Difference between revisions of "BIN-PHYLO-Tree analysis"
m |
m |
||
| Line 1: | Line 1: | ||
<div id="ABC"> | <div id="ABC"> | ||
| − | <div style="padding:5px; border:1px solid #000000; background-color:# | + | <div style="padding:5px; border:1px solid #000000; background-color:#f4d7b7; font-size:300%; font-weight:400; color: #000000; width:100%;"> |
Analysing Phylogenetic Trees | Analysing Phylogenetic Trees | ||
| − | <div style="padding:5px; margin-top:20px; margin-bottom:10px; background-color:# | + | <div style="padding:5px; margin-top:20px; margin-bottom:10px; background-color:#f4d7b7; font-size:30%; font-weight:200; color: #000000; "> |
(Species trees, gene trees and the importance of naming, Speciation and duplication signatures) | (Species trees, gene trees and the importance of naming, Speciation and duplication signatures) | ||
</div> | </div> | ||
| Line 10: | Line 10: | ||
| − | <div style="padding:5px; border:1px solid #000000; background-color:# | + | <div style="padding:5px; border:1px solid #000000; background-color:#f4d7b733; font-size:85%;"> |
<div style="font-size:118%;"> | <div style="font-size:118%;"> | ||
<b>Abstract:</b><br /> | <b>Abstract:</b><br /> | ||
| Line 42: | Line 42: | ||
<section begin=deliverables /> | <section begin=deliverables /> | ||
<!-- included from "./data/ABC-unit_components.txt", section: "deliverables-time_management" --> | <!-- included from "./data/ABC-unit_components.txt", section: "deliverables-time_management" --> | ||
| − | + | <li><b>Time management</b>: Before you begin, estimate how long it will take you to complete this unit. Then, record in your course journal: the number of hours you estimated, the number of hours you worked on the unit, and the amount of time that passed between start and completion of this unit.</li> | |
<!-- included from "./data/ABC-unit_components.txt", section: "deliverables-journal" --> | <!-- included from "./data/ABC-unit_components.txt", section: "deliverables-journal" --> | ||
| − | + | <li><b>Journal</b>: Document your progress in your [[FND-Journal|Course Journal]]. Some tasks may ask you to include specific items in your journal. Don't overlook these.</li> | |
<!-- included from "./data/ABC-unit_components.txt", section: "deliverables-insights" --> | <!-- included from "./data/ABC-unit_components.txt", section: "deliverables-insights" --> | ||
| − | + | <li><b>Insights</b>: If you find something particularly noteworthy about this unit, make a note in your [[ABC-Insights|'''insights!''' page]].</li> | |
<section end=deliverables /> | <section end=deliverables /> | ||
<!-- ============================ --> | <!-- ============================ --> | ||
| Line 53: | Line 53: | ||
<b>Prerequisites:</b><br /> | <b>Prerequisites:</b><br /> | ||
<!-- included from "./data/ABC-unit_components.txt", section: "notes-prerequisites" --> | <!-- included from "./data/ABC-unit_components.txt", section: "notes-prerequisites" --> | ||
| − | This unit builds on material covered in the following prerequisite units: | + | This unit builds on material covered in the following prerequisite units:<br /> |
*[[BIN-PHYLO-Tree_building|BIN-PHYLO-Tree_building (Building Phylogenetic Trees)]] | *[[BIN-PHYLO-Tree_building|BIN-PHYLO-Tree_building (Building Phylogenetic Trees)]] | ||
<section end=prerequisites /> | <section end=prerequisites /> | ||
| Line 62: | Line 62: | ||
| + | {{REVISE}} | ||
{{Smallvspace}} | {{Smallvspace}} | ||
Revision as of 12:39, 16 September 2020
Analysing Phylogenetic Trees
(Species trees, gene trees and the importance of naming, Speciation and duplication signatures)
Abstract:
The analysis of mixed gene trees.
|
Objectives:
|
Outcomes:
|
Deliverables:
Prerequisites:
This unit builds on material covered in the following prerequisite units:
Contents
Contents
Task:
- Read the introductory notes on analysing phylogenetic trees.
Analysis
Analysing your tree
In order to analyse your tree, you need a species tree as reference. This really is an absolute prerequisite to make your expectations about the observed tree explicit. Fortunately we have all species nicely documented in our database.
The reference species tree
Task:
To get a species tree, we make use of the smart and useful PhyloT service:
- Navigate to the PhyloT
- Execute the following R command to create a list of all taxonomy records for the species in your database (plus E. coli):
cat(paste(c(myDB$taxonomy$ID, "83333"), collapse=", "))
- Copy this list and paste it into the search field of the PhyloT page. Select: Scientific names; Internal Nodes collapsed; polytomy no; Format newick; Filename fungiTree.txt. Click on Generate tree. The file fungiTree.txt will be downloaded to your computer into your default download directory. Move it to your project directory. Then click on Visualize in iTOL and confirm the tree: the resulting tree should have twelve species names listed - ten "reference" fungi, E. coli (as the outgroup), and MYSPE. Make sure MYSPE is included! If it's not there, you did something wrong that needs to be fixed.
- Open fungiTree.txt in RStudio. This is a tree, specified in the so-called "Newick format". The topology of the tree is defined through the brackets, and the branch-lengths are all the same: this is a cladogram, not a phylogram.
Let's continue the analysis in R.
Task:
- Open RStudio and load the
ABC-unitsR project. If you have loaded it before, choose File → Recent projects → ABC-Units. If you have not loaded it before, follow the instructions in the RPR-Introduction unit. - Choose Tools → Version Control → Pull Branches to fetch the most recent version of the project from its GitHub repository with all changes and bug fixes included.
- Type
init()if requested. - Open the file
BIN-PHYLO-Tree_analysis.Rand follow the instructions.
Note: take care that you understand all of the code in the script. Evaluation in this course is cumulative and you may be asked to explain any part of code.
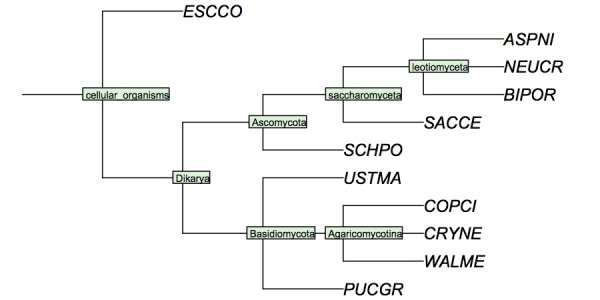
I have constructed a cladogram for many of the species we are analysing, based on data published for 1551 fungal ribosomal sequences. The six reference species are included. Such reference trees from rRNA data are a standard method of phylogenetic analysis, supported by the assumption that rRNA sequences are monophyletic and have evolved under comparable selective pressure in all species.
Cladogram of the "reference" fungi studied in the assignments. This cladogram is based on a tree returned by the NCBI Common Tree. It is thus a digest of cladistic relationships, not a representation of a specific molecular phylogeny.
Alternatively, you can look up your species in the latest version of the species tree for the fungi and add it to the tree by hand while resolving the trifurcations. See:
| Ebersberger et al. (2012) A consistent phylogenetic backbone for the fungi. Mol Biol Evol 29:1319-34. (pmid: 22114356) |
Task:
- Return to the RStudio project and continue with the script to its end. Note the deliverable at the end: to print out your trees and bring them to class.
Notes
Further reading, links and resources
| Szöllősi et al. (2015) Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos Trans R Soc Lond., B, Biol Sci 370:20140335. (pmid: 26323765) |
| Ebersberger et al. (2012) A consistent phylogenetic backbone for the fungi. Mol Biol Evol 29:1319-34. (pmid: 22114356) |
| Marcet-Houben & Gabaldón (2009) The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome. PLoS ONE 4:e4357. (pmid: 19190756) |
Also: Nature-Scitable (2008): Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups
If in doubt, ask! If anything about this learning unit is not clear to you, do not proceed blindly but ask for clarification. Post your question on the course mailing list: others are likely to have similar problems. Or send an email to your instructor.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2017-10-31
Version:
- 1.0
Version history:
- 1.0 First live version.
- 0.1 First stub
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.