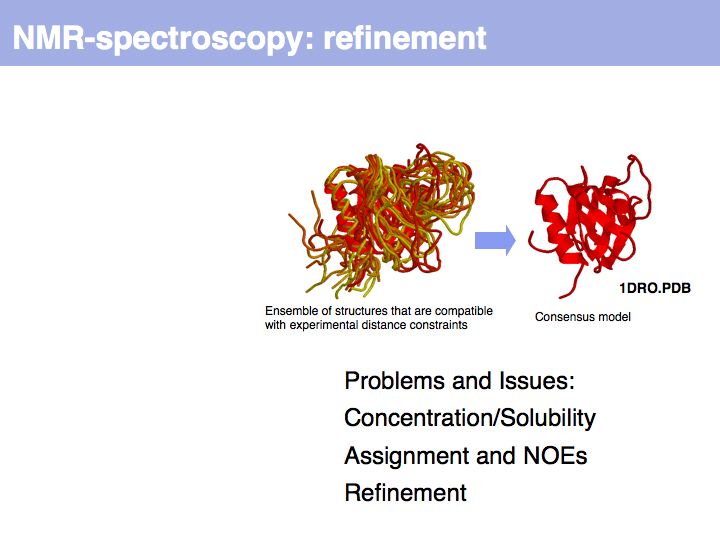
Slide 015
Slide 016

Lecture 09, Slide 016
*Procheck
*Procheck
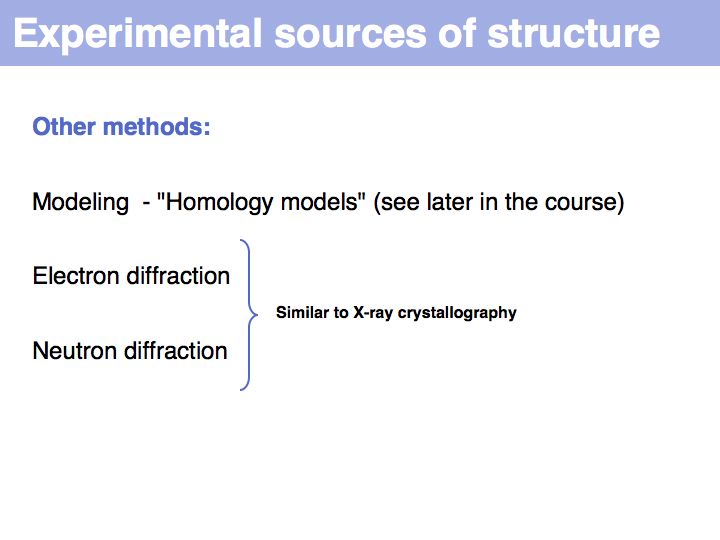
Slide 017
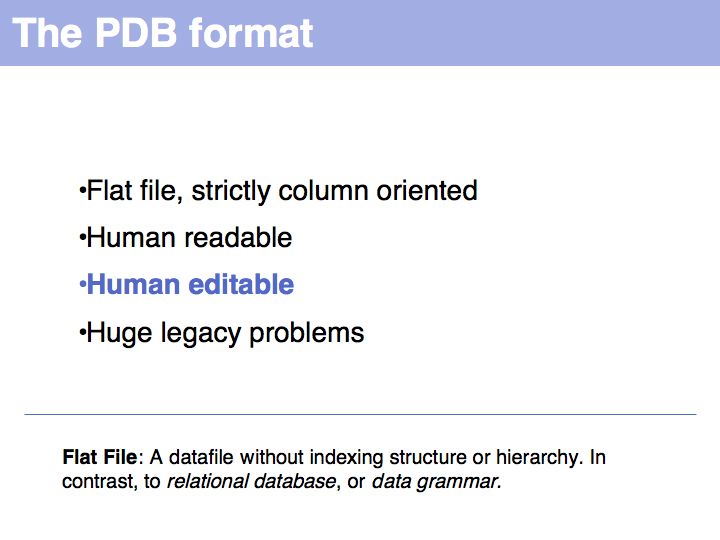
Structure abstractions can be held in databases
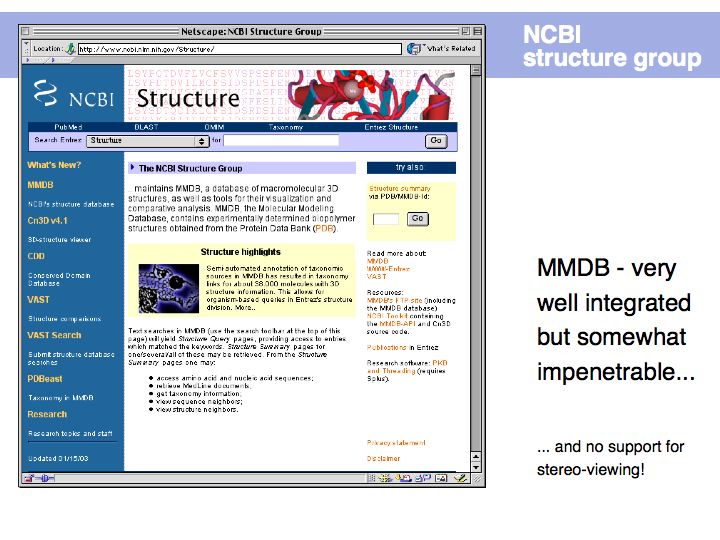
Slide 019
Slide 020

Lecture 09, Slide 020
The PDB
The PDB
Slide 021
Slide 022
Slide 023
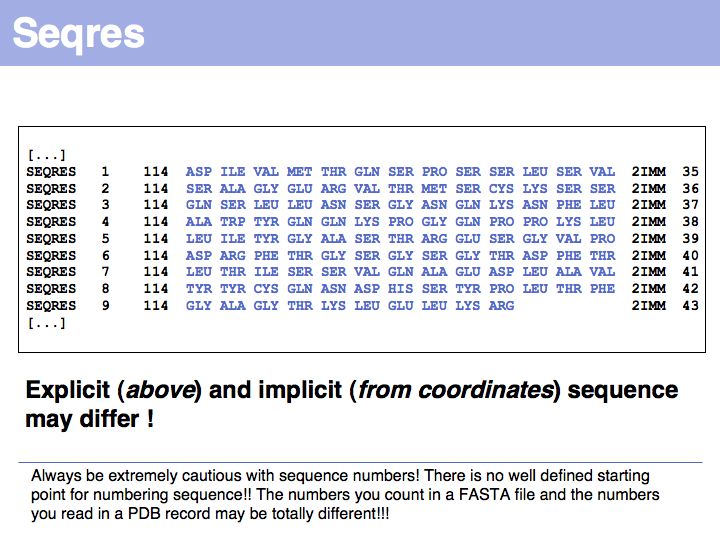
Slide 024

Lecture 09, Slide 024
Additional complications arise from "insertion codes". These are leters that allow the insertion of residues in a common numbering scheme of sequences that are compared between several homologes. In principle this is a good idea, since this makes comparison of residues much easier. But strings such as "23A" can no longer be tretaed as "sequence numbers" - thet are sequence labels and using them correctly can be a challenge.
Additional complications arise from "insertion codes". These are leters that allow the insertion of residues in a common numbering scheme of sequences that are compared between several homologes. In principle this is a good idea, since this makes comparison of residues much easier. But strings such as "23A" can no longer be tretaed as "sequence numbers" - thet are sequence labels and using them correctly can be a challenge.
Slide 025

Lecture 09, Slide 025
Potential pitfalls:
* Record type: changes not consistently applied for modifications
* Atom number: rarely used and a nuisance to update when changing.
* Item
* Atom name: careful about columns
* Amino acid type: selenocysteine. Some very old files use TRY for TRP
* Chain
* Alternate location
* Sequence number
* X,Y, and Z are given in Å (10<sup-10 m = 0.1 nm) values in a cartesian (i.e. orthogonal) coordinate system; but origin and orientation is arbitrary!
* Occupancy can describe: special locations, partially bound ligands, unobserved fragments of structure
* B-values, (also called temperature factors) are a measure of the volume of space around into which a
Read the Coordinate section of the PDB format specification (V 2.3)
Potential pitfalls:
* Record type: changes not consistently applied for modifications
* Atom number: rarely used and a nuisance to update when changing.
* Item
* Atom name: careful about columns
* Amino acid type: selenocysteine. Some very old files use TRY for TRP
* Chain
* Alternate location
* Sequence number
* X,Y, and Z are given in Å (10<sup-10 m = 0.1 nm) values in a cartesian (i.e. orthogonal) coordinate system; but origin and orientation is arbitrary!
* Occupancy can describe: special locations, partially bound ligands, unobserved fragments of structure
* B-values, (also called temperature factors) are a measure of the volume of space around into which a
Read the Coordinate section of the PDB format specification (V 2.3)
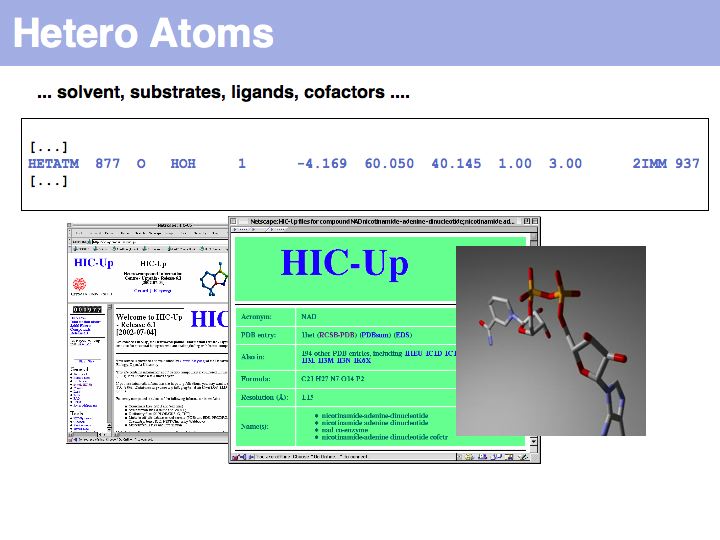
Slide 026

Lecture 09, Slide 026
See the HIC-Up database of hetero compounds in PDB files.
See the HIC-Up database of hetero compounds in PDB files.
Slide 027
Slide 028
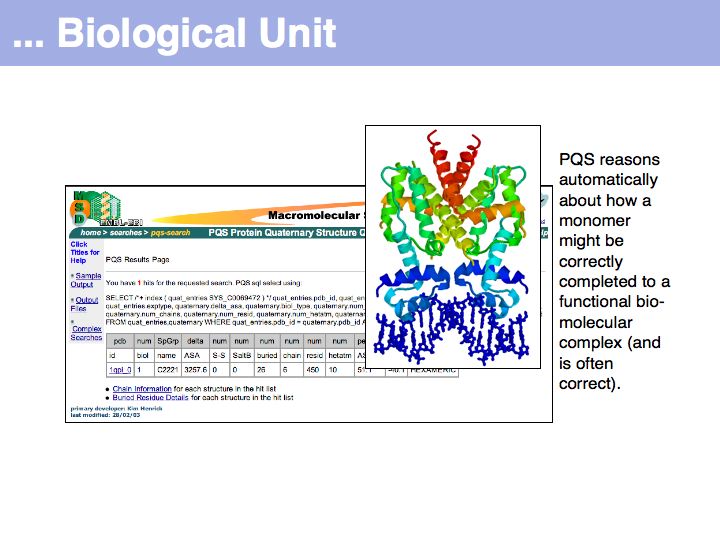
Slide 029

Lecture 09, Slide 029
The PQS service for Probable Quarternary Structures at the EB
The PQS service for Probable Quarternary Structures at the EB
Slide 030
Slide 031

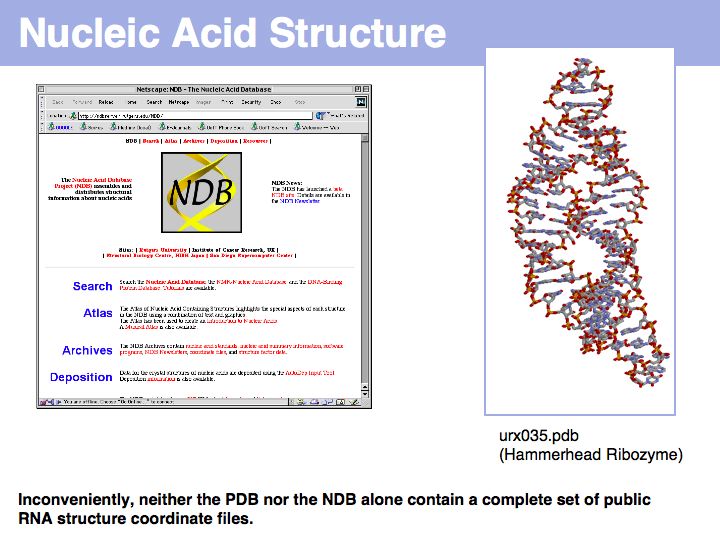
Lecture 09, Slide 031
The Nucleic Acid Structure Database
The Nucleic Acid Structure Database
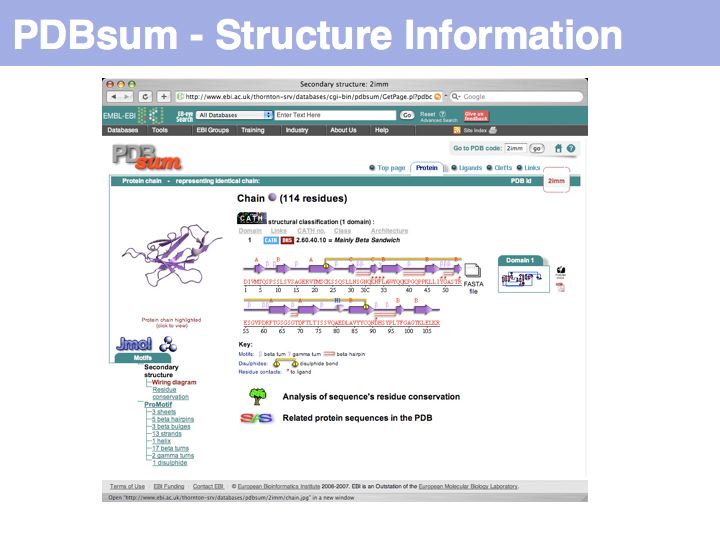
Slide 032

Lecture 09, Slide 032
PDBsum is a secondary database thst stores analyss and interpretation information for PDB coordinate sets.
PDBsum is a secondary database thst stores analyss and interpretation information for PDB coordinate sets.
Slide 033