Difference between revisions of "Lecture 17"
Jump to navigation
Jump to search
| Line 70: | Line 70: | ||
======Slide 014====== | ======Slide 014====== | ||
[[Image:L17_s014.jpg|frame|none|Lecture 17, Slide 014<br> | [[Image:L17_s014.jpg|frame|none|Lecture 17, Slide 014<br> | ||
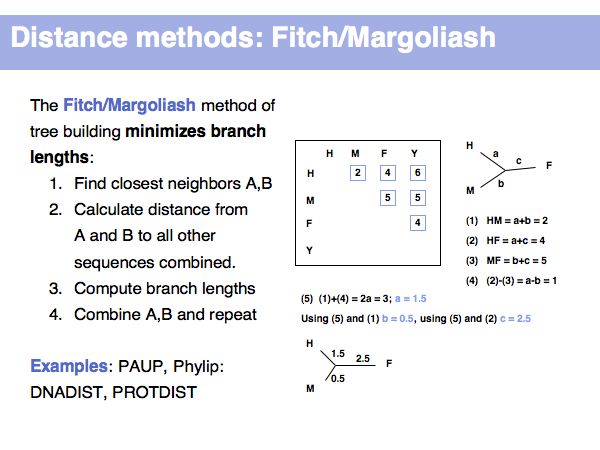
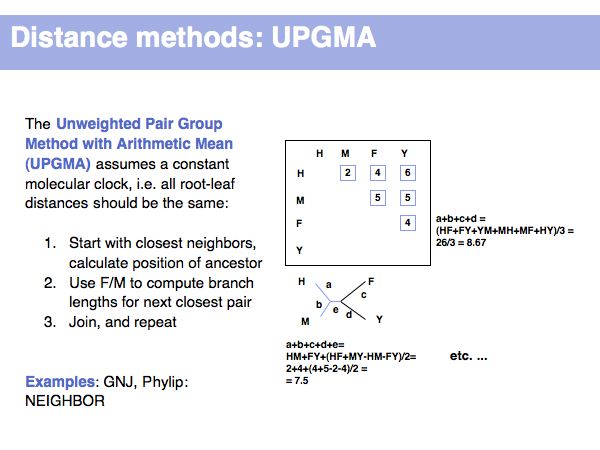
| + | Note that distance methods first reduce the explicit molecular data into a matrix of pairwise distances, then operate on that "summary" to construct the most plausible tree. They are fast, but less accurate than some alternatives. | ||
| + | ]] | ||
| − | |||
======Slide 015====== | ======Slide 015====== | ||
[[Image:L17_s015.jpg|frame|none|Lecture 17, Slide 015<br> | [[Image:L17_s015.jpg|frame|none|Lecture 17, Slide 015<br> | ||
Revision as of 19:02, 28 November 2006
(Previous lecture) ... (Next lecture)
Phylogenetic Analysis
...
Add:
- Summary points
- Exercises
- Further reading
Lecture Slides
Slide 001
Slide 002
Slide 003
Slide 004
Slide 005
Slide 006
Slide 007
Slide 008
Slide 009
Slide 010
Slide 011
Slide 012

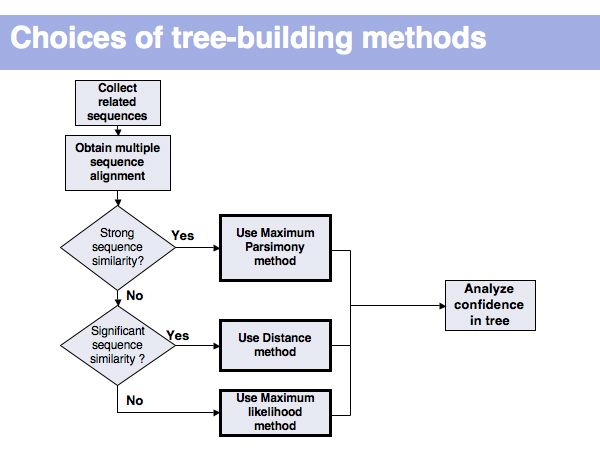
Lecture 17, Slide 012
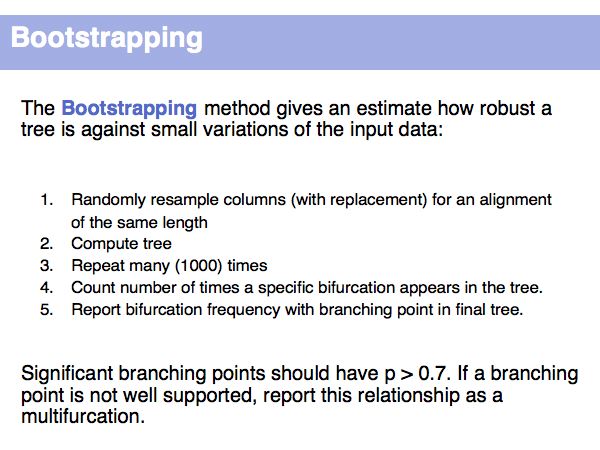
Usually, analysis of confidence implies a "bootstrapping" procedure: rerun the analysis many times with partial data and analyze which features of the tree (branching order -> topology!) are well conserved, and which ones depend critically on unreliable features of the input data.
Usually, analysis of confidence implies a "bootstrapping" procedure: rerun the analysis many times with partial data and analyze which features of the tree (branching order -> topology!) are well conserved, and which ones depend critically on unreliable features of the input data.