Difference between revisions of "Lecture 16"
Jump to navigation
Jump to search
| Line 57: | Line 57: | ||
======Slide 011====== | ======Slide 011====== | ||
[[Image:L16_s011.jpg|frame|none|Lecture 16, Slide 011<br> | [[Image:L16_s011.jpg|frame|none|Lecture 16, Slide 011<br> | ||
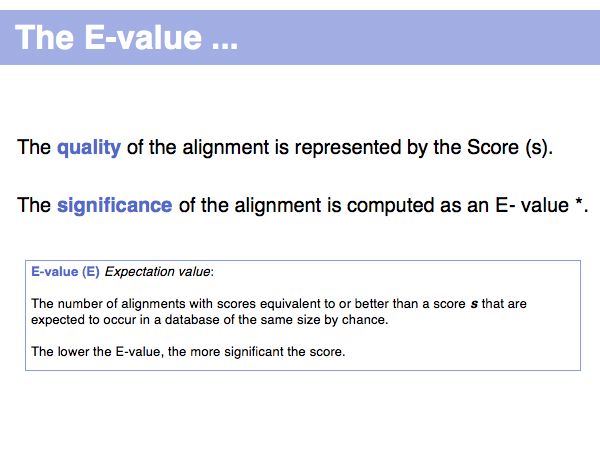
| + | (*) Computing E-values is possible for HSPs since the statistcs of gap-less alignments are analytically tractable. Similar conclusions in general cannot be drawn from gapped alignments. Note the E-values are not a statement about the retrieved hit, but a statement about an expected distribution of scores. Or, to rephrase this, a poor e-value does not mean that your hit is not a homologue, but it means that at that score an irrelevant sequence has a a high chance of scoring well due to chance similarities. | ||
| + | ]] | ||
| − | |||
======Slide 012====== | ======Slide 012====== | ||
[[Image:L16_s012.jpg|frame|none|Lecture 16, Slide 012<br> | [[Image:L16_s012.jpg|frame|none|Lecture 16, Slide 012<br> | ||
Revision as of 15:22, 28 November 2006
(Previous lecture) ... (Next lecture)
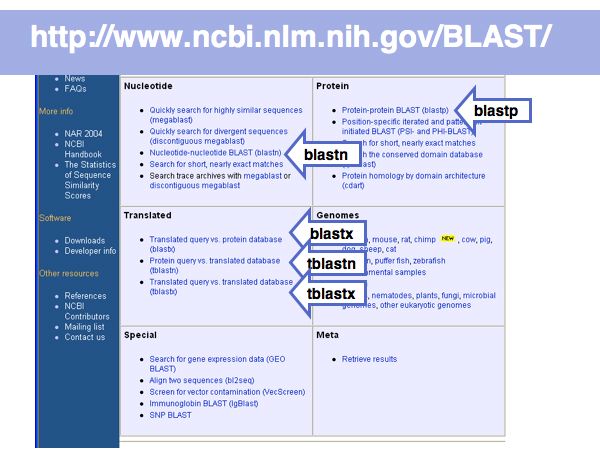
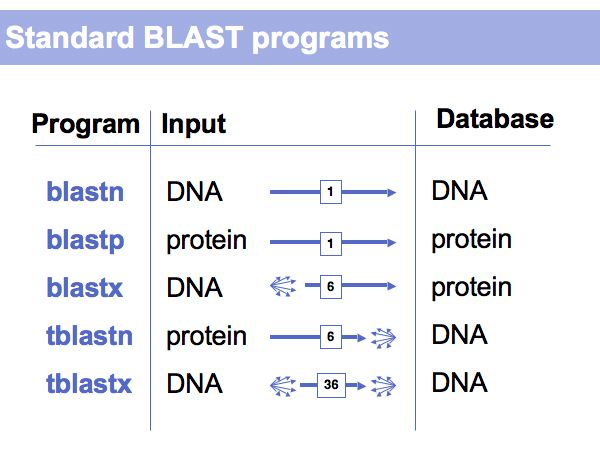
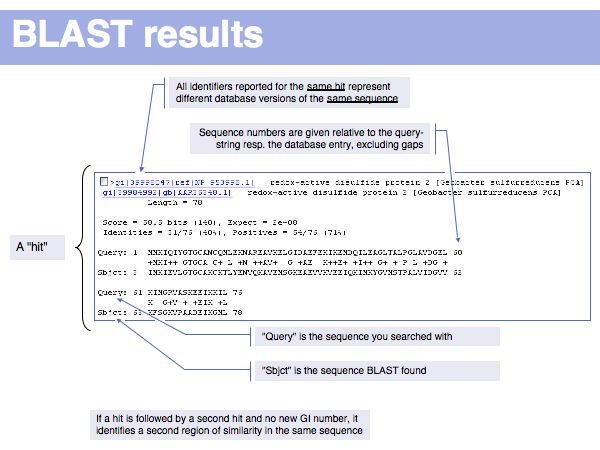
Fast Sequence Database Searches (BLAST)
...
Add:
- Summary points
- Exercises
- Further reading
Lecture Slides
Slide 001
Slide 002
Slide 003
Slide 004
Slide 005
Slide 006
Slide 007
Slide 008
Slide 009
Slide 010
Slide 011

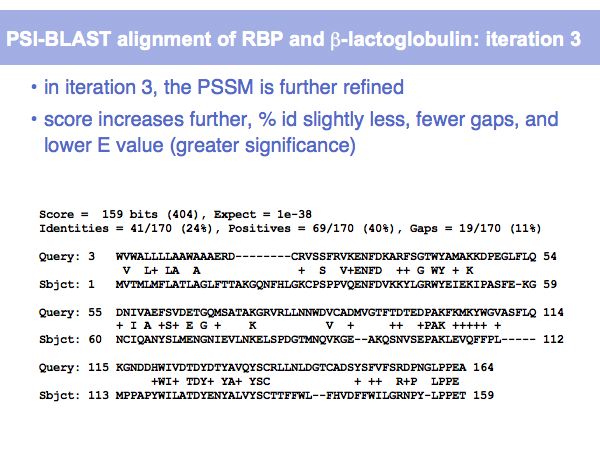
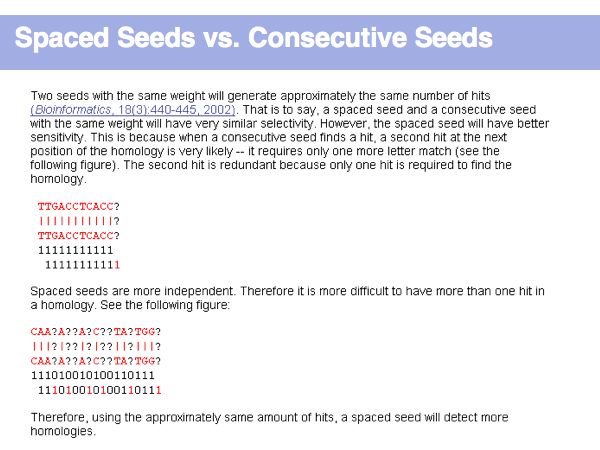
Lecture 16, Slide 011
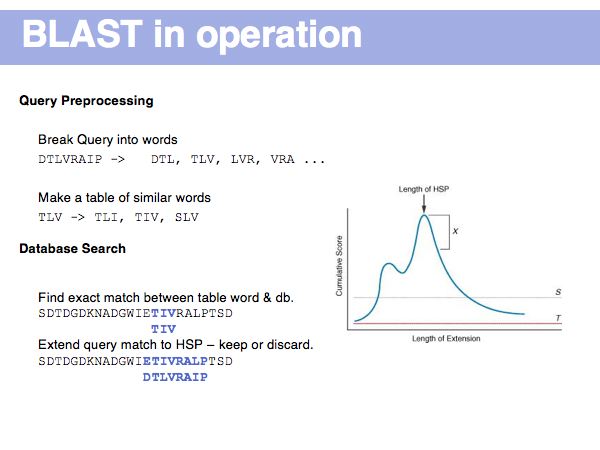
(*) Computing E-values is possible for HSPs since the statistcs of gap-less alignments are analytically tractable. Similar conclusions in general cannot be drawn from gapped alignments. Note the E-values are not a statement about the retrieved hit, but a statement about an expected distribution of scores. Or, to rephrase this, a poor e-value does not mean that your hit is not a homologue, but it means that at that score an irrelevant sequence has a a high chance of scoring well due to chance similarities.
(*) Computing E-values is possible for HSPs since the statistcs of gap-less alignments are analytically tractable. Similar conclusions in general cannot be drawn from gapped alignments. Note the E-values are not a statement about the retrieved hit, but a statement about an expected distribution of scores. Or, to rephrase this, a poor e-value does not mean that your hit is not a homologue, but it means that at that score an irrelevant sequence has a a high chance of scoring well due to chance similarities.