Difference between revisions of "Lecture 04"
Jump to navigation
Jump to search
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | <div style="padding: 5px; background: #FF4560; border:solid 2px #000000;"> | + | <!-- div style="padding: 5px; background: #FF4560; border:solid 2px #000000;"> |
'''Update Warning!''' | '''Update Warning!''' | ||
This page has not been revised yet for the 2007 Fall term. Some of the slides may be reused, but please consider the page as a whole out of date as long as this warning appears here. | This page has not been revised yet for the 2007 Fall term. Some of the slides may be reused, but please consider the page as a whole out of date as long as this warning appears here. | ||
| − | </div> | + | </div --> |
| | ||
| Line 12: | Line 12: | ||
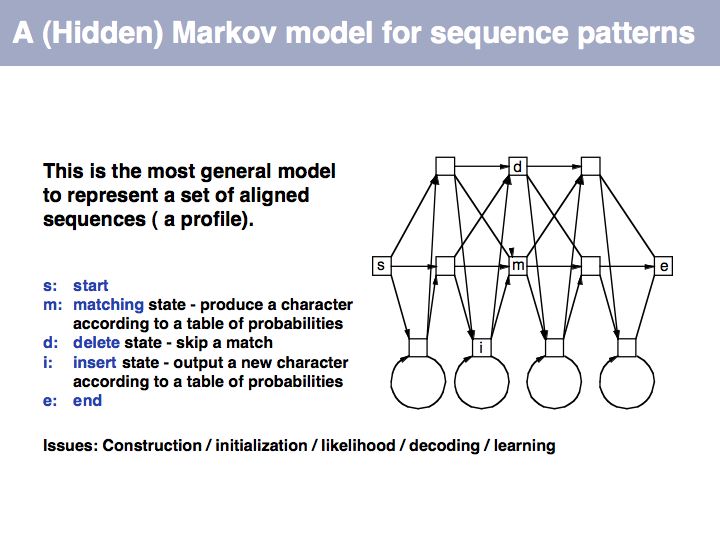
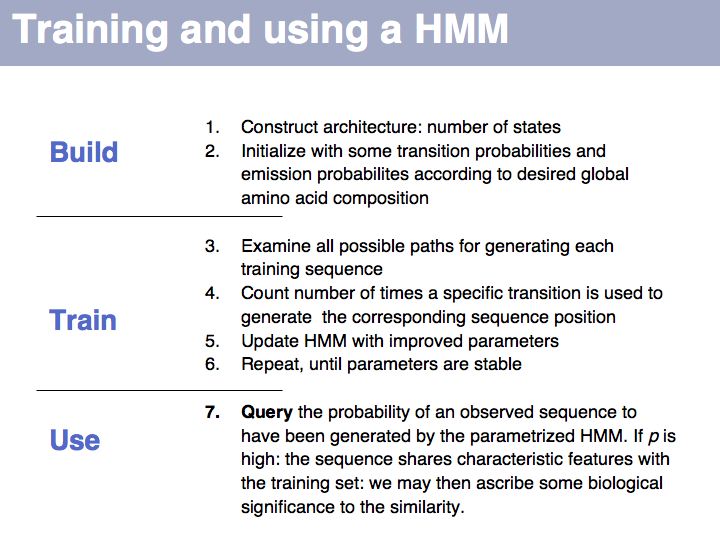
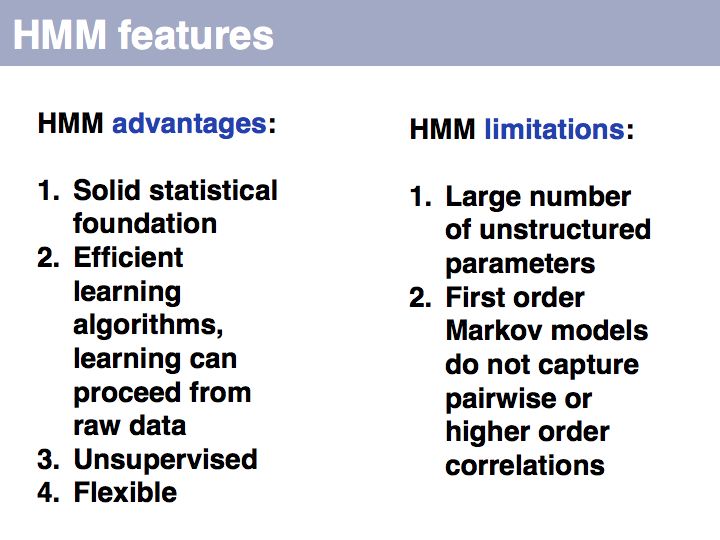
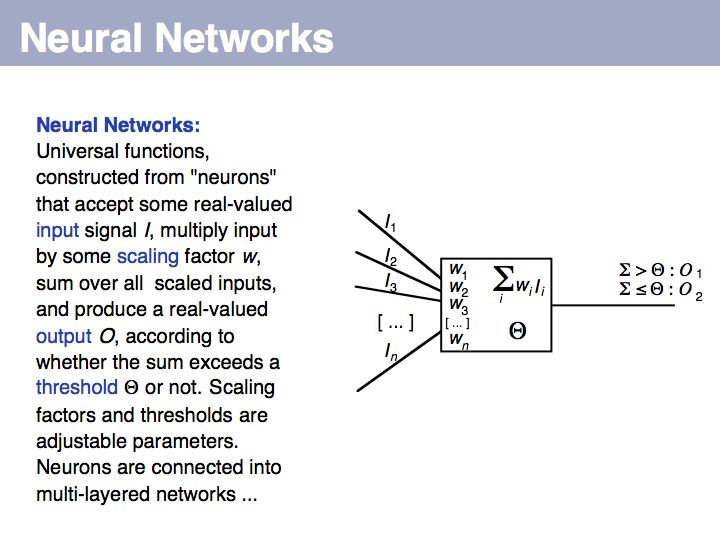
==Sequence Analysis== | ==Sequence Analysis== | ||
| − | + | ;What you should take home from this part of the course: | |
| − | + | *Understand key concepts in probabilistic pattern representation and matching, especially PSSMs. Understand that machine-learning tools such as HMMs (Hidden Markov Models) and NN (Neural Networks) can be used for probabilistic pattern matching and classification. | |
| − | * | + | *Understand the concept of a sequence logo. |
| − | * Exercises | + | *Be familiar with the SignalP Web server. |
| − | * | + | *Know basic concepts of statistics and probability theory, key terms of descriptive statistics; |
| + | *Understand probability tables in principle; | ||
| + | *Have encountered important probability distributions; | ||
| + | *Understand different error types; | ||
| + | *Understand the terms: significance, confidence interval and statistical test. | ||
| + | *Be familiar with the concepts and strategy of simulation testing and understand why its simplicity is making an important contribution to computational biology. | ||
| + | | ||
| + | |||
| + | ;Links summary: | ||
| + | *[http://weblogo.berkeley.edu/ WebLogo] | ||
| + | *[http://www.lecb.ncifcrf.gov/~toms/sequencelogo.html Tom Schneider's Sequence Logo pages] (and introductions to information theory) | ||
| + | *[http://www.cbs.dtu.dk/services/SignalP/ The SignalP server] | ||
| + | | ||
| + | |||
| + | ;Exercises | ||
| + | |||
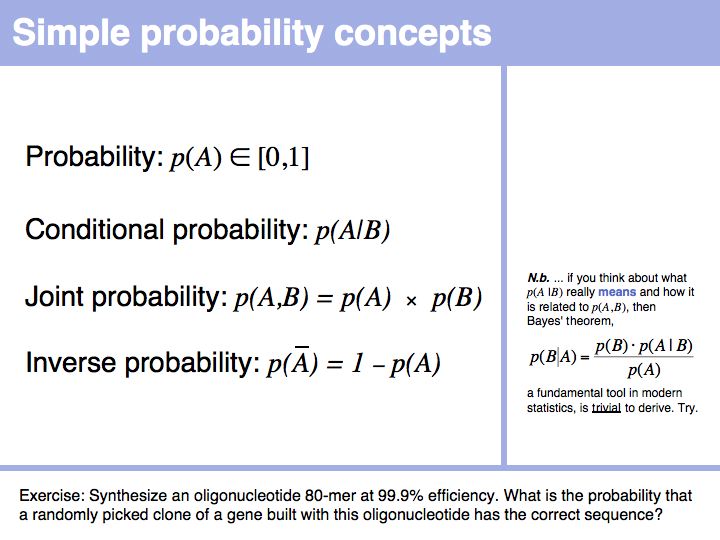
| + | *If you assume that an 80-mer oligonucleotide can be synthesized with 99.9% coupling efficiency per step and a 0.2% chance of coupling a leftover nucleotide from the previous synthesis step, what is the probability that a randomly picked clone of a gene built with this oligonucleotide has the correct sequence? | ||
| + | *In a recent doctoral thesis defence the candidate claimed that in a microarray expression analysis he was able to show reciprocal regulation of two genes (one related to immune stimulation, the other related to immune suppression): this would mean whenever one gene is regulated up, the other is downregulated, and ''vice versa''. The claim was based on observing this effect in eight of ten experiments. Expression levels were scored semiquantitatively on a scale of (++,+,0,-, and --). Given that such experiments have experimental error as well as biological variability, '''sketch''' a simulation test that would analyse whether in fact a significant (anti)correlation had been observed, or whether this result could just as well be due to meaningless fluctuations. | ||
| + | |||
| + | | ||
| + | | ||
==Lecture Slides== | ==Lecture Slides== | ||
======Slide 001====== | ======Slide 001====== | ||
| − | [[Image:L04_s001.jpg|frame|none|Lecture 04, Slide 001]] | + | [[Image:L04_s001.jpg|frame|none|Lecture 04, Slide 001<br> |
| + | From the [http://sciencenow.sciencemag.org/cgi/content/full/2007/914/2 Science News, Sept. 14]. As far as systems biology complexities go, this one must be near the top: intimate interactions between human's most- and second-most complex systems. The key method here is a bioinformatics approach to classifying genes: pattern searches in the promoter regions. (''NB. Not studying in isolation but forming study groups is an excellent idea!''). Are you more lonely than average ? Check with the [http://www3.shastacollege.edu/lvalvatne/psych15/ucla_loneliness_scale.htm UCLA loneliness scale]. | ||
| + | ]] | ||
======Slide 002====== | ======Slide 002====== | ||
| − | [[Image:L04_s002.jpg|frame|none|Lecture 04, Slide 002]] | + | [[Image:L04_s002.jpg|frame|none|Lecture 04, Slide 002<br> |
| + | |||
| + | ]] | ||
======Slide 003====== | ======Slide 003====== | ||
| − | [[Image:L04_s003.jpg|frame|none|Lecture 04, Slide 003]] | + | [[Image:L04_s003.jpg|frame|none|Lecture 04, Slide 003<br> |
| + | |||
| + | ]] | ||
======Slide 004====== | ======Slide 004====== | ||
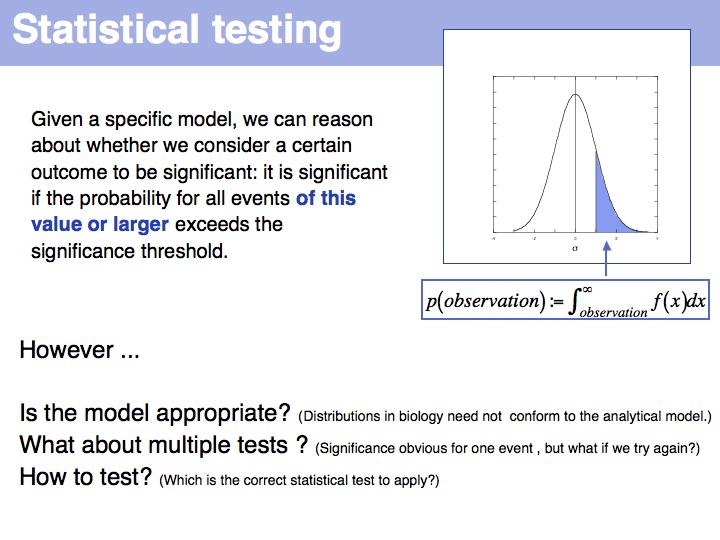
| − | [[Image:L04_s004.jpg|frame|none|Lecture 04, Slide 004]] | + | [[Image:L04_s004.jpg|frame|none|Lecture 04, Slide 004<br> |
| + | |||
| + | ]] | ||
======Slide 005====== | ======Slide 005====== | ||
| − | [[Image:L04_s005.jpg|frame|none|Lecture 04, Slide 005]] | + | [[Image:L04_s005.jpg|frame|none|Lecture 04, Slide 005<br> |
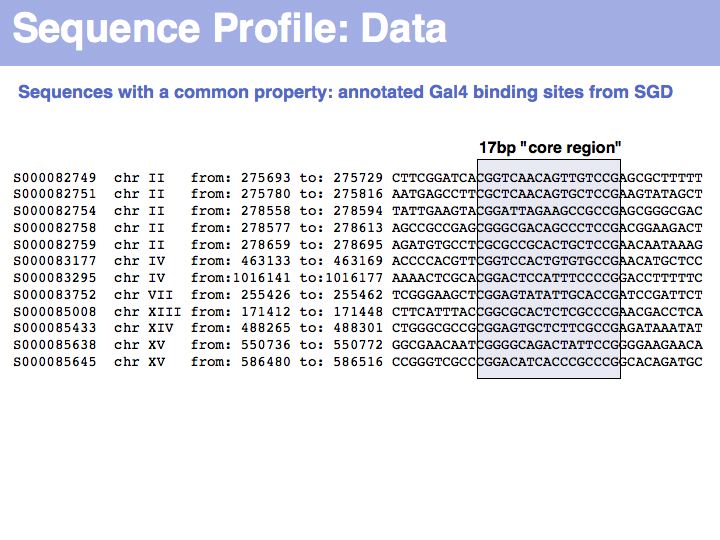
| + | To generate this collection of sequences, the feature "Gal4-binding-site" was searched in the [''Saccharomyces'' Genome Database SGD]; in the resulting [http://db.yeastgenome.org/cgi-bin/gbrowse/scgenome/?name=GAL4_binding_site overview page] binding site annotations recorded by [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=15343339 Harbison ''et al.'' (2004)] were shown for all occurrences; the actual sequences were retrieved by specifying the genome coordinates in the [http://db.yeastgenome.org/cgi-bin/getSeq?chr=2&beg=275703&end=275719 appropriate search form] of the database. I have added ten bases upstream and downstream of the core binding region. This procedure '''could''' be done by hand in about the same time it took me to write the small ''screen-scraping'' program to fetch the sequences. Depending on your programming proficiency, you will find that some tasks can efficiently be done manually, for some tasks it is more efficient to spend the time to search for a better way to achieve them on the Web and only for a comparatively small number of tasks it is worthwhile (or mandatory) to write your own code. | ||
| + | ]] | ||
======Slide 006====== | ======Slide 006====== | ||
| − | [[Image:L04_s006.jpg|frame|none|Lecture 04, Slide 006]] | + | [[Image:L04_s006.jpg|frame|none|Lecture 04, Slide 006<br> |
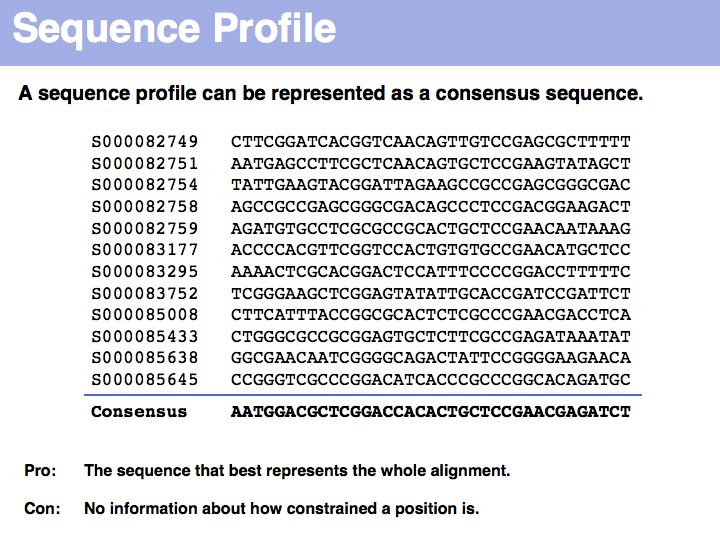
| + | A consensus sequence simply lists the most frequent amino acid or nucleotide at each position, or a random one if there is more than one with the highest frequency. The consensus sequence is the one that you would synthesize to make an idealized representative of the set. It is likely to bind more tightly or to be more stable than each of the individual sequences in the alignment. | ||
| + | ]] | ||
======Slide 007====== | ======Slide 007====== | ||
| − | [[Image:L04_s007.jpg|frame|none|Lecture 04, Slide 007]] | + | [[Image:L04_s007.jpg|frame|none|Lecture 04, Slide 007<br> |
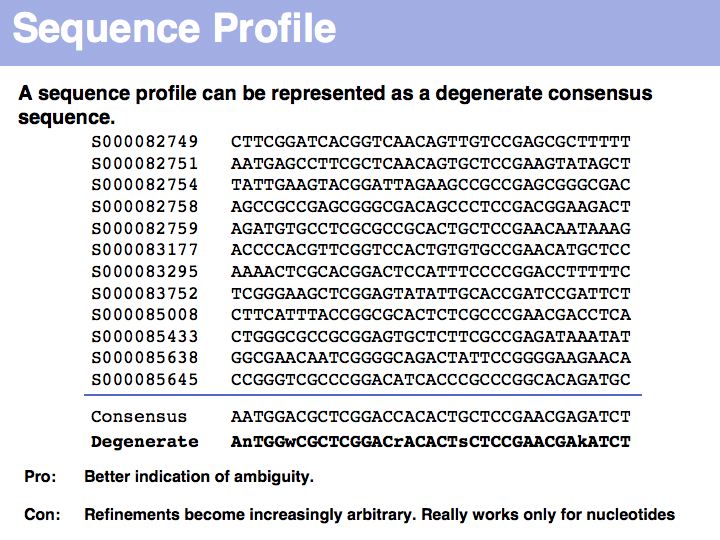
| + | Introducing nucleotide ambiguity codes to represent situations in which more than one nucleotide has the highest frequency improves the situation a bit, but there is also ambiguity. Consider the situation after the conserved '''<tt>CCG</tt>''' pattern: 9 '''<tt>A</tt>'''s and 3'''<tt>G</tt>'''s: should we report the consensus'''<tt>A</tt>'''A, or ist it more interesting to report that the only observed alternative is another purine base and write '''<tt>Y</tt>''' instead? | ||
| + | ]] | ||
======Slide 008====== | ======Slide 008====== | ||
| − | [[Image:L04_s008.jpg|frame|none|Lecture 04, Slide 008]] | + | [[Image:L04_s008.jpg|frame|none|Lecture 04, Slide 008<br> |
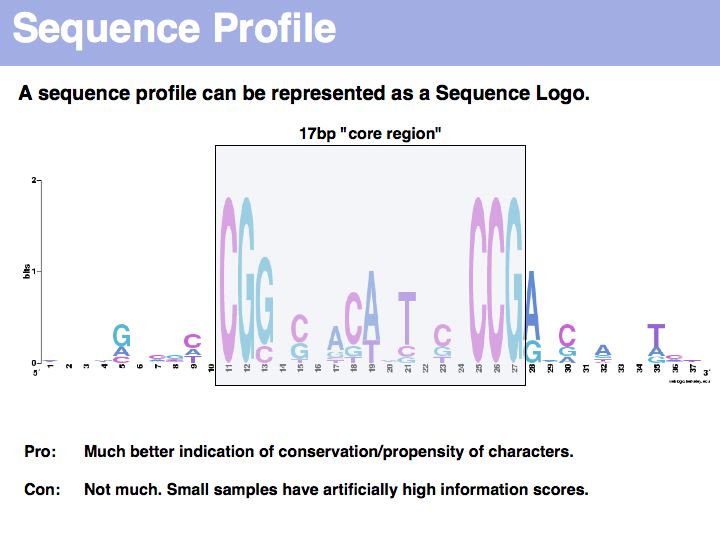
| + | <!-- used the following colors: A=698BD1 C=69B5D1 G=BA69D1 T=BA69D1 -->Sequence logo of Gal4 binding sites with 10 nucleotides flanking bases. Created with [http://weblogo.berkeley.edu/ '''WebLogo''']. A Sequence Logo is a graphical representation of aligned sequences where at each position the height of a column is proportional to the (Shannon) information of that position and the relative size of each character is proportional to its frequency within the column. Sequence Logos were pioneered by [http://www.lecb.ncifcrf.gov/~toms/sequencelogo.html Tom Schneider] who maintains an informative Website about their use and theoretical foundations. Note that there is considerable additional information in the flanking sequences that are not included in the published description of the core binding pattern; it is advantageous if you are able to rerun such analyses, rather than rely on someone else's opinion. | ||
| + | ]] | ||
======Slide 009====== | ======Slide 009====== | ||
| − | [[Image:L04_s009.jpg|frame|none|Lecture 04, Slide 009]] | + | [[Image:L04_s009.jpg|frame|none|Lecture 04, Slide 009<br> |
| + | |||
| + | ]] | ||
======Slide 010====== | ======Slide 010====== | ||
| − | [[Image:L04_s010.jpg|frame|none|Lecture 04, Slide 010]] | + | [[Image:L04_s010.jpg|frame|none|Lecture 04, Slide 010<br> |
| + | |||
| + | ]] | ||
======Slide 011====== | ======Slide 011====== | ||
| − | [[Image:L04_s011.jpg|frame|none|Lecture 04, Slide 011]] | + | [[Image:L04_s011.jpg|frame|none|Lecture 04, Slide 011<br> |
| + | Since log(0) is not defined, we have to introduce an arbitrary correction for unobserved characters. In this example I have simply added 0.1 to each character frequency before calculating log odds. | ||
| + | ]] | ||
======Slide 012====== | ======Slide 012====== | ||
| − | [[Image:L04_s012.jpg|frame|none|Lecture 04, Slide 012]] | + | [[Image:L04_s012.jpg|frame|none|Lecture 04, Slide 012<br> |
| + | In this informal example, I have simply counted matches with the consensus sequence (excluding "N"). We can slide the PSSM over the entire chromosome, and calculate scores for each position. Only the middle sequence is an annotated binding site. Whatever method we use for probabilistic pattern matching, we will '''always''' get a score. It is then '''our '''problem to decide what the score means. | ||
| + | ]] | ||
======Slide 013====== | ======Slide 013====== | ||
| − | [[Image:L04_s013.jpg|frame|none|Lecture 04, Slide 013]] | + | [[Image:L04_s013.jpg|frame|none|Lecture 04, Slide 013<br> |
| + | |||
| + | ]] | ||
======Slide 014====== | ======Slide 014====== | ||
| − | [[Image:L04_s014.jpg|frame|none|Lecture 04, Slide 014]] | + | [[Image:L04_s014.jpg|frame|none|Lecture 04, Slide 014<br> |
| + | |||
| + | ]] | ||
======Slide 015====== | ======Slide 015====== | ||
| − | [[Image:L04_s015.jpg|frame|none|Lecture 04, Slide 015]] | + | [[Image:L04_s015.jpg|frame|none|Lecture 04, Slide 015<br> |
| + | This ''first order Markov model'' depends only on the current state. Higher-order models take increasing lengths of "history" into account, how the system arrived in its current state. Note that the exit probabilities fo a state always have to sum to 1.0. The so called "stationary probability" over a long period of time for p(rain) is 0.167 - this is determined by the combined effects of all individual transition probabilities. The stationary probabilities for two- or three consecutive rainy days are 4.2% and 2.1%, respectively. | ||
| + | ]] | ||
======Slide 016====== | ======Slide 016====== | ||
| − | [[Image:L04_s016.jpg|frame|none|Lecture 04, Slide 016]] | + | [[Image:L04_s016.jpg|frame|none|Lecture 04, Slide 016<br> |
| + | Hidden Markov Model: on [http://en.wikipedia.org/wiki/Hidden_Markov_model '''Wikipedia''']. | ||
| + | ]] | ||
======Slide 017====== | ======Slide 017====== | ||
| − | [[Image:L04_s017.jpg|frame|none|Lecture 04, Slide 017]] | + | [[Image:L04_s017.jpg|frame|none|Lecture 04, Slide 017<br> |
| + | |||
| + | ]] | ||
======Slide 018====== | ======Slide 018====== | ||
| − | [[Image:L04_s018.jpg|frame|none|Lecture 04, Slide 018]] | + | [[Image:L04_s018.jpg|frame|none|Lecture 04, Slide 018<br> |
| + | |||
| + | ]] | ||
======Slide 019====== | ======Slide 019====== | ||
| − | [[Image:L04_s019.jpg|frame|none|Lecture 04, Slide 019]] | + | [[Image:L04_s019.jpg|frame|none|Lecture 04, Slide 019<br> |
| + | |||
| + | ]] | ||
======Slide 020====== | ======Slide 020====== | ||
| − | [[Image:L04_s020.jpg|frame|none|Lecture 04, Slide 020]] | + | [[Image:L04_s020.jpg|frame|none|Lecture 04, Slide 020<br> |
| + | |||
| + | ]] | ||
======Slide 021====== | ======Slide 021====== | ||
| − | [[Image:L04_s021.jpg|frame|none|Lecture 04, Slide 021]] | + | [[Image:L04_s021.jpg|frame|none|Lecture 04, Slide 021<br> |
| + | |||
| + | ]] | ||
======Slide 022====== | ======Slide 022====== | ||
| − | [[Image:L04_s022.jpg|frame|none|Lecture 04, Slide 022]] | + | [[Image:L04_s022.jpg|frame|none|Lecture 04, Slide 022<br> |
| + | |||
| + | ]] | ||
======Slide 023====== | ======Slide 023====== | ||
| − | [[Image:L04_s023.jpg|frame|none|Lecture 04, Slide 023]] | + | [[Image:L04_s023.jpg|frame|none|Lecture 04, Slide 023<br> |
| + | [http://en.wikipedia.org/wiki/Signal_peptide Signal peptide] example for recognition of sequence features with HMMs or NNs: common features in gram-negative signal-peptide sequences are shown in a [http://weblogo.berkeley.edu Sequence Logo]. Sequences were aligned on the signal-peptidase cleavage site. Their common features include a positively charged N-terminus, a hydrophobic helical stretch and a small residue that precedes the actual cleavage site. | ||
| + | ]] | ||
======Slide 024====== | ======Slide 024====== | ||
| − | [[Image:L04_s024.jpg|frame|none|Lecture 04, Slide 024]] | + | [[Image:L04_s024.jpg|frame|none|Lecture 04, Slide 024<br> |
| + | [http://www.cbs.dtu.dk/services/SignalP/ SignalP] is the premier Web server to detect signal sequences. | ||
| + | ]] | ||
======Slide 025====== | ======Slide 025====== | ||
| − | [[Image:L04_s025.jpg|frame|none|Lecture 04, Slide 025]] | + | [[Image:L04_s025.jpg|frame|none|Lecture 04, Slide 025<br> |
| + | |||
| + | ]] | ||
======Slide 026====== | ======Slide 026====== | ||
| − | [[Image:L04_s026.jpg|frame|none|Lecture 04, Slide 026]] | + | [[Image:L04_s026.jpg|frame|none|Lecture 04, Slide 026<br> |
| + | ... however: machine learning will find correlations, not causalities. It cannot replace your biological insight to distinguish a statistical anomaly from a biologically meaningful result! | ||
| + | ]] | ||
======Slide 027====== | ======Slide 027====== | ||
| − | + | <small>deleted</small> | |
| + | |||
======Slide 028====== | ======Slide 028====== | ||
| − | [[Image:L04_s028.jpg|frame|none|Lecture 04, Slide 028]] | + | [[Image:L04_s028.jpg|frame|none|Lecture 04, Slide 028<br> |
| + | |||
| + | ]] | ||
======Slide 029====== | ======Slide 029====== | ||
| − | [[Image:L04_s029.jpg|frame|none|Lecture 04, Slide 029]] | + | [[Image:L04_s029.jpg|frame|none|Lecture 04, Slide 029<br> |
| + | |||
| + | ]] | ||
======Slide 030====== | ======Slide 030====== | ||
| − | [[Image:L04_s030.jpg|frame|none|Lecture 04, Slide 030]] | + | [[Image:L04_s030.jpg|frame|none|Lecture 04, Slide 030<br> |
| + | This is why statistics is cool. | ||
| + | ]] | ||
======Slide 031====== | ======Slide 031====== | ||
| − | [[Image:L04_s031.jpg|frame|none|Lecture 04, Slide 031]] | + | [[Image:L04_s031.jpg|frame|none|Lecture 04, Slide 031<br> |
| + | The distinction is somewhat artificial in practice: parameter estimations from probability theory can be excellent descriptors! | ||
| + | ]] | ||
======Slide 032====== | ======Slide 032====== | ||
| − | [[Image:L04_s032.jpg|frame|none|Lecture 04, Slide 032]] | + | [[Image:L04_s032.jpg|frame|none|Lecture 04, Slide 032<br> |
| + | You should be familiar with these most fundamental descriptors, they come up time- and time again in the literature. Here is a series of highly readable reviews on topics of medical statistics by Jonathan Ball and Coauthors:<br> | ||
| + | <br> | ||
| + | *(1)[http://ccforum.com/content/6/1/66 Presenting and summarising data]<br> | ||
| + | *(2) [http://ccforum.com/content/6/2/143 Samples and populations]<br> | ||
| + | *(3) [http://ccforum.com/content/6/3/222 Hypothesis testing and P values]<br> | ||
| + | *(4) [http://ccforum.com/content/6/4/335 Sample size calculations]<br> | ||
| + | *(5) [http://ccforum.com/content/6/5/424 Comparison of means]<br> | ||
| + | *(6) [http://ccforum.com/content/6/6/509 Nonparametric methods]<br> | ||
| + | *(7) [http://ccforum.com/content/7/6/451 Correlation and regression]<br> | ||
| + | *(8) [http://ccforum.com/content/8/1/46 Qualitative data - tests of association]<br> | ||
| + | *(9) [http://ccforum.com/content/8/2/130 One-way analysis of variance]<br> | ||
| + | *(10) [http://ccforum.com/content/8/3/196 Further nonparametric methods]<br> | ||
| + | *(11) [http://ccforum.com/content/8/4/287 Assessing risk]<br> | ||
| + | *(12) [http://ccforum.com/content/8/5/389 Survival analysis]<br> | ||
| + | *(13) [http://ccforum.com/content/8/6/508 Receiver operating characteristic curves]<br> | ||
| + | *(14) [http://ccforum.com/content/9/1/112 Logistic regression] | ||
| + | ]] | ||
| + | |||
======Slide 033====== | ======Slide 033====== | ||
| − | [[Image:L04_s033.jpg|frame|none|Lecture 04, Slide 033]] | + | [[Image:L04_s033.jpg|frame|none|Lecture 04, Slide 033<br> |
| + | |||
| + | ]] | ||
======Slide 034====== | ======Slide 034====== | ||
| − | [[Image:L04_s034.jpg|frame|none|Lecture 04, Slide 034]] | + | [[Image:L04_s034.jpg|frame|none|Lecture 04, Slide 034<br> |
| + | Statistical model: on [http://en.wikipedia.org/wiki/Statistical_model '''Wikipedia''']. | ||
| + | ]] | ||
======Slide 035====== | ======Slide 035====== | ||
| − | [[Image:L04_s035.jpg|frame|none|Lecture 04, Slide 035]] | + | [[Image:L04_s035.jpg|frame|none|Lecture 04, Slide 035<br> |
| + | Probability: on [http://en.wikipedia.org/wiki/Probability '''Wikipedia'''] and on [http://mathworld.wolfram.com/Probability.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 036====== | ======Slide 036====== | ||
| − | [[Image:L04_s036.jpg|frame|none|Lecture 04, Slide 036]] | + | [[Image:L04_s036.jpg|frame|none|Lecture 04, Slide 036<br> |
| + | (*) <small>Worst error in the clipart: no two faces of a die have the same number of dots. Three more errors: opposing sides must add to seven. The ones should be at the bottom if the sixes face up.</small> | ||
| + | ]] | ||
======Slide 037====== | ======Slide 037====== | ||
| − | [[Image:L04_s037.jpg|frame|none|Lecture 04, Slide 037]] | + | [[Image:L04_s037.jpg|frame|none|Lecture 04, Slide 037<br> |
| + | |||
| + | ]] | ||
======Slide 038====== | ======Slide 038====== | ||
| − | [[Image:L04_s038.jpg|frame|none|Lecture 04, Slide 038]] | + | [[Image:L04_s038.jpg|frame|none|Lecture 04, Slide 038<br> |
| + | |||
| + | ]] | ||
======Slide 039====== | ======Slide 039====== | ||
| − | [[Image:L04_s039.jpg|frame|none|Lecture 04, Slide 039]] | + | [[Image:L04_s039.jpg|frame|none|Lecture 04, Slide 039<br> |
| + | |||
| + | ]] | ||
======Slide 040====== | ======Slide 040====== | ||
| − | [[Image:L04_s040.jpg|frame|none|Lecture 04, Slide 040]] | + | [[Image:L04_s040.jpg|frame|none|Lecture 04, Slide 040<br> |
| + | Still not convinced? Try the simulation [http://www.stat.sc.edu/~west/javahtml/LetsMakeaDeal.html '''here''']. | ||
| + | ]] | ||
======Slide 041====== | ======Slide 041====== | ||
| − | [[Image:L04_s041.jpg|frame|none|Lecture 04, Slide 041]] | + | [[Image:L04_s041.jpg|frame|none|Lecture 04, Slide 041<br> |
| + | Probability distribution: on [http://en.wikipedia.org/wiki/Probability_distribution '''Wikipedia'''] and on [http://mathworld.wolfram.com/StatisticalDistribution.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 042====== | ======Slide 042====== | ||
| − | [[Image:L04_s042.jpg|frame|none|Lecture 04, Slide 042]] | + | [[Image:L04_s042.jpg|frame|none|Lecture 04, Slide 042<br> |
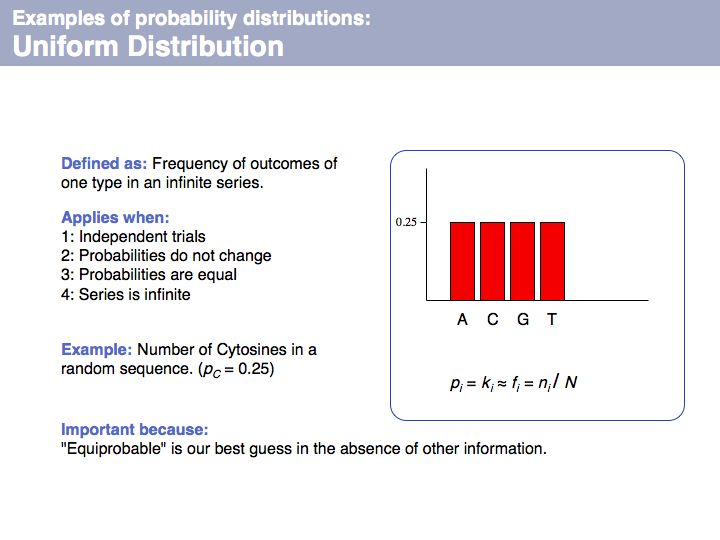
| + | Uniform distribution: on [http://en.wikipedia.org/wiki/Uniform_distribution_%28discrete%29 '''Wikipedia'''] and on [http://mathworld.wolfram.com/UniformDistribution.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 043====== | ======Slide 043====== | ||
| − | [[Image:L04_s043.jpg|frame|none|Lecture 04, Slide 043]] | + | [[Image:L04_s043.jpg|frame|none|Lecture 04, Slide 043<br> |
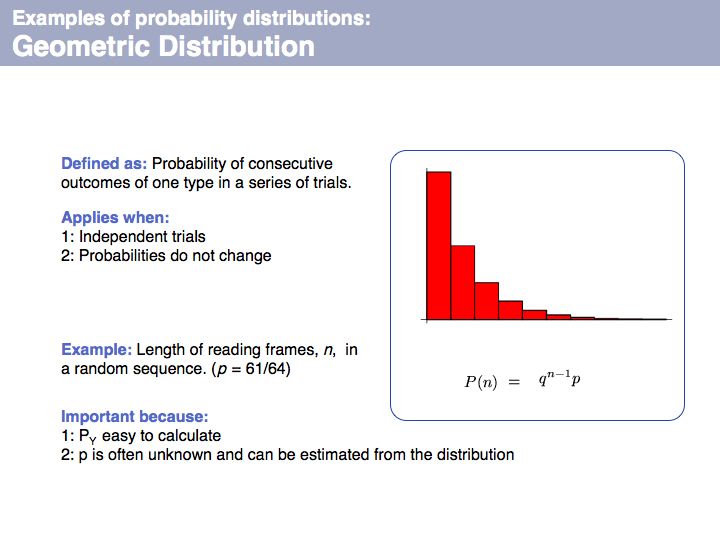
| + | Geometric distribution: on [http://en.wikipedia.org/wiki/Geometric_distribution '''Wikipedia'''] and on [http://mathworld.wolfram.com/GeometricDistribution.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 044====== | ======Slide 044====== | ||
| − | [[Image:L04_s044.jpg|frame|none|Lecture 04, Slide 044]] | + | [[Image:L04_s044.jpg|frame|none|Lecture 04, Slide 044<br> |
| + | Binomial distribution: on [http://en.wikipedia.org/wiki/Binomial_distribution '''Wikipedia'''] and on [http://mathworld.wolfram.com/BinomialDistribution.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 045====== | ======Slide 045====== | ||
| − | [[Image:L04_s045.jpg|frame|none|Lecture 04, Slide 045]] | + | [[Image:L04_s045.jpg|frame|none|Lecture 04, Slide 045<br> |
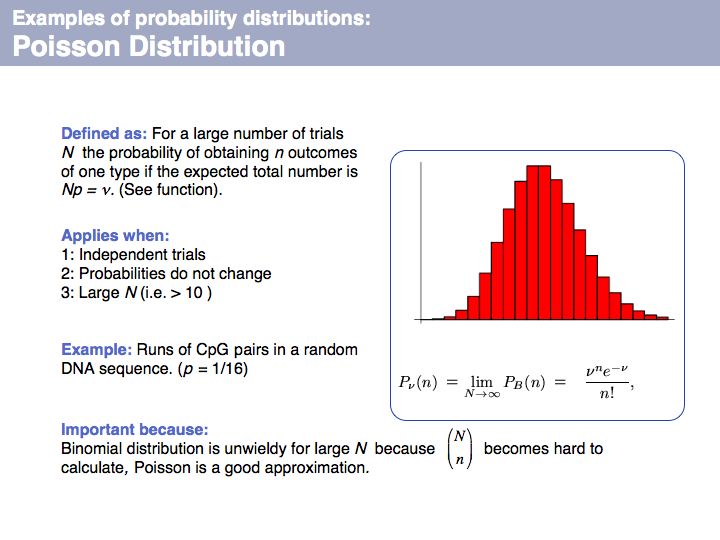
| + | Poisson distribution: on [http://en.wikipedia.org/wiki/Poisson_distribution '''Wikipedia'''] and on [http://mathworld.wolfram.com/PoissonDistribution.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 046====== | ======Slide 046====== | ||
| − | [[Image:L04_s046.jpg|frame|none|Lecture 04, Slide 046]] | + | [[Image:L04_s046.jpg|frame|none|Lecture 04, Slide 046<br> |
| + | Normal distribution: on [http://en.wikipedia.org/wiki/Normal_distribution '''Wikipedia'''] and on [http://mathworld.wolfram.com/NormalDistribution.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 047====== | ======Slide 047====== | ||
| − | [[Image:L04_s047.jpg|frame|none|Lecture 04, Slide 047]] | + | [[Image:L04_s047.jpg|frame|none|Lecture 04, Slide 047<br> |
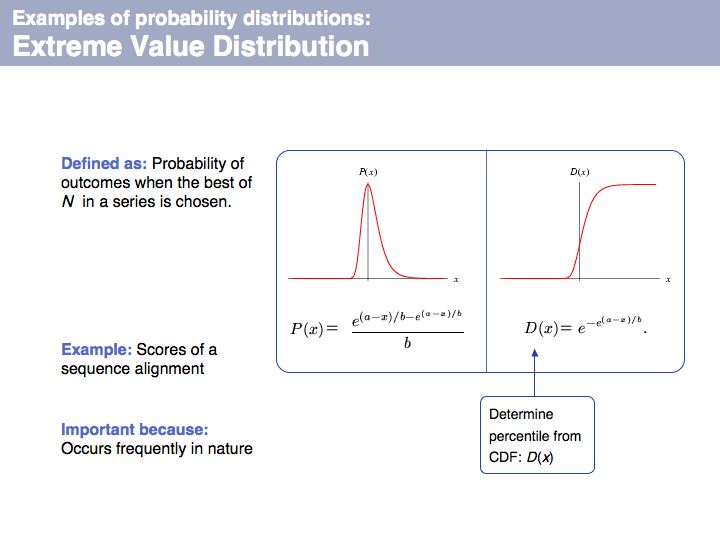
| + | Extreme value distribution: on [http://en.wikipedia.org/wiki/Generalized_extreme_value_distribution '''Wikipedia'''] and on [http://mathworld.wolfram.com/ExtremeValueDistribution.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 048====== | ======Slide 048====== | ||
| − | [[Image:L04_s048.jpg|frame|none|Lecture 04, Slide 048]] | + | [[Image:L04_s048.jpg|frame|none|Lecture 04, Slide 048<br> |
| + | |||
| + | ]] | ||
======Slide 049====== | ======Slide 049====== | ||
| − | [[Image:L04_s049.jpg|frame|none|Lecture 04, Slide 049]] | + | [[Image:L04_s049.jpg|frame|none|Lecture 04, Slide 049<br> |
| + | Don't use: Type I error - say "False positive". Don't use: Type II error - say "False negative". | ||
| + | ]] | ||
======Slide 050====== | ======Slide 050====== | ||
| − | [[Image:L04_s050.jpg|frame|none|Lecture 04, Slide 050]] | + | [[Image:L04_s050.jpg|frame|none|Lecture 04, Slide 050<br> |
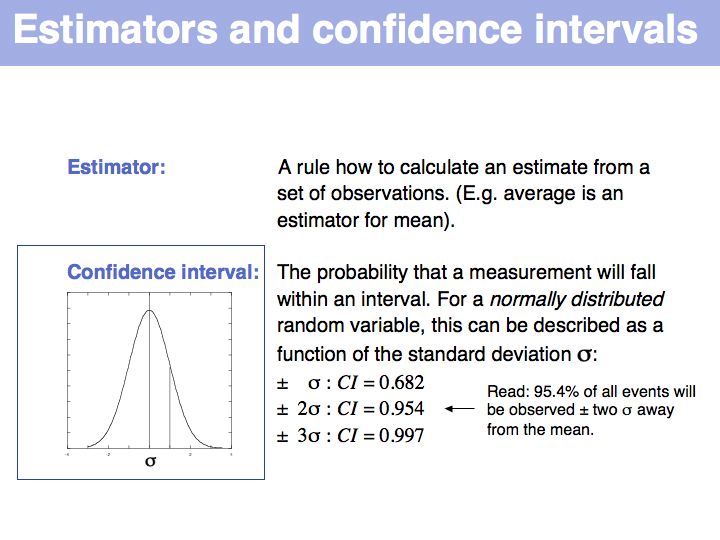
| + | Estimator: on [http://en.wikipedia.org/wiki/Estimator '''Wikipedia'''] and on [http://mathworld.wolfram.com/Estimator.html '''MathWorld'''].<br>Confidence interval: on [http://en.wikipedia.org/wiki/Confidence_interval '''Wikipedia'''] and on [http://mathworld.wolfram.com/ConfidenceInterval.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 051====== | ======Slide 051====== | ||
| − | [[Image:L04_s051.jpg|frame|none|Lecture 04, Slide 051]] | + | [[Image:L04_s051.jpg|frame|none|Lecture 04, Slide 051<br> |
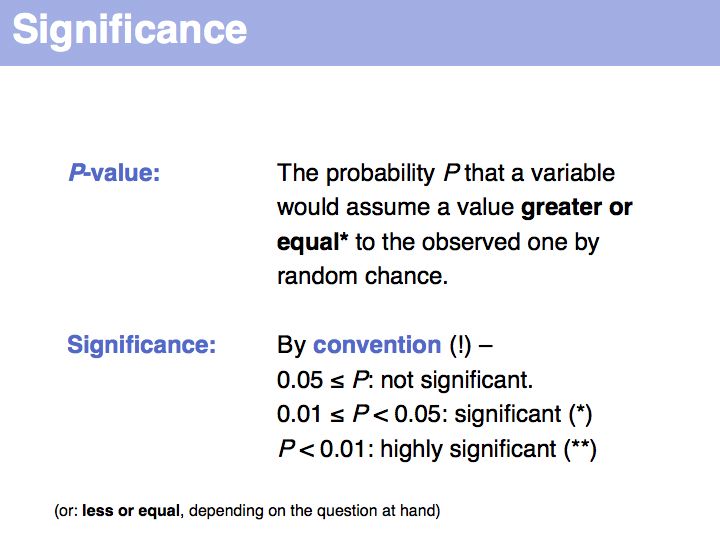
| + | '''Significance''' is really the key concept of our entire discussion. This is one term you '''must''' be completely familiar with. Significance: on [http://en.wikipedia.org/wiki/Statistical_significance '''Wikipedia'''] and on [http://mathworld.wolfram.com/Significance.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 052====== | ======Slide 052====== | ||
| − | [[Image:L04_s052.jpg|frame|none|Lecture 04, Slide 052]] | + | [[Image:L04_s052.jpg|frame|none|Lecture 04, Slide 052<br> |
| + | Note that a P-value is not the probability of a particular event occurring - that probability could be arbitrarily small, depending on the resolution of the measurement. It is the probability that an event occurs that is equal to or larger than the observed one. | ||
| + | ]] | ||
======Slide 053====== | ======Slide 053====== | ||
| − | [[Image:L04_s053.jpg|frame|none|Lecture 04, Slide 053]] | + | [[Image:L04_s053.jpg|frame|none|Lecture 04, Slide 053<br> |
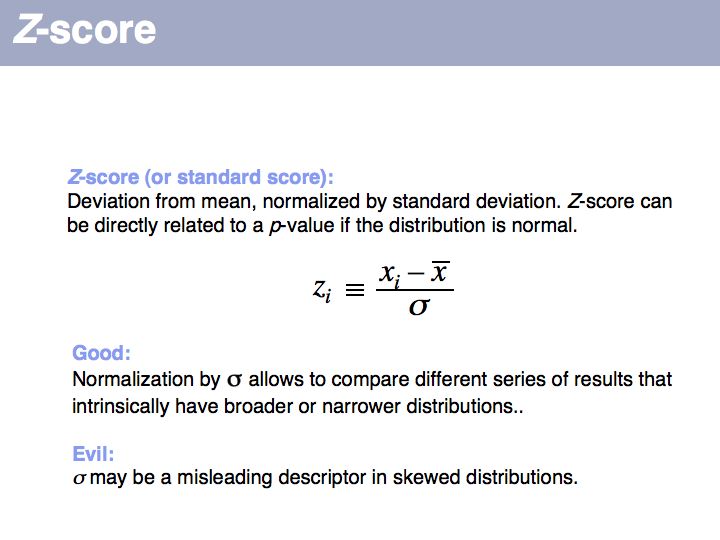
| + | ''Z''-score: on [http://en.wikipedia.org/wiki/Z_score '''Wikipedia'''] and on [http://mathworld.wolfram.com/z-Score.html '''MathWorld''']. | ||
| + | ]] | ||
======Slide 054====== | ======Slide 054====== | ||
| − | [[Image:L04_s054.jpg|frame|none|Lecture 04, Slide 054]] | + | [[Image:L04_s054.jpg|frame|none|Lecture 04, Slide 054<br> |
| + | Multiple testing: on [http://en.wikipedia.org/wiki/Multiple_testing '''Wikipedia'''] | ||
| + | ]] | ||
======Slide 055====== | ======Slide 055====== | ||
| − | [[Image:L04_s055.jpg|frame|none|Lecture 04, Slide 055]] | + | [[Image:L04_s055.jpg|frame|none|Lecture 04, Slide 055<br> |
| + | |||
| + | ]] | ||
======Slide 056====== | ======Slide 056====== | ||
| − | [[Image:L04_s056.jpg|frame|none|Lecture 04, Slide 056]] | + | [[Image:L04_s056.jpg|frame|none|Lecture 04, Slide 056<br> |
| + | Hypothesis testing: on [http://en.wikipedia.org/wiki/Statistical_hypothesis_testing '''Wikipedia'''] and on [http://mathworld.wolfram.com/HypothesisTesting.html '''MathWorld'''] | ||
| + | ]] | ||
======Slide 057====== | ======Slide 057====== | ||
| − | [[Image:L04_s057.jpg|frame|none|Lecture 04, Slide 057]] | + | [[Image:L04_s057.jpg|frame|none|Lecture 04, Slide 057<br> |
| + | |||
| + | ]] | ||
======Slide 058====== | ======Slide 058====== | ||
| − | [[Image:L04_s058.jpg|frame|none|Lecture 04, Slide 058]] | + | [[Image:L04_s058.jpg|frame|none|Lecture 04, Slide 058<br> |
| + | |||
| + | ]] | ||
======Slide 059====== | ======Slide 059====== | ||
| − | [[Image:L04_s059.jpg|frame|none|Lecture 04, Slide 059]] | + | [[Image:L04_s059.jpg|frame|none|Lecture 04, Slide 059<br> |
| + | |||
| + | ]] | ||
======Slide 060====== | ======Slide 060====== | ||
| − | [[Image:L04_s060.jpg|frame|none|Lecture 04, Slide 060]] | + | [[Image:L04_s060.jpg|frame|none|Lecture 04, Slide 060<br> |
| + | |||
| + | ]] | ||
| + | ======Slide 061====== | ||
| + | [[Image:L04_s061.jpg|frame|none|Lecture 04, Slide 061<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 062====== | ||
| + | [[Image:L04_s062.jpg|frame|none|Lecture 04, Slide 062<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 063====== | ||
| + | [[Image:L04_s063.jpg|frame|none|Lecture 04, Slide 063<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 064====== | ||
| + | [[Image:L04_s064.jpg|frame|none|Lecture 04, Slide 064<br> | ||
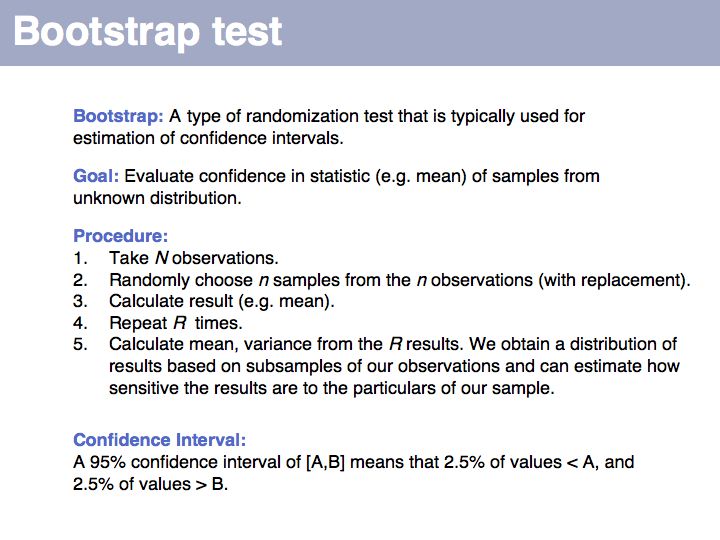
| + | Bootstrapping is a type of simulation testing. The resampling procedure simulates possible samples that we ''could'' have observed and establishes how sensitive our conclusions are to the precise composition of the sample. More on [http://en.wikipedia.org/wiki/Bootstrapping_%28statistics%29 '''Wikipedia'''] and on [http://mathworld.wolfram.com/BootstrapMethods.html '''MathWorld'''] | ||
| + | ]] | ||
| + | ======Slide 065====== | ||
| + | [[Image:L04_s065.jpg|frame|none|Lecture 04, Slide 065<br> | ||
| + | We can describe a set of observations as a distribution, and we can express this distribution as a vector if we define each element of the vector to represent a particular amino acid. This gives us a convenient and intuitive way to define a metric to compare two distributions - by considering the difference between all components of the two distributions. If we interpret this geometrically, the distribution of ''n''-elements corresponds to a <b>point in an <i>n</i>-dimensional space</b>and the difference we are using here is the '''distance''' between the two points defined by the two distributions. We could use different metrics, but this one (the ''vector norm'') is intuitive and convenient. The comparison between the frequency distribution of all amino acids in the sequence database (''f''<sub>exp</sub>, the '''expected''' distribution for a random sample of amino acids ) | ||
| + | ]] | ||
| + | ======Slide 066====== | ||
| + | [[Image:L04_s066.jpg|frame|none|Lecture 04, Slide 066<br> | ||
| + | We can apply the same metric to a set of the same number of simulated amino acids, in which the probability of picking an amino acid is given by its expectation value, f<sub>exp</sub>. If we do this many times, we will obtain a distribution of ''d'' values that tells us how different the relative frequencies of amino acids are, when they are generated by our simulator, relative to what we see in the database. Note that under many simulations we still gat an error every time, simply because the number of amino acids in every single run is small (20, in our example) and thus do what we want, the sample can never exactly reproduce the database distribution. This is important to understand: we are not simulating the distribution, we are simulating the influence of a limited-size sample! | ||
| + | ]] | ||
| + | ======Slide 067====== | ||
| + | [[Image:L04_s067.jpg|frame|none|Lecture 04, Slide 067<br> | ||
| + | Once we have simulated the experiment many times, we can compare the '''observed''' outcome with the one that would be '''expected''' if the amino acids had been randomly picked from a database distribution. In our example, the result deviates considerably from what we would expect, but not as much so that it meet a significance level of 95%. | ||
| + | ]] | ||
| + | ======Slide 068====== | ||
| + | [[Image:L04_s068.jpg|frame|none|Lecture 04, Slide 068<br> | ||
| + | Arbitrary probability distributions can be easily generated in the computer if the target probabilities are mapped on the interval [0,1|. Standard random number generators generate uniformly distributed random numbers. Thus when you check into which interval one has fallen, this designates a choice with the right target probability so in that interval will then pick a | ||
| + | ]] | ||
| + | ======Slide 069====== | ||
| + | [[Image:L04_s069.jpg|frame|none|Lecture 04, Slide 069<br> | ||
| + | If we want to simulate events according to a particular probability distribution, we can use the procedure given above. The procedure is not very efficient, since many values will be discarded if the interval is large. For each particular distribution there will be more efficient, specialized ways to generate it. However this procedure is completely general and it is trivial to change the target probability distribution's parameters; all you need is the definition of the distribution. | ||
| + | ]] | ||
| + | ======Slide 070====== | ||
| + | [[Image:L04_s070.jpg|frame|none|Lecture 04, Slide 070<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 071====== | ||
| + | [[Image:L04_s071.jpg|frame|none|Lecture 04, Slide 071<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 072====== | ||
| + | [[Image:L04_s072.jpg|frame|none|Lecture 04, Slide 072<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 073====== | ||
| + | [[Image:L04_s073.jpg|frame|none|Lecture 04, Slide 073<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 074====== | ||
| + | [[Image:L04_s074.jpg|frame|none|Lecture 04, Slide 074<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 075====== | ||
| + | [[Image:L04_s075.jpg|frame|none|Lecture 04, Slide 075<br> | ||
| + | |||
| + | ]] | ||
| + | |||
| + | | ||
| + | |||
| + | ---- | ||
| + | <small>[[Lecture_03|(Previous lecture)]] ... [[Lecture_05|(Next lecture)]]</small> | ||
Latest revision as of 23:51, 22 September 2007
(Previous lecture) ... (Next lecture)
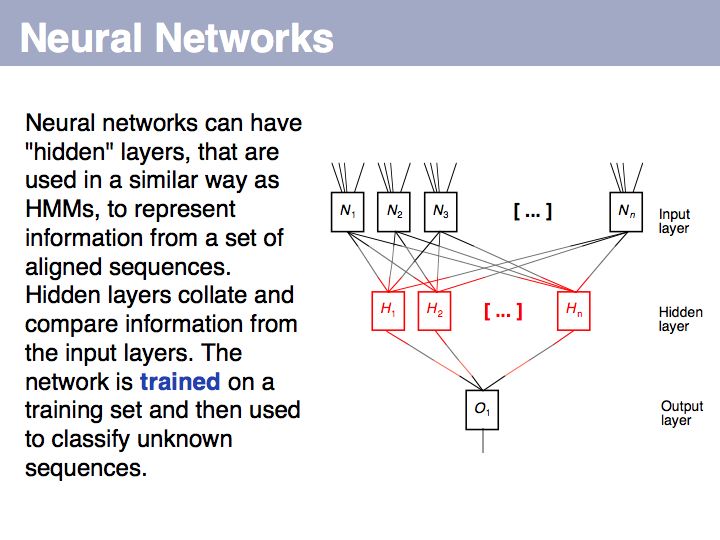
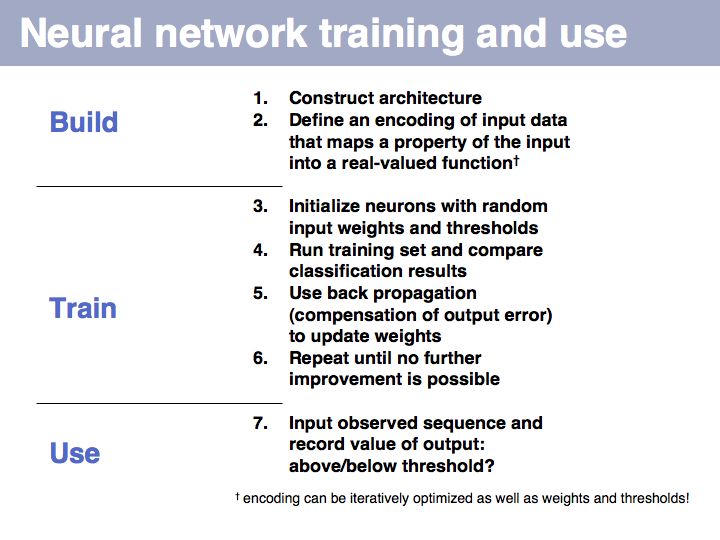
Sequence Analysis
- What you should take home from this part of the course
- Understand key concepts in probabilistic pattern representation and matching, especially PSSMs. Understand that machine-learning tools such as HMMs (Hidden Markov Models) and NN (Neural Networks) can be used for probabilistic pattern matching and classification.
- Understand the concept of a sequence logo.
- Be familiar with the SignalP Web server.
- Know basic concepts of statistics and probability theory, key terms of descriptive statistics;
- Understand probability tables in principle;
- Have encountered important probability distributions;
- Understand different error types;
- Understand the terms: significance, confidence interval and statistical test.
- Be familiar with the concepts and strategy of simulation testing and understand why its simplicity is making an important contribution to computational biology.
- Links summary
- WebLogo
- Tom Schneider's Sequence Logo pages (and introductions to information theory)
- The SignalP server
- Exercises
- If you assume that an 80-mer oligonucleotide can be synthesized with 99.9% coupling efficiency per step and a 0.2% chance of coupling a leftover nucleotide from the previous synthesis step, what is the probability that a randomly picked clone of a gene built with this oligonucleotide has the correct sequence?
- In a recent doctoral thesis defence the candidate claimed that in a microarray expression analysis he was able to show reciprocal regulation of two genes (one related to immune stimulation, the other related to immune suppression): this would mean whenever one gene is regulated up, the other is downregulated, and vice versa. The claim was based on observing this effect in eight of ten experiments. Expression levels were scored semiquantitatively on a scale of (++,+,0,-, and --). Given that such experiments have experimental error as well as biological variability, sketch a simulation test that would analyse whether in fact a significant (anti)correlation had been observed, or whether this result could just as well be due to meaningless fluctuations.
Lecture Slides
Slide 001

Lecture 04, Slide 001
From the Science News, Sept. 14. As far as systems biology complexities go, this one must be near the top: intimate interactions between human's most- and second-most complex systems. The key method here is a bioinformatics approach to classifying genes: pattern searches in the promoter regions. (NB. Not studying in isolation but forming study groups is an excellent idea!). Are you more lonely than average ? Check with the UCLA loneliness scale.
From the Science News, Sept. 14. As far as systems biology complexities go, this one must be near the top: intimate interactions between human's most- and second-most complex systems. The key method here is a bioinformatics approach to classifying genes: pattern searches in the promoter regions. (NB. Not studying in isolation but forming study groups is an excellent idea!). Are you more lonely than average ? Check with the UCLA loneliness scale.
Slide 002
Slide 003
Slide 004
Slide 005

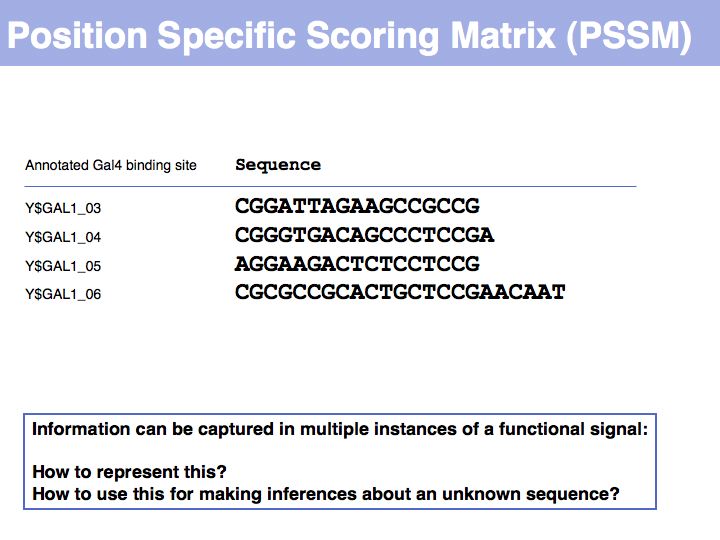
Lecture 04, Slide 005
To generate this collection of sequences, the feature "Gal4-binding-site" was searched in the [Saccharomyces Genome Database SGD]; in the resulting overview page binding site annotations recorded by Harbison et al. (2004) were shown for all occurrences; the actual sequences were retrieved by specifying the genome coordinates in the appropriate search form of the database. I have added ten bases upstream and downstream of the core binding region. This procedure could be done by hand in about the same time it took me to write the small screen-scraping program to fetch the sequences. Depending on your programming proficiency, you will find that some tasks can efficiently be done manually, for some tasks it is more efficient to spend the time to search for a better way to achieve them on the Web and only for a comparatively small number of tasks it is worthwhile (or mandatory) to write your own code.
To generate this collection of sequences, the feature "Gal4-binding-site" was searched in the [Saccharomyces Genome Database SGD]; in the resulting overview page binding site annotations recorded by Harbison et al. (2004) were shown for all occurrences; the actual sequences were retrieved by specifying the genome coordinates in the appropriate search form of the database. I have added ten bases upstream and downstream of the core binding region. This procedure could be done by hand in about the same time it took me to write the small screen-scraping program to fetch the sequences. Depending on your programming proficiency, you will find that some tasks can efficiently be done manually, for some tasks it is more efficient to spend the time to search for a better way to achieve them on the Web and only for a comparatively small number of tasks it is worthwhile (or mandatory) to write your own code.
Slide 006

Lecture 04, Slide 006
A consensus sequence simply lists the most frequent amino acid or nucleotide at each position, or a random one if there is more than one with the highest frequency. The consensus sequence is the one that you would synthesize to make an idealized representative of the set. It is likely to bind more tightly or to be more stable than each of the individual sequences in the alignment.
A consensus sequence simply lists the most frequent amino acid or nucleotide at each position, or a random one if there is more than one with the highest frequency. The consensus sequence is the one that you would synthesize to make an idealized representative of the set. It is likely to bind more tightly or to be more stable than each of the individual sequences in the alignment.
Slide 007

Lecture 04, Slide 007
Introducing nucleotide ambiguity codes to represent situations in which more than one nucleotide has the highest frequency improves the situation a bit, but there is also ambiguity. Consider the situation after the conserved CCG pattern: 9 As and 3Gs: should we report the consensusAA, or ist it more interesting to report that the only observed alternative is another purine base and write Y instead?
Introducing nucleotide ambiguity codes to represent situations in which more than one nucleotide has the highest frequency improves the situation a bit, but there is also ambiguity. Consider the situation after the conserved CCG pattern: 9 As and 3Gs: should we report the consensusAA, or ist it more interesting to report that the only observed alternative is another purine base and write Y instead?
Slide 008

Lecture 04, Slide 008
Sequence logo of Gal4 binding sites with 10 nucleotides flanking bases. Created with WebLogo. A Sequence Logo is a graphical representation of aligned sequences where at each position the height of a column is proportional to the (Shannon) information of that position and the relative size of each character is proportional to its frequency within the column. Sequence Logos were pioneered by Tom Schneider who maintains an informative Website about their use and theoretical foundations. Note that there is considerable additional information in the flanking sequences that are not included in the published description of the core binding pattern; it is advantageous if you are able to rerun such analyses, rather than rely on someone else's opinion.
Sequence logo of Gal4 binding sites with 10 nucleotides flanking bases. Created with WebLogo. A Sequence Logo is a graphical representation of aligned sequences where at each position the height of a column is proportional to the (Shannon) information of that position and the relative size of each character is proportional to its frequency within the column. Sequence Logos were pioneered by Tom Schneider who maintains an informative Website about their use and theoretical foundations. Note that there is considerable additional information in the flanking sequences that are not included in the published description of the core binding pattern; it is advantageous if you are able to rerun such analyses, rather than rely on someone else's opinion.
Slide 009
Slide 010
Slide 011
Slide 012

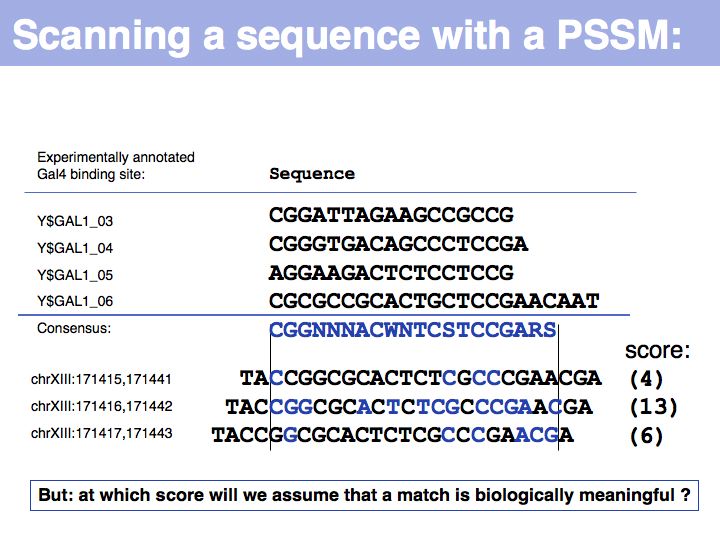
Lecture 04, Slide 012
In this informal example, I have simply counted matches with the consensus sequence (excluding "N"). We can slide the PSSM over the entire chromosome, and calculate scores for each position. Only the middle sequence is an annotated binding site. Whatever method we use for probabilistic pattern matching, we will always get a score. It is then our problem to decide what the score means.
In this informal example, I have simply counted matches with the consensus sequence (excluding "N"). We can slide the PSSM over the entire chromosome, and calculate scores for each position. Only the middle sequence is an annotated binding site. Whatever method we use for probabilistic pattern matching, we will always get a score. It is then our problem to decide what the score means.
Slide 013
Slide 014
Slide 015

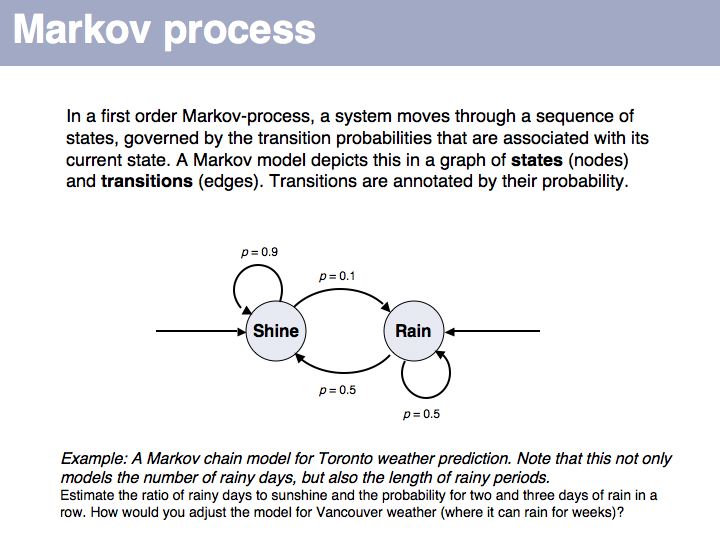
Lecture 04, Slide 015
This first order Markov model depends only on the current state. Higher-order models take increasing lengths of "history" into account, how the system arrived in its current state. Note that the exit probabilities fo a state always have to sum to 1.0. The so called "stationary probability" over a long period of time for p(rain) is 0.167 - this is determined by the combined effects of all individual transition probabilities. The stationary probabilities for two- or three consecutive rainy days are 4.2% and 2.1%, respectively.
This first order Markov model depends only on the current state. Higher-order models take increasing lengths of "history" into account, how the system arrived in its current state. Note that the exit probabilities fo a state always have to sum to 1.0. The so called "stationary probability" over a long period of time for p(rain) is 0.167 - this is determined by the combined effects of all individual transition probabilities. The stationary probabilities for two- or three consecutive rainy days are 4.2% and 2.1%, respectively.
Slide 016

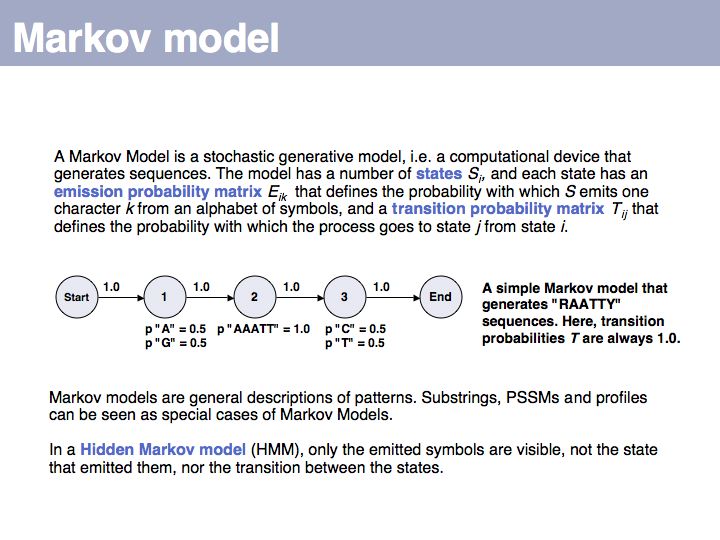
Lecture 04, Slide 016
Hidden Markov Model: on Wikipedia.
Hidden Markov Model: on Wikipedia.
Slide 017
Slide 018
Slide 019
Slide 020
Slide 021
Slide 022
Slide 023

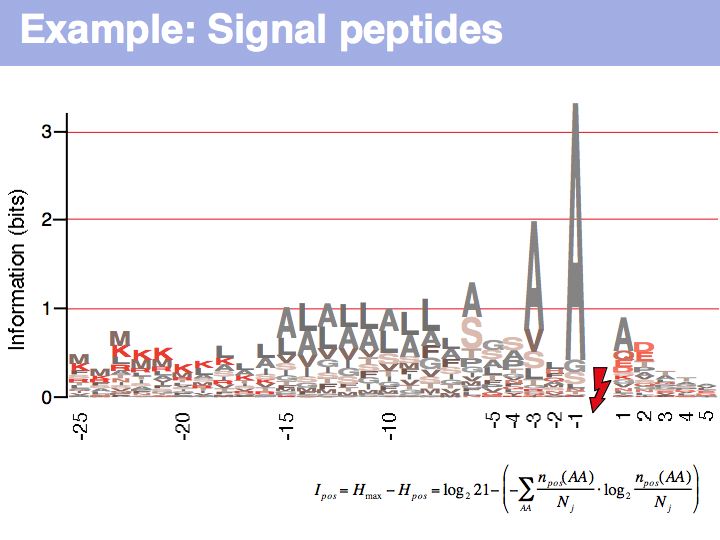
Lecture 04, Slide 023
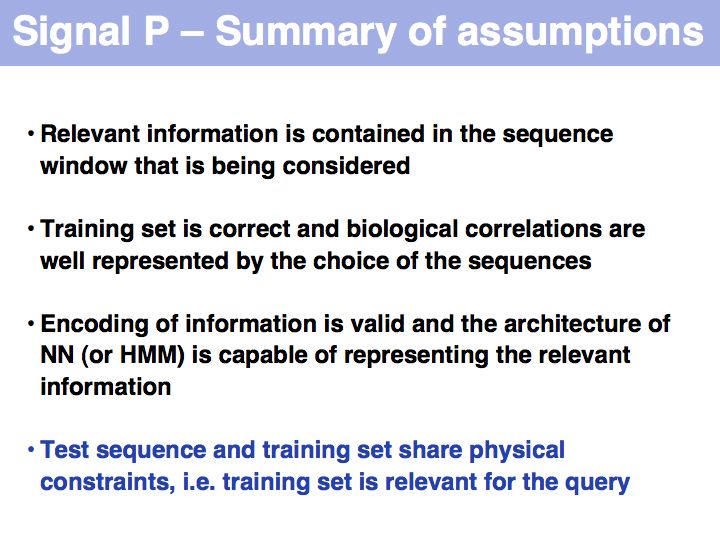
Signal peptide example for recognition of sequence features with HMMs or NNs: common features in gram-negative signal-peptide sequences are shown in a Sequence Logo. Sequences were aligned on the signal-peptidase cleavage site. Their common features include a positively charged N-terminus, a hydrophobic helical stretch and a small residue that precedes the actual cleavage site.
Signal peptide example for recognition of sequence features with HMMs or NNs: common features in gram-negative signal-peptide sequences are shown in a Sequence Logo. Sequences were aligned on the signal-peptidase cleavage site. Their common features include a positively charged N-terminus, a hydrophobic helical stretch and a small residue that precedes the actual cleavage site.
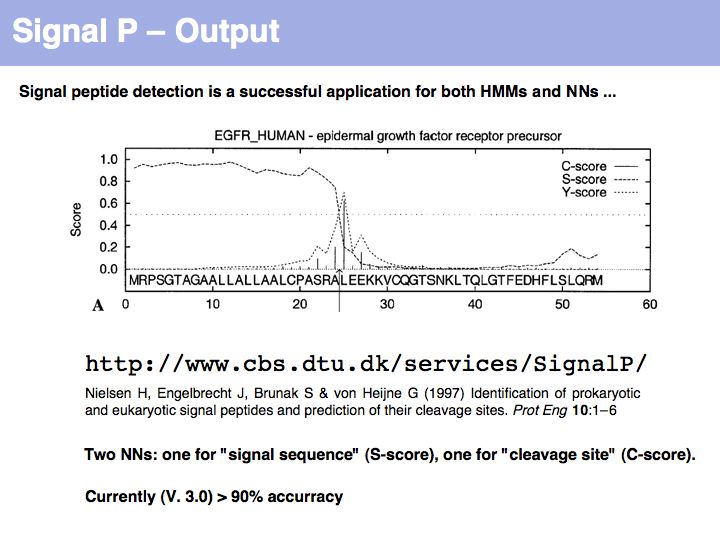
Slide 024

Lecture 04, Slide 024
SignalP is the premier Web server to detect signal sequences.
SignalP is the premier Web server to detect signal sequences.
Slide 025
Slide 026
Slide 027
deleted
Slide 028
Slide 029
Slide 030
Slide 031
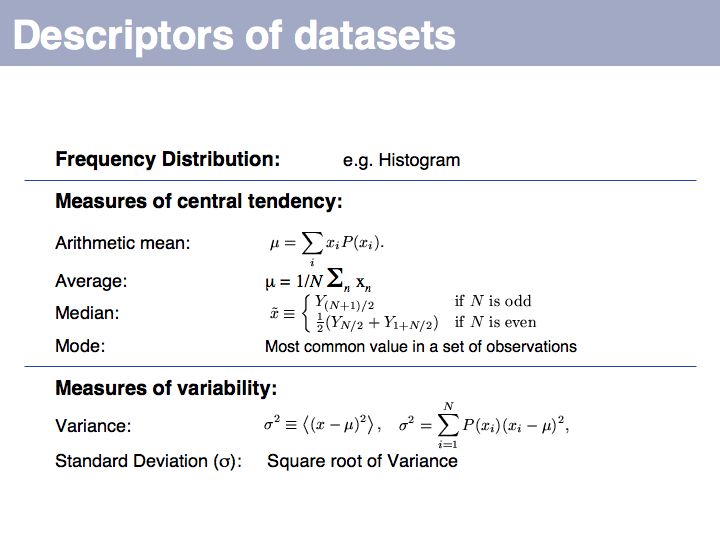
Slide 032

Lecture 04, Slide 032
You should be familiar with these most fundamental descriptors, they come up time- and time again in the literature. Here is a series of highly readable reviews on topics of medical statistics by Jonathan Ball and Coauthors:
*(1)Presenting and summarising data
*(2) Samples and populations
*(3) Hypothesis testing and P values
*(4) Sample size calculations
*(5) Comparison of means
*(6) Nonparametric methods
*(7) Correlation and regression
*(8) Qualitative data - tests of association
*(9) One-way analysis of variance
*(10) Further nonparametric methods
*(11) Assessing risk
*(12) Survival analysis
*(13) Receiver operating characteristic curves
*(14) Logistic regression
You should be familiar with these most fundamental descriptors, they come up time- and time again in the literature. Here is a series of highly readable reviews on topics of medical statistics by Jonathan Ball and Coauthors:
*(1)Presenting and summarising data
*(2) Samples and populations
*(3) Hypothesis testing and P values
*(4) Sample size calculations
*(5) Comparison of means
*(6) Nonparametric methods
*(7) Correlation and regression
*(8) Qualitative data - tests of association
*(9) One-way analysis of variance
*(10) Further nonparametric methods
*(11) Assessing risk
*(12) Survival analysis
*(13) Receiver operating characteristic curves
*(14) Logistic regression
Slide 033
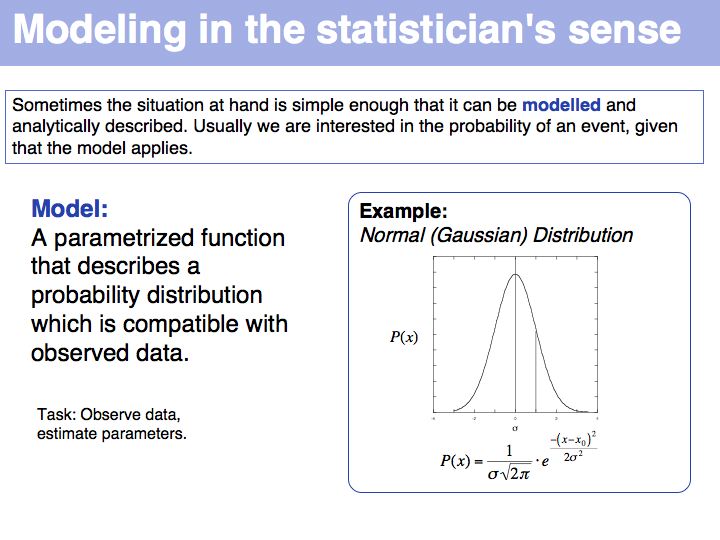
Slide 034

Lecture 04, Slide 034
Statistical model: on Wikipedia.
Statistical model: on Wikipedia.
Slide 035
Slide 036
Slide 037
Slide 038
Slide 039
Slide 040

Lecture 04, Slide 040
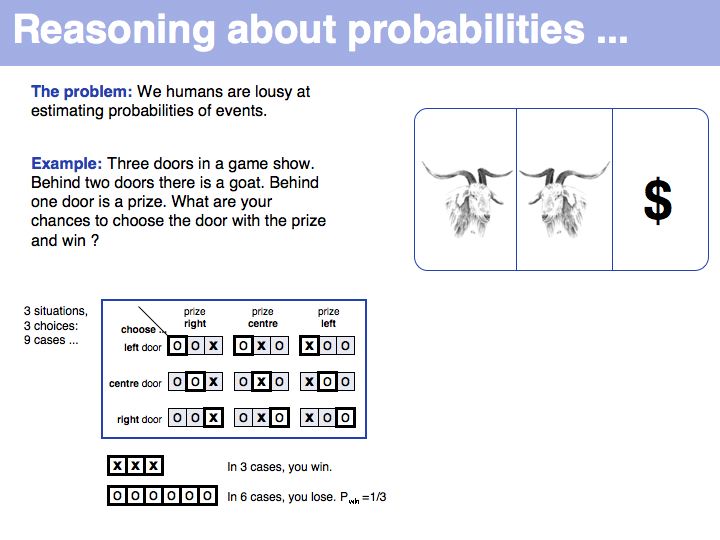
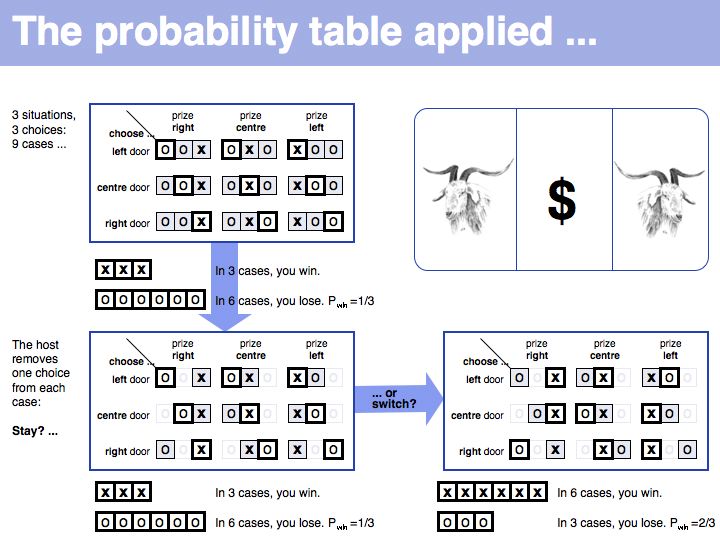
Still not convinced? Try the simulation here.
Still not convinced? Try the simulation here.
Slide 041
Slide 042
Slide 043
Slide 044
Slide 045
Slide 046
Slide 047
Slide 048
Slide 049
Slide 050
Slide 051
Slide 052
Slide 053
Slide 054

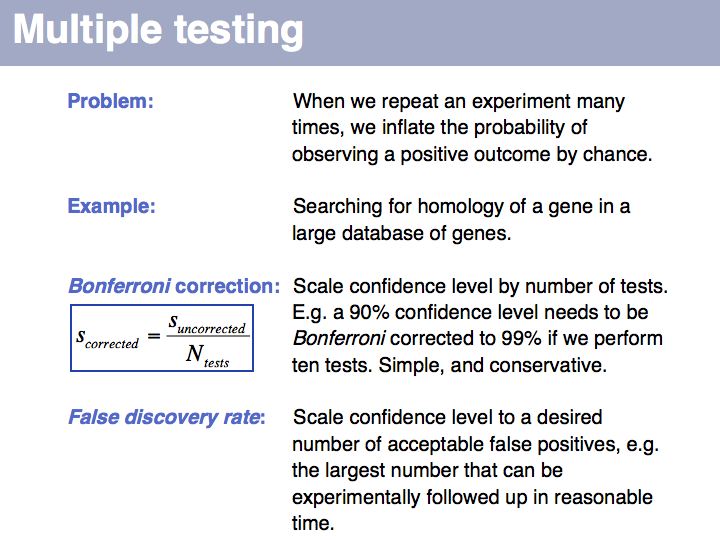
Lecture 04, Slide 054
Multiple testing: on Wikipedia
Multiple testing: on Wikipedia
Slide 055
Slide 056
Slide 057
Slide 058
Slide 059
Slide 060
Slide 061
Slide 062
Slide 063
Slide 064
Slide 065

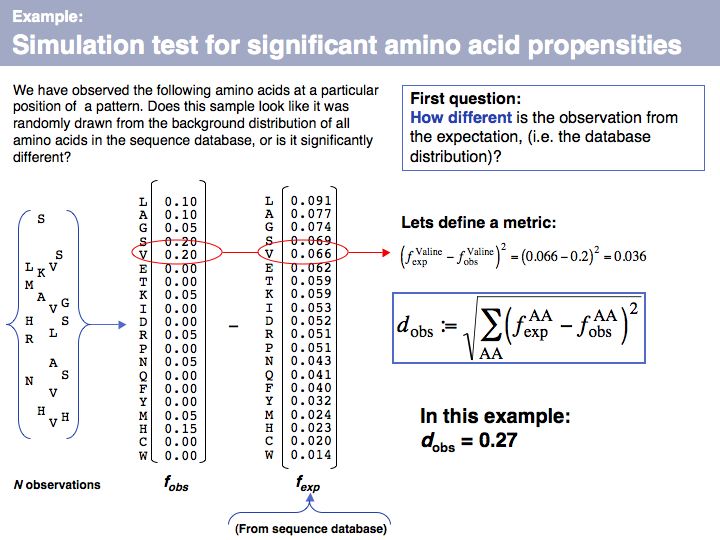
Lecture 04, Slide 065
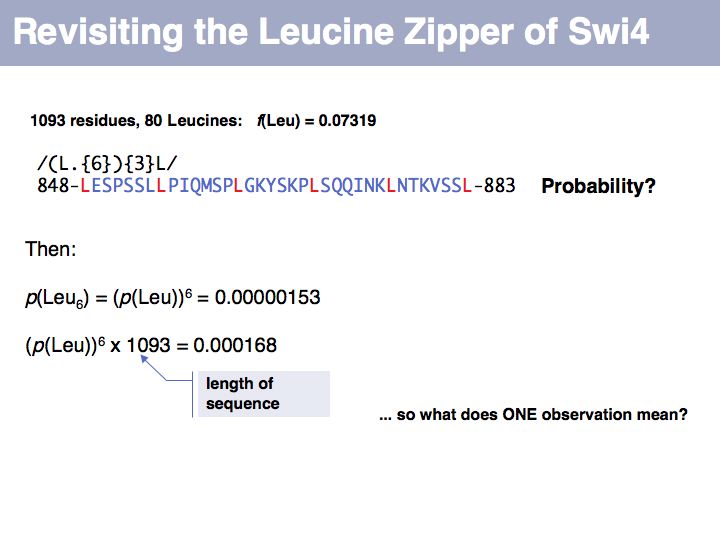
We can describe a set of observations as a distribution, and we can express this distribution as a vector if we define each element of the vector to represent a particular amino acid. This gives us a convenient and intuitive way to define a metric to compare two distributions - by considering the difference between all components of the two distributions. If we interpret this geometrically, the distribution of n-elements corresponds to a point in an n-dimensional spaceand the difference we are using here is the distance between the two points defined by the two distributions. We could use different metrics, but this one (the vector norm) is intuitive and convenient. The comparison between the frequency distribution of all amino acids in the sequence database (fexp, the expected distribution for a random sample of amino acids )
We can describe a set of observations as a distribution, and we can express this distribution as a vector if we define each element of the vector to represent a particular amino acid. This gives us a convenient and intuitive way to define a metric to compare two distributions - by considering the difference between all components of the two distributions. If we interpret this geometrically, the distribution of n-elements corresponds to a point in an n-dimensional spaceand the difference we are using here is the distance between the two points defined by the two distributions. We could use different metrics, but this one (the vector norm) is intuitive and convenient. The comparison between the frequency distribution of all amino acids in the sequence database (fexp, the expected distribution for a random sample of amino acids )
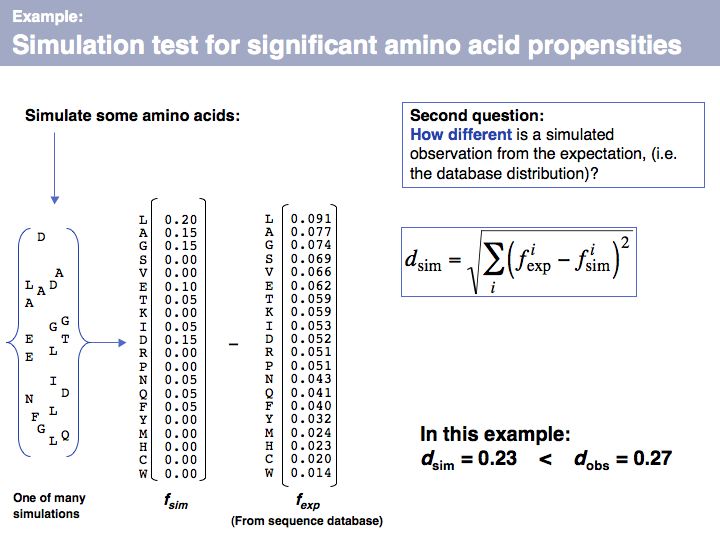
Slide 066

Lecture 04, Slide 066
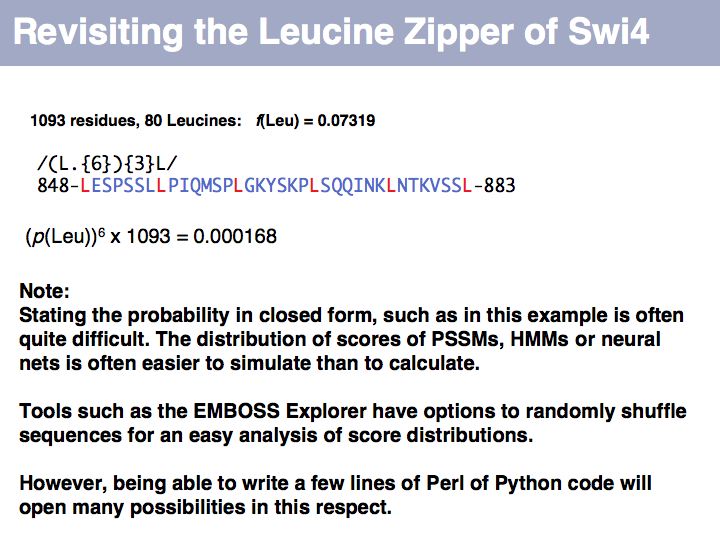
We can apply the same metric to a set of the same number of simulated amino acids, in which the probability of picking an amino acid is given by its expectation value, fexp. If we do this many times, we will obtain a distribution of d values that tells us how different the relative frequencies of amino acids are, when they are generated by our simulator, relative to what we see in the database. Note that under many simulations we still gat an error every time, simply because the number of amino acids in every single run is small (20, in our example) and thus do what we want, the sample can never exactly reproduce the database distribution. This is important to understand: we are not simulating the distribution, we are simulating the influence of a limited-size sample!
We can apply the same metric to a set of the same number of simulated amino acids, in which the probability of picking an amino acid is given by its expectation value, fexp. If we do this many times, we will obtain a distribution of d values that tells us how different the relative frequencies of amino acids are, when they are generated by our simulator, relative to what we see in the database. Note that under many simulations we still gat an error every time, simply because the number of amino acids in every single run is small (20, in our example) and thus do what we want, the sample can never exactly reproduce the database distribution. This is important to understand: we are not simulating the distribution, we are simulating the influence of a limited-size sample!
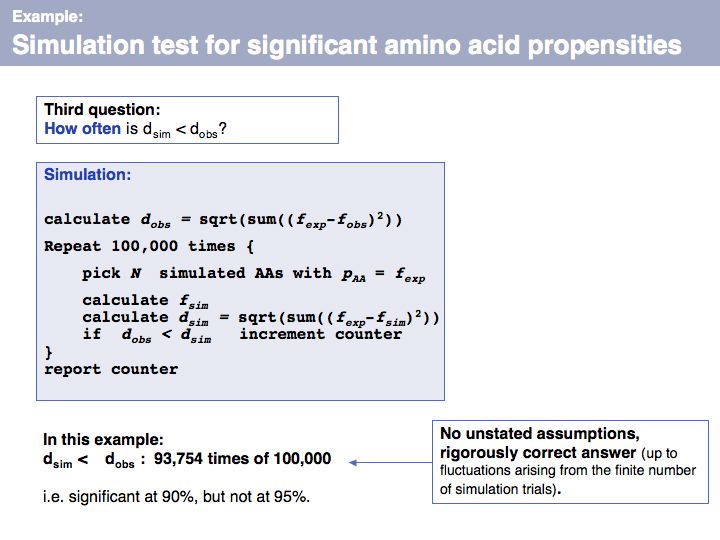
Slide 067

Lecture 04, Slide 067
Once we have simulated the experiment many times, we can compare the observed outcome with the one that would be expected if the amino acids had been randomly picked from a database distribution. In our example, the result deviates considerably from what we would expect, but not as much so that it meet a significance level of 95%.
Once we have simulated the experiment many times, we can compare the observed outcome with the one that would be expected if the amino acids had been randomly picked from a database distribution. In our example, the result deviates considerably from what we would expect, but not as much so that it meet a significance level of 95%.
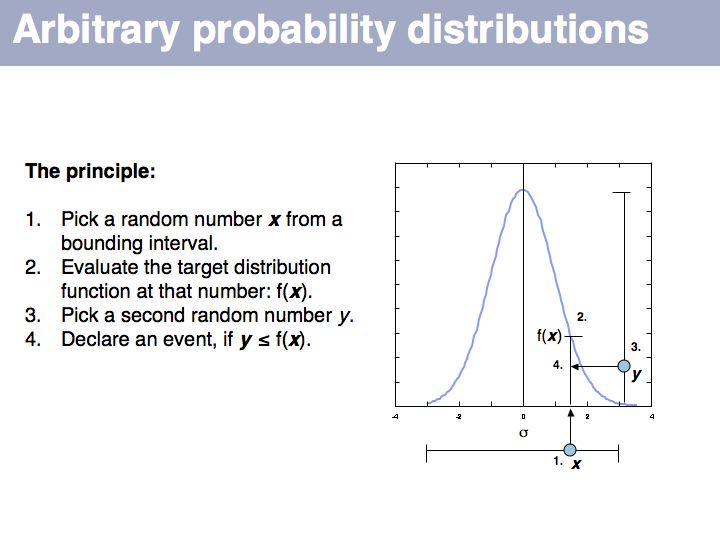
Slide 068
Slide 069

Lecture 04, Slide 069
If we want to simulate events according to a particular probability distribution, we can use the procedure given above. The procedure is not very efficient, since many values will be discarded if the interval is large. For each particular distribution there will be more efficient, specialized ways to generate it. However this procedure is completely general and it is trivial to change the target probability distribution's parameters; all you need is the definition of the distribution.
If we want to simulate events according to a particular probability distribution, we can use the procedure given above. The procedure is not very efficient, since many values will be discarded if the interval is large. For each particular distribution there will be more efficient, specialized ways to generate it. However this procedure is completely general and it is trivial to change the target probability distribution's parameters; all you need is the definition of the distribution.
Slide 070
Slide 071
Slide 072
Slide 073
Slide 074
Slide 075