Sequence data
Contents
- 1 Objectives
- 2 Links
- 3 Exercises
- 4 Slides
- 4.1 Slide 0007
- 4.2 Abstraction
- 4.2.1 Slide 0009
- 4.2.2 Slide 0010
- 4.2.3 Slide 0012
- 4.2.4 Slide 0013
- 4.2.5 Slide 0014
- 4.2.6 Slide 0015
- 4.2.7 Slide 0016
- 4.2.8 Slide 0017
- 4.2.9 Slide 0018
- 4.2.10 Slide 0019
- 4.2.11 Slide 0020
- 4.2.12 Slide 0021
- 4.2.13 Slide 0022
- 4.2.14 Slide 0023
- 4.2.15 Slide 0024
- 4.2.16 Slide 0025
- 4.2.17 Slide 0026
- 4.2.18 Slide 0027
- 4.2.19 Slide 0028
- 4.2.20 Slide 0029
- 4.2.21 Slide 0030
- 4.2.22 Slide 0031
- 4.2.23 Slide 0032
- 4.2.24 Slide 0033
- 4.2.25 Slide 0034
- 4.3 Representation
- 4.4 Databases
- 4.4.1 Slide 0043
- 4.4.2 Slide 0044
- 4.4.3 Slide 0045
- 4.4.4 Slide 0046
- 4.4.5 Slide 0047
- 4.4.6 Slide 0048
- 4.4.7 Slide 0049
- 4.4.8 Slide 0050
- 4.4.9 Slide 0051
- 4.4.10 Slide 0052
- 4.4.11 Slide 0053
- 4.4.12 Slide 0054
- 4.4.13 Slide 0055
- 4.4.14 Slide 0056
- 4.4.15 Slide 0057
- 4.4.16 Slide 0058
- 4.4.17 Slide 0059
- 4.4.18 Slide 0060
- 4.4.19 Slide 0061
- 4.4.20 Slide 0062
- 4.4.21 Slide 0063
- 4.4.22 Slide 0064
- 4.4.23 Slide 0065
- 4.4.24 Slide 0066
- 4.4.25 Slide 0067
Sequence Abstraction and Databases
Objectives
- Know the one-letter code and key properties of all 20 proteinogenic aminoacids;
- Understand the benefits and limitations of the sequence abstraction;
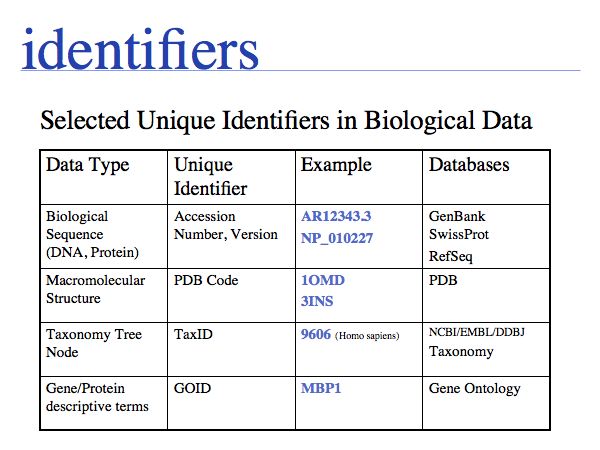
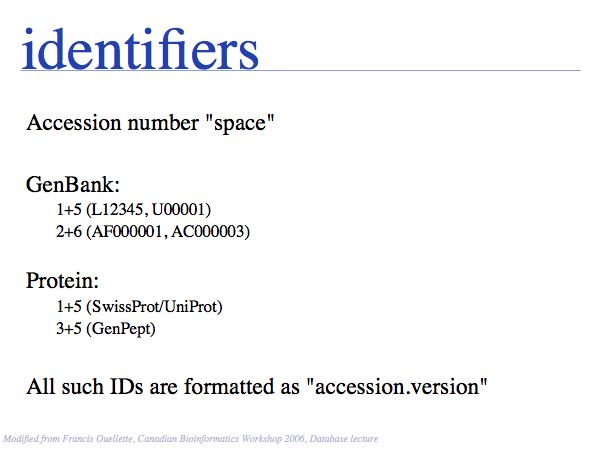
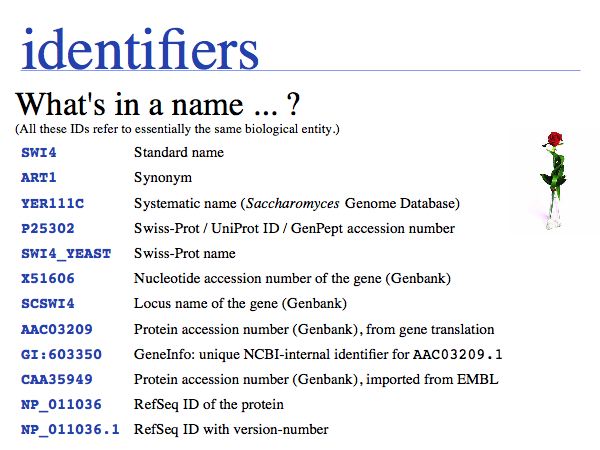
- Recognize common sequence identifiers, utilize them confidently;
- Know about the contents of key sequence databases;
- Be able to retrieve sequence data.
- Know about and confidently use the fields in GenBank and GenPept records.
Links
- IUPAC
- SMILES
- IUPAC one letter codes
- Amino acids (Wikipedia)
- Gene Ontology
- Open Biology Ontologies
- FASTA format
- a GenBank record example
- a GenPept record example
- Genbank Overview
- The PDB
Exercises
- Find a protein that contains a selenocysteine residue (e.g. human glutathione peroxidase). Check the Genbank record to see how this residue is represented in the sequence and in the record. Find and compare the corresponding SwissProt record.
- Find a secreted protein such as E. coli beta-lactamase. Look into the Genbank record whether you can identify the signal-peptide that is post-translationally removed. Find the corresponding SwissProt entry and look for the annotation.
- Human mitochondrial proteins are translated according to a different genetic code from human nuclear proteins. Looking at the CDS of mitochondrial Cytochrome B (NC_001807), how would you know?
Slides
Slide 0007
In Shakespeare's classic tragedy of romantic love and family allegiance, Juliet encapsulates the play's central struggle in this phrase by claiming that Romeo's family name is an artificial and meaningless convention. Just like in the world of the sequence abstraction, this is only partially true: the problems are not just based in the fact that Romeo is called a Montague, but that he is in fact a member of that family. Even if a Thing does not change if it's abstract label changes, such labels rarely exist in isolation: other Things might be referred to by the same label and changing one changes the composition of the entire set. (Or, to remain with our example, as soon as Romeo renounces his name and thus his family, the family would be no longer the same.) Even worse - and this is something we encounter every day in bioinformatics - if identifiers are not stable over time, cross-references to that identifier fail. If you decide you'll call a rose a skunk, people would become very confused.
Abstraction
Slide 0009
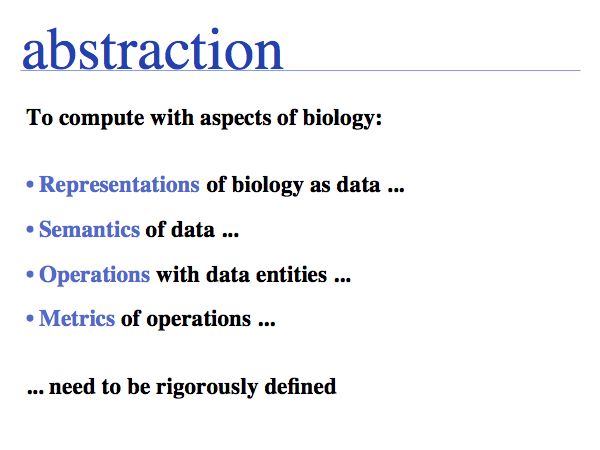
It's worthwhile to deconstruct the above statement: first, we need to consider what it means to compute. Then we need to consider the best representations. To represent a natural object always includes omitting some less-important features. But what is
important and what is not is not the same in all cases.
Slide 0010
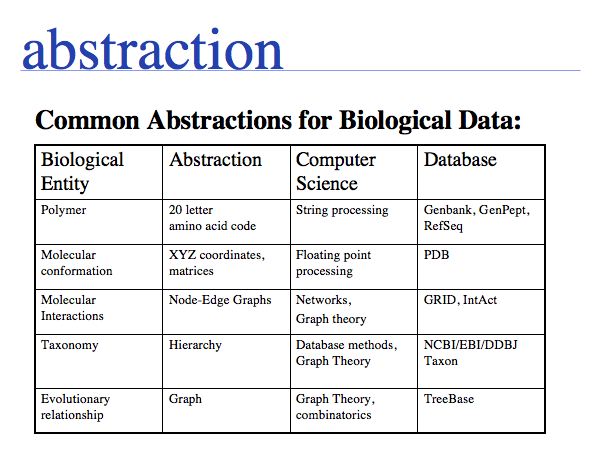
In order to make biology computable, we have to rigorously define our system of objects and their relationships. This is useful even beyond the requirements of bioinformatics. It is an exercise in clarifying the conceptual foundations of biology itself. In many instances, definitions in current, common use are deficient, either because our current state of knowledge has gone beyond the original ideas we were trying to subsume with a term (e.g. gene, or pathway), or because an inconsistent formal and colloquial meaning of terms lead to ambiguities (e.g. function), or because the technical meaning of terms is poorly understood and generally misused (e.g. homology).
Slide 0012
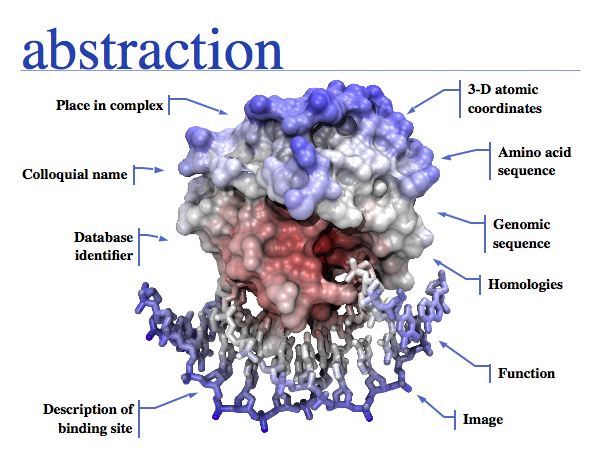
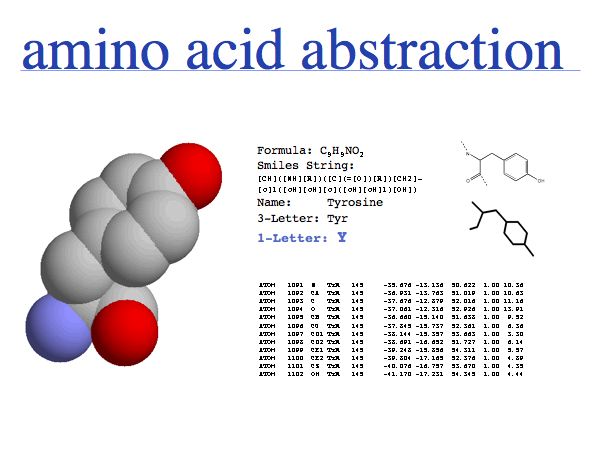
Think about the following question: this image represents a particular biomolecule. What is the best abstraction ?
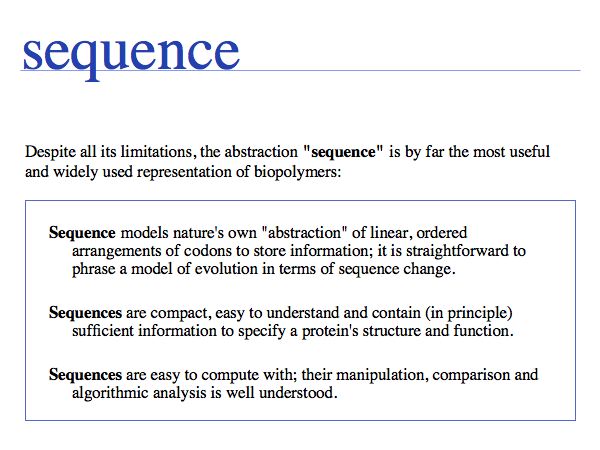
Asking about the best abstraction is an ill-posed question if the purpose is not specified. The best for what? There are many possible abstractions, each serving different purposes. Each of the abstractions above can serve as a representation of the biomolecule, each one emphasizes a different perspective and can serve a different purpose. Some are more suitable than others for any given purpose. Even though abstractions help us model nature by focussing on particular aspects, we must be aware that real molecules have many more properties and features than can be captured in any single model.
Slide 0013
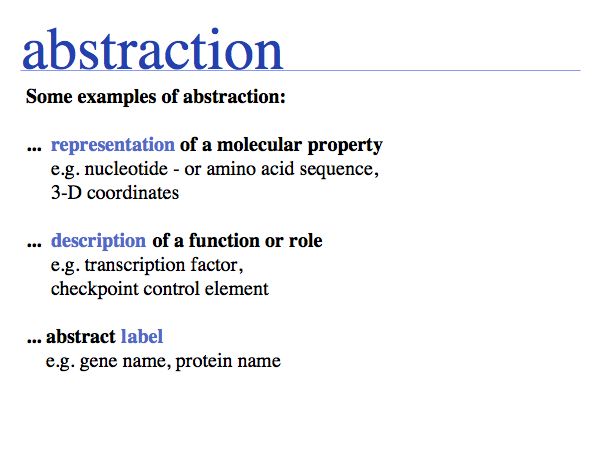
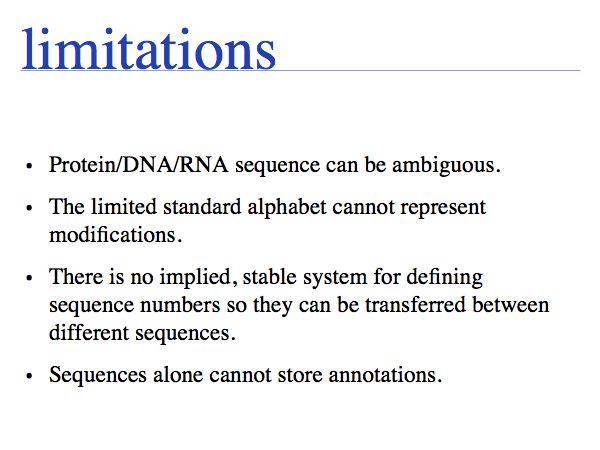
Working with abstractions implies we are no longer manipulating the biological entity, but it's representation. This distinction becomes crucial, when we start computing with representations to infer facts about the original entities. Inferences must be related back to biology! Common problems include (a) that the abstraction may not be rich enough to capture the property we are investigating (e.g. one-letter sequence codes cannot represent amino acid modifications or sequence numbers), or (b) that the abstraction may be ambiguous (e.g. one protein may have more than one homologue in a related organism, thus the relationship between gene IDs is ambiguous) or (c) that the abstraction may not be unique (e.g. one protein may have more than one function, the same protein name may refer to unrelated proteins in different species).
Slide 0014
Slide 0015
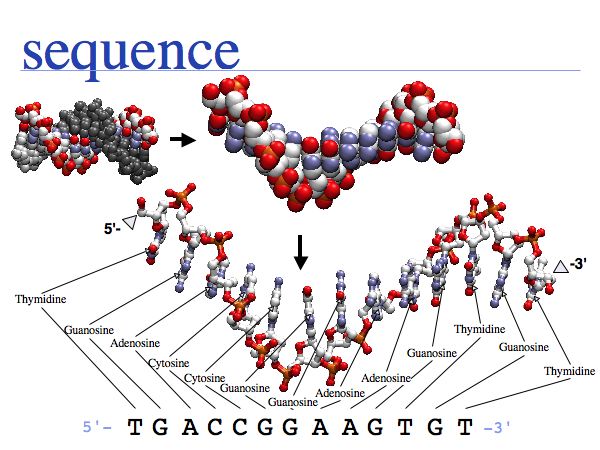
Nucleic acids can form heterocopolymers as DNA or RNA, thus their structural formula can be described (to a close approximation) simply by listing the nucleotide bases in a defined order. A one-letter code has been defined as a shorthand notation for this. By convention, DNA or RNA sequence is written in the direction from the bond with the 5' carbon of the ribose (or deoxyribose), to the bond with the 3'-carbon. Since the 5'-carbon carries a free 5'-phosphate at the terminus of the polynucleotide and the 3'-carbon carries a free OH, this direction happens to be the same direction that nucleic acid polymers are replicated by the DNA-polymerase: a phosphate of a single nucleotide is attached to the free -OH of the polynucleotide. This direction is also the direction of transcription of the RNA polymerase (for the same reason) and (incidentally) the direction of translation by the ribosome. Due to base-pair complementarity, only one strand of a double stranded sequence needs to be recorded since the sequence of the complementary strand is implied: it is simply the reverse complement. The one letter abbreviations are defined by the International Union of Pure and Applied Chemistry (IUPAC) as follows:
- A = adenine
- C = cytosine
- G = guanine
- T = thymine
- R = G A (purine)
- Y = T C (pyrimidine)
- K = G T (keto)
- M = A C (amino)
- S = G C (strong bonds)
- W = A T (weak bonds)
- B = G T C (not A)
- D = G A T (not C)
- H = A C T (not G)
- V = G C A (not T)
- N = A G C T (any)
Slide 0016
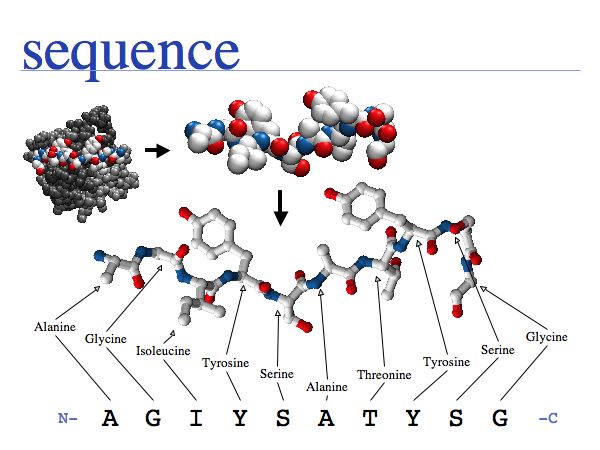
Proteins are amino acid heterocopolymers, thus their structural formula can be described (to a close approximation) simply by listing their constituent amino acid residues in a defined order. The one-letter code is a shorthand notation for this. By convention, protein sequence is written from aminoterminus (N-) to carboxyterminus (-C). This happens to be the same direction in which the protein is synthesized on the ribosome.
Slide 0017
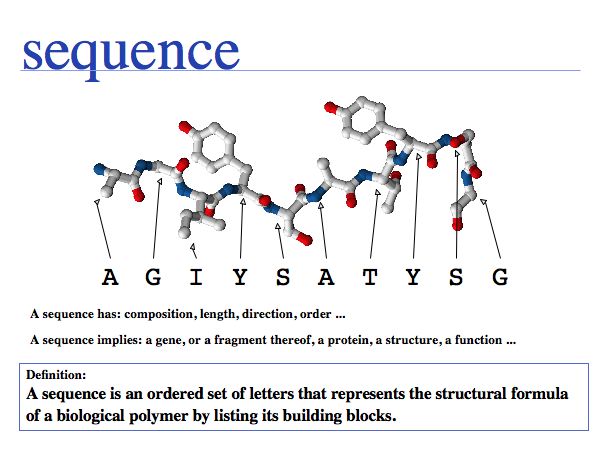
A sequence refers to, or represents, a biomolecule.
Slide 0018
An ordered set of letters to represent biomolecules has some obvious limitations.
Slide 0019
An amino acid is a molecule. A number of abstractions are in common use for this molecule: its chemical formula simply describes the elemental composition, a so called SMILES string (see also here) captures bonding topology as well, its information is equivalent to a chemical graph. A set of records of 3D coordinates can describe the three-dimensional conformation, this can in turn be displayed in a number of different image options - like a simple line drawing or a set of spheres, color coded by element, with relative sizes corresponding to the elements' Van der Waals radii.
Slide 0020

... see IUPAC one letter codes.
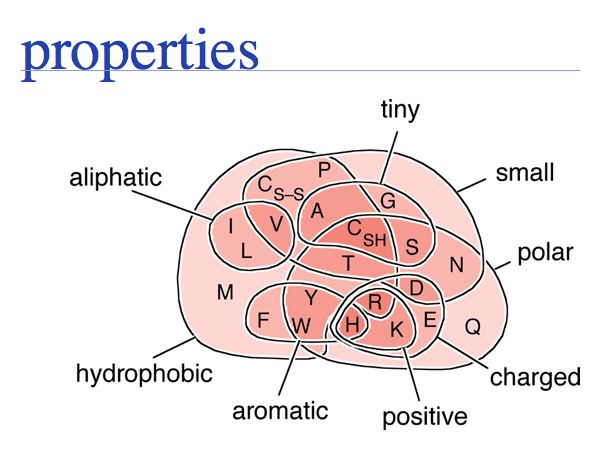
Sequence is the most important abstraction in biology; you need to know your amino acids in order to relate a sequence back to the biopolymer. Required knowledge is: the structural formula, the one- and three- letter codes and key properties (such as charge, relative size, polarity) for all 20 proteinogenic amino acids. A resource that summarizes amino acid properties is at http://speedy.embl-heidelberg.de/aas/, another good entry and overview page is at Wikipedia.
Slide 0021
Of course, the precise role of a particular amino acid depends on its context in a folded protein, however this crude mental map providses a good first approach to estimate amino acid similarity.
Slide 0022
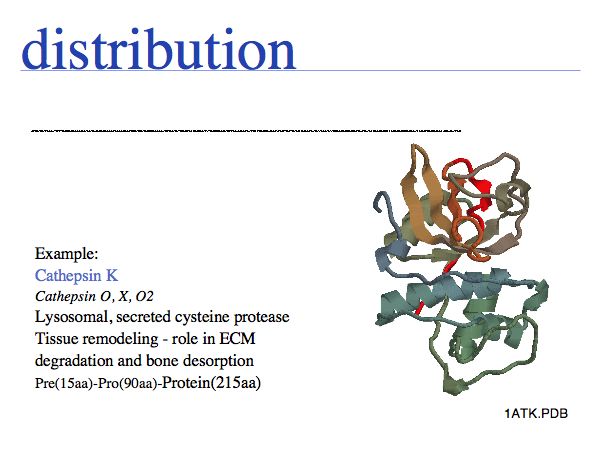
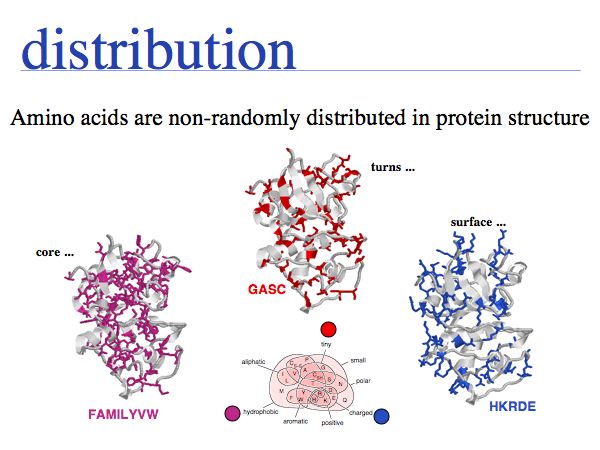
The physicochemical properties of amino acids determines their role in e.g. a folded protein structure. For example, consider the amino acid distribution in a typical enzyme, such as cathepsin K (1ATK).
Slide 0023
- Hydrophobic amino acids - the group FAMILYVW - are found predominantly in the core of a protein.
- Small amino acids such as GASC are often found in turns, at the boundaries of secondary structure elements
- charged amino acid sidechains - (+):KRH and (-)DE - are almost exclusively found on the surface; the energetic requirements for desolvation of the sidechain makes their incorporation into the core unfavourable.
Slide 0024
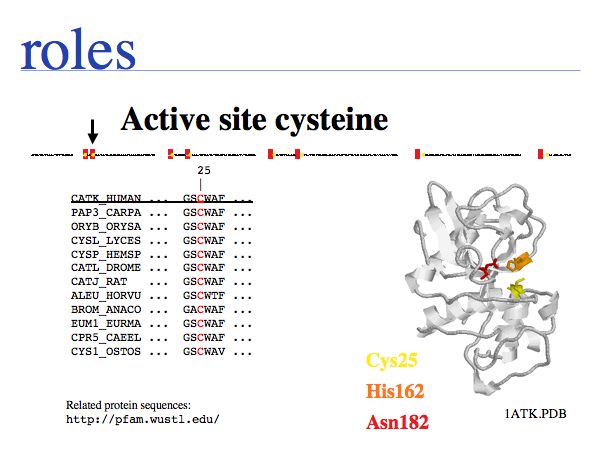
Cysteine can take on a number of different roles, depending on its context. Here, cysteine forms part of the active site, it is the nucleophile in the catalytic triad C-H-N; cathepsin is thus an example of a cysteine-protease. This particular cysteine is absolutely conserved in related proteins since its substitution would lead to (nearly) complete loss of enzymatic function.
Slide 0025
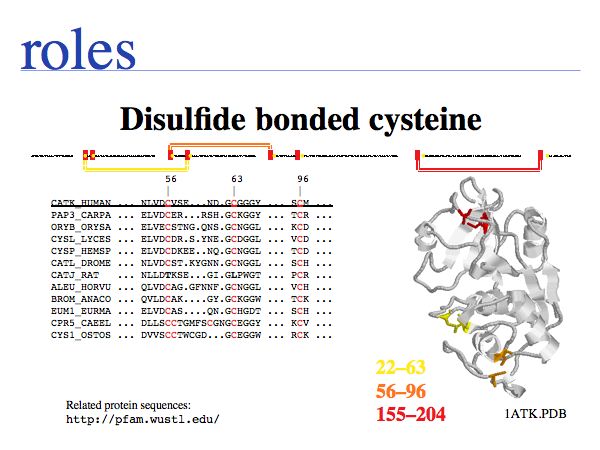
In secreted proteins only, cysteine often forms structural disulfide bridges in which two thiol groups oxidize to a covalent disulfide bond. These cysteines usually are highly conserved. Proteins that are localized in the reducing environment of the cytoplasm do not form structural disulfide bridges.
Slide 0026
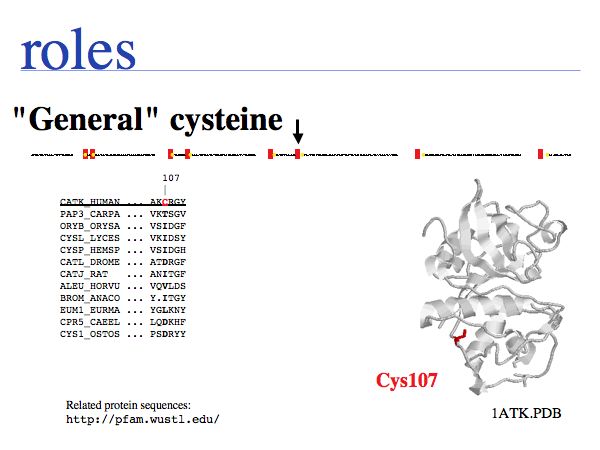
Cysteine can also be found in a very general role, simply as a somewhat polar, small residue. Cysteines in such a general role are only seen infrequently in secreted proteins since the unpaired cysteines can interfere with the formation of the correct disulfide topology; this can lead to slow folding and generally makes the protein sensitive to oxidation. Such cysteines are poorly conserved in related proteins.
Slide 0027
Slide 0028
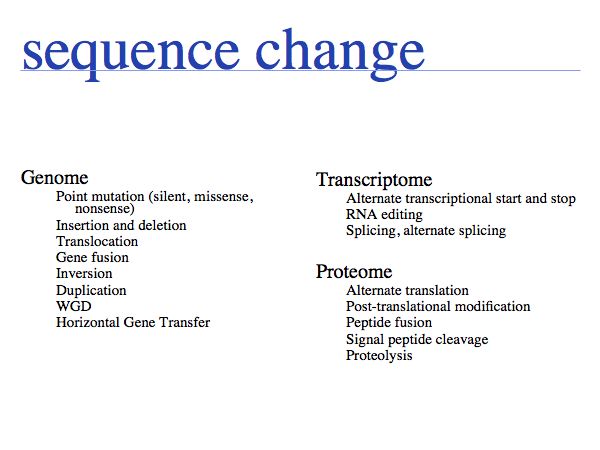
Sequences in biology are not static and a large number of processes act to modify sequences in the course of evolution as well as during the normal function of the cell. Some of these processes generate problems for representing sequences; for example sequence numbers may depend on alternate splicing, or cleavage of pre- or pro- sequences. As well, the very common post-translational modifications cannot be mapped to the 20-letter code.
Slide 0029
Slide 0030
Slide 0031
Slide 0032
Often synonym constrained controlled vocabularies (CVs) are presented as option lists on Web forms. If the CV list is too long for this to be practical, defining the correct form becomes a challenge. In well engineered databases a lot of effort is spent on properly mapping terms to the CV; typically a large dictionary of synonym mappings is employed in some form.
Slide 0033
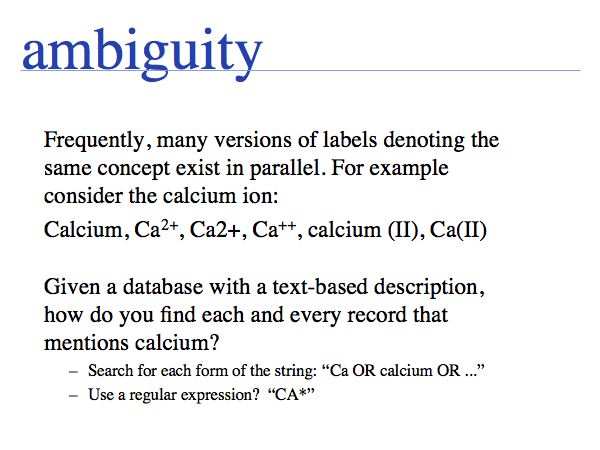
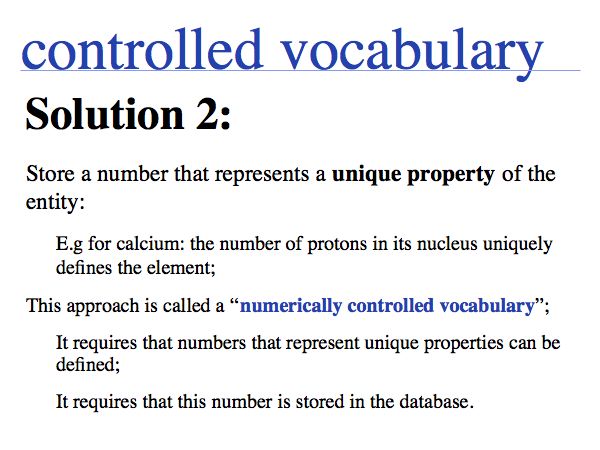
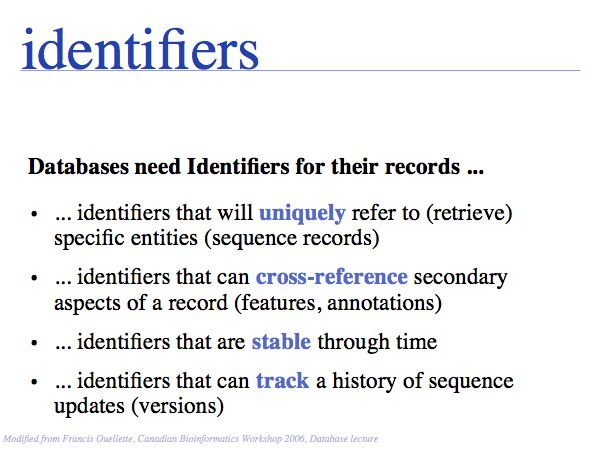
NB. The situation that a unique property of an entity can be concisely described is the ideal case: in that case the identifier captures the most fundamental aspect of the molecule. For calcium, the element does not just have the atomic number 20, it is the element with 20 protons. Similarly oxytocin does not just have the sequence CYIQNCPLG, it is the peptide with that sequence. However, these are favourable exceptions and in general we have to define unique, abstract labels. These are usually called identifiers.
Slide 0034
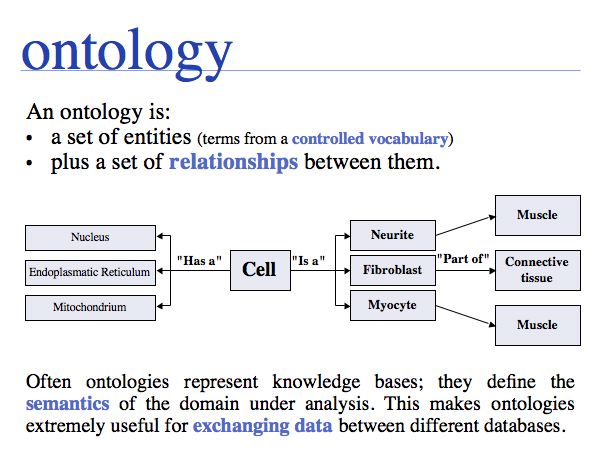
An ontology is a structured CV, with various kinds of relationships defined among them. The "Is a" relationship is probably the most common, but there is really no limit on the types of relationships that may be useful to describe a domain of knowledge. Read more about the Gene Ontology project and the Open Biology Ontologies.
Representation
Slide 0036
In the computer, all data is represented as bits. It depends on the context whether the bits are interpreted as an element of data - integer, floating point number or text -, a complex data structure such as an image or a formatted word-processor document, or even an instruction to the processor.
Slide 0037
Read more about the FASTA format. This is a simple, readable representation but it usually does not contain extensive annotations. Because of it's simplicity, it is something like the lingua franca of file-formats; most bioinformatics tools that operate with sequences are able to read and write FASTA formatted sequences.
Slide 0038
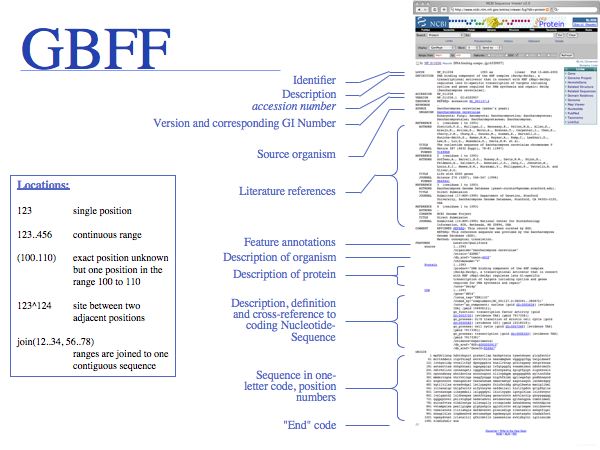
Genbank has its own GenBank Flat file Format (GBFF). Go here for a GenBank record example; go here for a GenPept record example. The amount of annotation can be extensive, providing cross-references, lists of features, sequence translations etc. You should be familiar with the syntax of location identifiers and you should be familiar with the standard contents of a Genbank record.
Slide 0039
One would wish for precise format specifications published with every data record, one would wish for self-describing formats, one would wish for ontologies, and controlled vocabularies and parser code being made available by the databases ... in reality one can't publish such features - or quality assurance efforts in general - and one can't strengthen one's grant proposals with careful, detail-oriented work and thus it doesn't get funded and doesn't get done. The economical and intellectual damage due to this situation is vast. Several lifetimes of graduate student man- and womanpower have been heedlessly wasted through the need to periodically update BLAST output file parsers. And it is said that there is practically not a single parser that can correctly read all information in all PDB files.
Slide 0040
Just like in human language, rigorous syntax rules enforce that you can't use bad grammar and get away with it.
Slide 0041
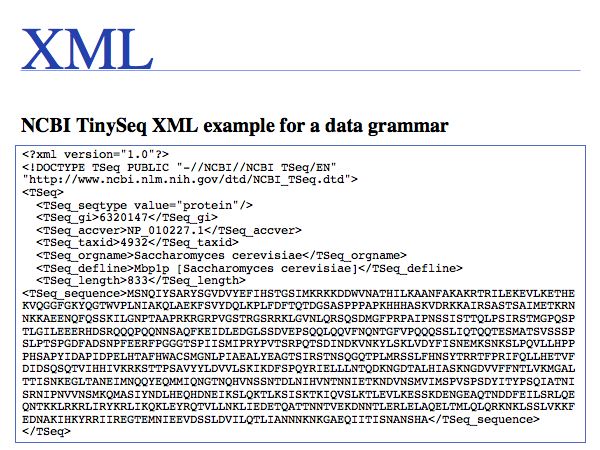
XML formatted files are human readable - in principle. The abundance of tags can make this challenging in practice. The loss of readability is a trade-off for the gain in rigour.
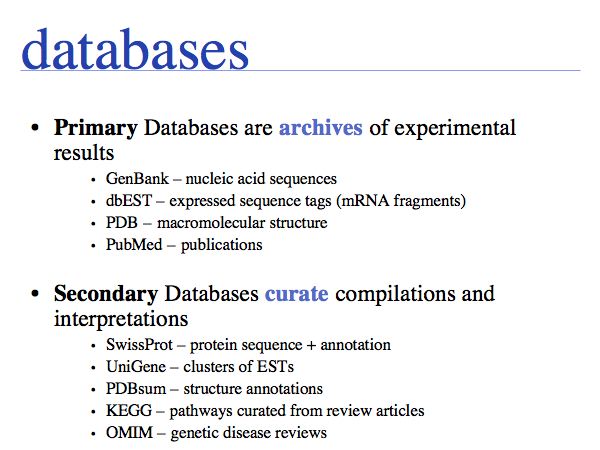
Databases
Slide 0043
Slide 0044
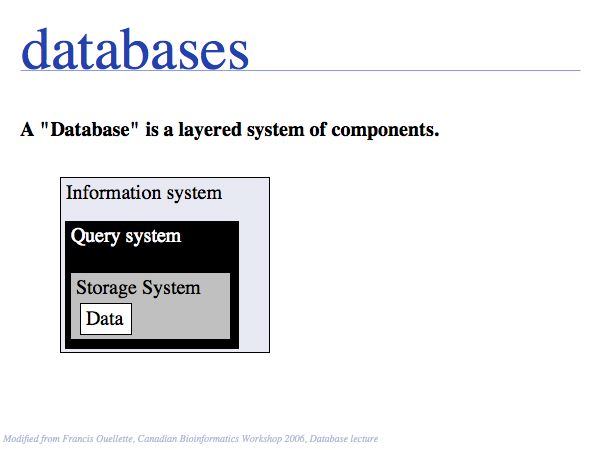
At the core of the database is the data and of course the data should be the primary focus of attention of the database providers. Issues are
- the correctness of the data,
- mechanisms to validate and maintain records,
- frequency of updates,
- consistency
etc.
Slide 0045
Slide 0046
Slide 0047
Slide 0048
Slide 0049
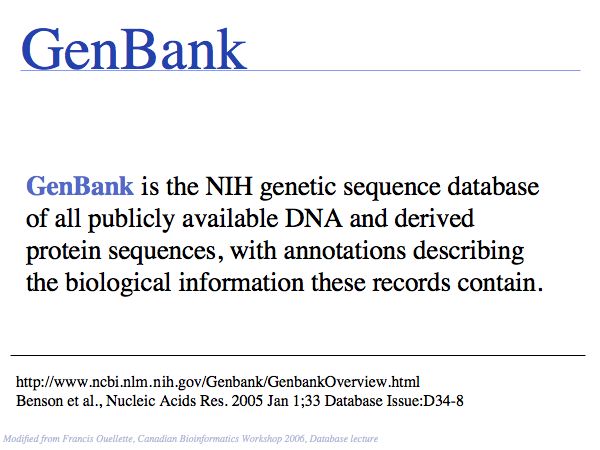
see: the Genbank Overview.
Slide 0050
The PDB
Slide 0051
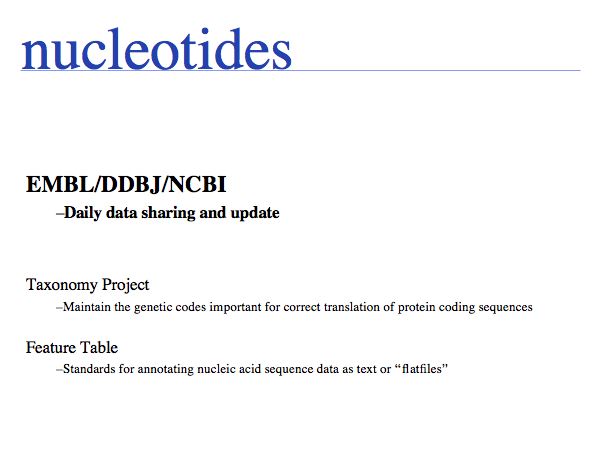
The Sequence Database Consortium ensures that public data is synchronized and made available world-wide.
Slide 0052
Slide 0053
Slide 0054
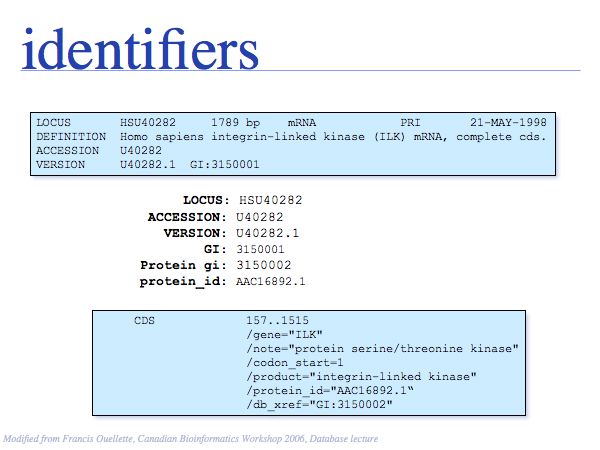
You should be clear on what these terms mean. See the Glossary for more information.
Slide 0055
Slide 0056
Slide 0057
Slide 0058
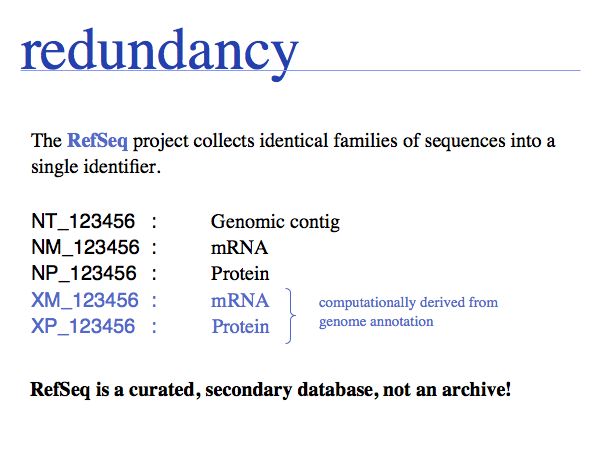
Redundancy is currently a major problem in sequence database searches, because irrelevant duplicates can completely swamp interesting similarities. (cf:[ http://www.ncbi.nlm.nih.gov/RefSeq/ the RefSeq project])
Slide 0059
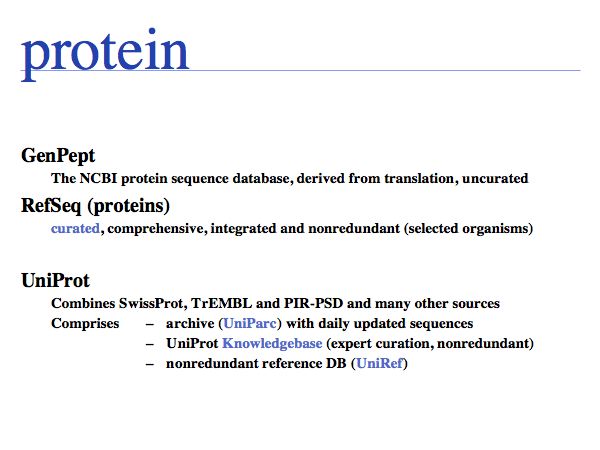
The NCBI has written up an explanation of the differences between GenBank, RefSeq and Uniprot.
Slide 0060
UniProt is arguably the more comprehensive resource, however it is not integrated with the GenBank world, although that would be reasonably straightforward to do. For example, neither does the NCBI record for the Swi4 protein contain a reference to the UniProt accession number, nor does the UniProt record for the same protein contain an NCBI accession number or a GI. National database politics are not always in the best interest of the worldwide scientific community.
Slide 0061
Slide 0062
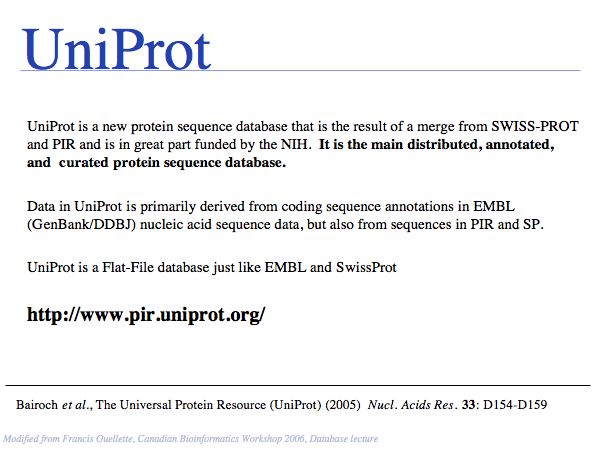
SwissProt records - a subset of the UniProt Knowledge Base - are the highest standard, manually curated, non-redundant protein records available. Unfortunately the growth of the sequence databases far exceeds any human curation capabilities!
Slide 0063
With all the stored and available sequence data, the challenges to cross-reference information have become very apparent.
Slide 0064
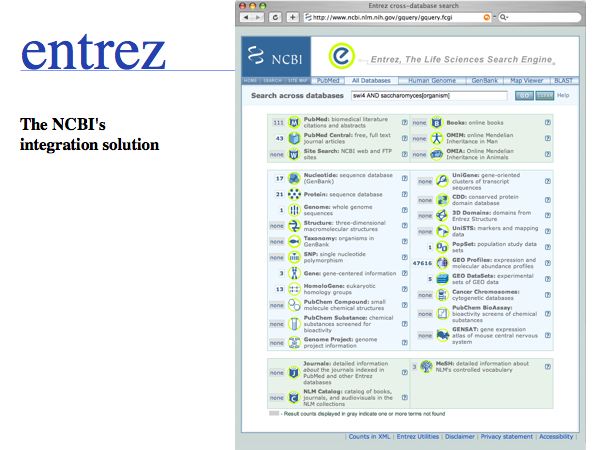
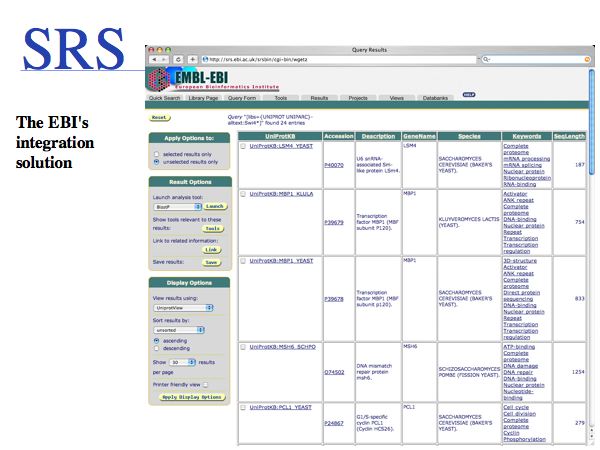
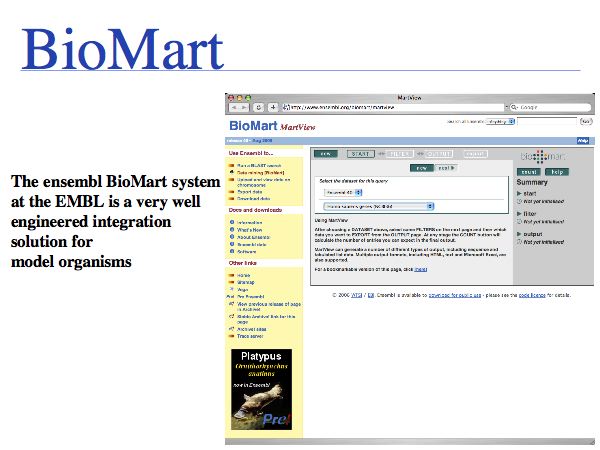
Several strategies for integration have been pursued. For the purposes of Web based bioinformatics, cross-referencing through shared identifiers is one approach. However in practice, researchers often simply perform sequence similarity searches across the databases.
Slide 0065
Slide 0066
Slide 0067