Difference between revisions of "Lecture 09"
Jump to navigation
Jump to search
| Line 37: | Line 37: | ||
======Slide 006====== | ======Slide 006====== | ||
[[Image:L09_s006.jpg|frame|none|Lecture 09, Slide 006<br> | [[Image:L09_s006.jpg|frame|none|Lecture 09, Slide 006<br> | ||
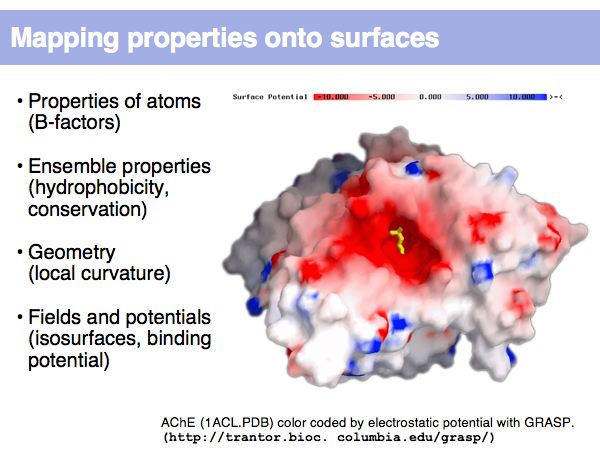
| + | Surfaces represent the parts of a protein that interact with the surroundings. In this example, the electrostatic potential mapping shows how an electrostatic potential gradient attracts the substrate molecule into Acetylcholine-esterase's active site. | ||
| + | ]] | ||
| − | |||
======Slide 007====== | ======Slide 007====== | ||
[[Image:L09_s007.jpg|frame|none|Lecture 09, Slide 007<br> | [[Image:L09_s007.jpg|frame|none|Lecture 09, Slide 007<br> | ||
Revision as of 00:35, 27 November 2006
(Previous lecture) ... (Next lecture)
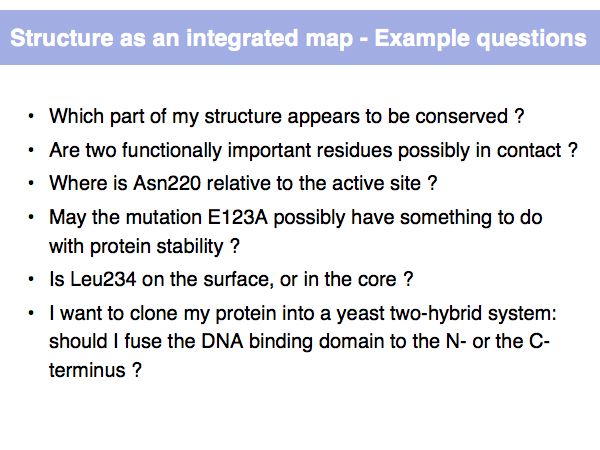
Structure Analysis
...
Add:
- Summary points
- Exercises
- Further reading
Lecture Slides
Slide 001
Slide 002
Slide 003
Slide 004
Slide 005

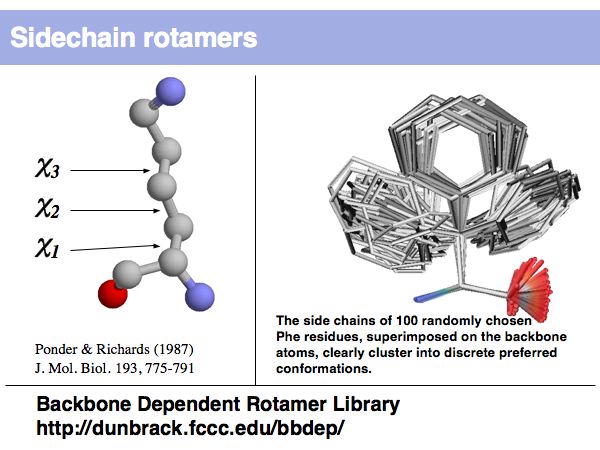
Lecture 09, Slide 005
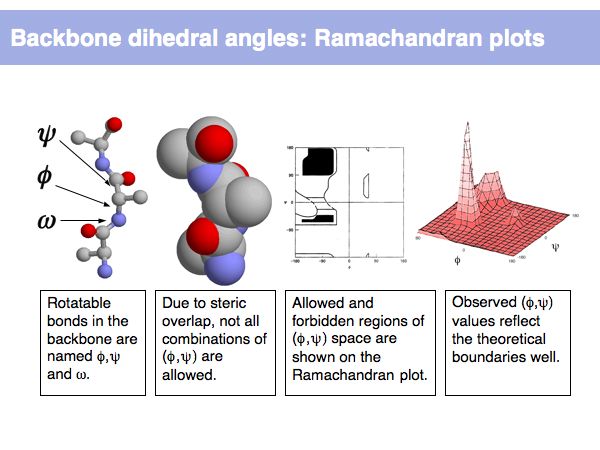
Rotamers are low-energy conformations of side-chain dihedral angles. Only a small number of rotamer states and combinations are significantly populated in natural proteins. This tremendously simplifies protein structure modelling and prediction problems. However it also guides analysis, e.g. in enzyme active sites the rotamers often exist in strained, rare conformations.
Rotamers are low-energy conformations of side-chain dihedral angles. Only a small number of rotamer states and combinations are significantly populated in natural proteins. This tremendously simplifies protein structure modelling and prediction problems. However it also guides analysis, e.g. in enzyme active sites the rotamers often exist in strained, rare conformations.