BIN-ALI-BLAST
BLAST heuristic sequence alignment
(BLAST algorithm and Web interface, interpretation of BLAST alignments)
Abstract:
This unit introduces the BLAST algorithm and practices a BLAST search via the Web interface, and scripted in R.
|
Objectives:
|
Outcomes:
|
Deliverables:
Prerequisites:
This unit builds on material covered in the following prerequisite units:
Contents
Evaluation
Evaluation: NA
Contents
BLAST is by a margin the most important computational tool of molecular biology. It is so important, that we couldn't even begin our explorations here without it, and thus we have already used BLAST in the BIN-Storing_data unit, to find the most similar sequence to MBP1_SACCE in MYSPE.
Task:
- Read the introductory notes on the BLAST algorithm for fast, heuristic, database-scale sequence alignment to discover homologues.
A good, detailed introduction to finding homologues on a database scale - and recognizing whether similar sequences are indeed homologous - is Pearson (2013)[1].
Reciprocal Best Matches
In this unit we will use BLAST to perform Reciprocal Best Matches.
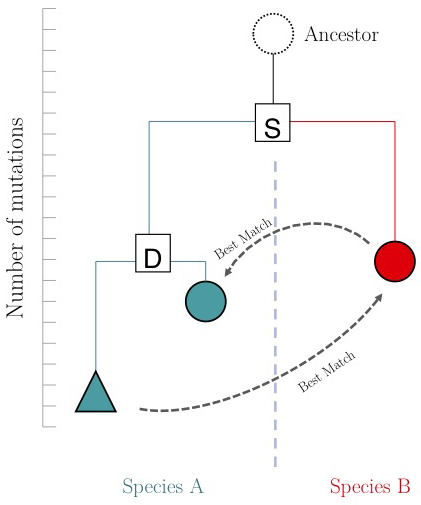
One of the important questions of model-organism based inference is: which genes perform the same function in two different organisms. In the absence of other information, our best guess is that these are the two genes that are mutually most similar. The keyword here is mutually. If MBP1_SACCE from S. cerevisiae is the best match to RES2_SCHPO in S. pombe, the two proteins are only mutually most similar if RES2_SCHPO is more similar to MBP1_SACCE than to any other S. cerevisiae protein. We call this a Reciprocal Best Match, or "RBM"[2].
The argument is summarized in the figure on the right: genes that evolve under continuos selective pressure on their function have relatively lower mutation rates and are thus more similar to each other, than genes that undergo neo- or sub-functionalization after duplication.
However, there is a catch: proteins are often composed of multiple domains that implement distinct roles of their function. Under the assumptions above we could hypothesize:
- a gene in MYSPE that has the "same" function as the Mbp1 cell-cycle checkpoint switch in yeast should be an RBM to Mbp1;
- a gene that binds to the same DNA sites as Mbp1 should have a DNA-binding domain that is an RBM to the DNA binding domain of Mbp1.
Thus we'll compare RBMs in MYSPE for full-length Mbp1_SACCE and its DNA-binding domain, and see if the results are the same.
Full-length RBM
You have already performed the first half of the experiment: matching from S. cerevisiae to MYSPE. The backward match is simple.
Task:
- Access BLAST and follow the link to the protein blast program.
- Enter the RefSeq ID for
MBP1_MYSPEin the Query sequence field. - Select
refseq_proteinas the database to search in, and enterSaccharomyces cerevisiae (taxid:4932)to restrict the organism for which hits are reported. - Run BLAST. Examine the results.
If your top-hit is NP_010227, you have confirmed the RBM between Mbp1_SACCE and Mbp1_MYSPE. If it is not, let me know. I expect this to be the same and would like to verify your results if it is not[3].
RBM for the DNA binding domain
The DNA-binding domain of Mbp1_SACCE is called an APSES domain. If the RBM between Saccharomyces cerevisiae Mbp1 and MYSPE is truly an orthologue, we expect all of the protein's respective domains to have the RBM property as well. But let's not simply assume what we can easily test. We'll define the sequence of the APSES domain in MBP1_SACCE and MYSPE and see how these definitions reflect in a BLAST search.
Defining the range of the APSES domain annotation
The APSES domain is a well-defined type of DNA-binding domain that is ubiquitous in fungi and unique in that kingdom. Structurally it is a member of the Winged Helix-Turn-Helix family. Recently it was found that it is homologous to the somewhat shorter, prokaryotic KilA-N domain; thus the APSES domain was retired from pFam and instances were merged into the KilA-N family. However InterPro has a KilA-N entry but still recognizes the APSES domain.
KilA-N domain boundaries in Mbp1 can be derived from the results of a CDD search with the ID 1BM8_A (the Mbp1 DNA binding domain crystal structure). The KilA-N superfamily domain alignment is returned.
- (pfam 04383): KilA-N domain; The amino-terminal module of the D6R/N1R proteins defines a novel, conserved DNA-binding domain (the KilA-N domain) that is found in a wide range of proteins of large bacterial and eukaryotic DNA viruses. The KilA-N domain family also includes the previously defined APSES domain. The KilA-N and APSES domains may also share a common fold with the nucleic acid-binding modules of the LAGLIDADG nucleases and the amino-terminal domains of the tRNA endonuclease.
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
1BM8A 16 IHSTGSIMKRKKDDWVNATHILKAANFAKaKRTRILEKEVLKETHEKVQ---------------GGFGKYQGTWVPLNIA 80
Cdd:pfam04383 3 YNDFEIIIRRDKDGYINATKLCKAAGETK-RFRNWLRLESTKELIEELSeennvdkseiiigrkGKNGRLQGTYVHPDLA 81
90
....*....|....
1BM8A 81 KQLA----EKFSVY 90
Cdd:pfam04383 82 LAIAswisPEFALK 95
Note that CDD and SMART are not consistent in how they apply pFam 04383 to the Mbp1 sequence. See annotation below.
The CDD KilA-N domain definition begins at position 16 of the 1BM8 sequence. But virtually all fungal APSES domains have a longer, structurally defined, conserved N-terminus. Blindly applying the KilA-N domain definition to these proteins would lose important information. For most purposes we will prefer the sequence spanned by the 1BM8_A structure. The sequence is given below, the KilA-N domain is coloured dark green. By this definition the APSES domain is 99 amino acids long and comprises residues 4 to 102 of the NP_010227 sequence.
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
1BM8A 1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKETHEKVQGGFGKYQGTWVPLNIA 80
90
....*....|....*....
1BM8A 81 KQLAEKFSVYDQLKPLFDF 99
- Yeast APSES domain sequence in FASTA format
>APSES_MBP1 Residues 4-102 of S. cerevisiae Mbp1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF
- Synopsis of ranges
| Domain | Link | Length | Boundary | Range (Mbp1) | Range (1BM8) |
| KilA-N: pfam04383 (CDD) | CDD alignment | 72 | STGSI ... KFSVY | 21 - 93 | 18 - 90 |
| KilA-N: pfam04383 (SMART) | Smart main page | 79 | IHSTG ... YDQLK | 19 - 97 | 16 - 94 |
| KilA-N: SM01252 (SMART) | Smart main page | 84 | TGSIM ... DFTQT | 22 - 105 | 19 - 99... |
| APSES: Interpro IPR003163 | (Interpro) | 130 | QIYSA ... IRSAS | 3 - 133 | 1 - 99... |
| APSES (1BM8) | – | 99 | QIYSA ... PLFDF | 4 - 102 | 1 - 99 |
Executing the forward search
Task:
- Access BLAST and follow the link to the protein blast program.
- Forward search:
- Paste only the APSES domain sequence for
MBP1_SACCEin the Query sequence field (copy the sequence from above). - Select
refseq_proteinas the database to search in, and enter the correct taxonomy ID for MYSPE in the Organism field. - Run BLAST. Examine the results.
- If the top hit is the same protein you have already seen, oK. If it's not add it to your protein database in RStudio.
- Paste only the APSES domain sequence for
With this we have confirmed the sequence with the most highly conserved APSES domain in MYSPE. Can we take the sequence for the reverse search from the alignment that BLAST returns? Actually, that is not a good idea. The BLAST alignment is not guaranteed to be optimal. We should do an optimal sequnece alignment instead. That is: we use two different tools here for two different purposes: we use BLAST to identify one protein as the most similar among many alternatives and we use optimal sequence alignment to determine the best alignment between two sequences. That best alignment is what we will annotate as the MYSPE APSES domain.
We will execute the sequence alignment and the reverse search in R.
Task:
- Open RStudio and load the
ABC-unitsR project. If you have loaded it before, choose File → Recent projects → ABC-Units. If you have not loaded it before, follow the instructions in the RPR-Introduction unit. - Choose Tools → Version Control → Pull Branches to fetch the most recent version of the project from its GitHub repository with all changes and bug fixes included.
- Type
init()if requested. - Open the file
BIN-ALI-BLAST.Rand follow the instructions.
Note: take care that you understand all of the code in the script. Evaluation in this course is cumulative and you may be asked to explain any part of code.
Further reading, links and resources
- Contents of the BLAST report
| Boratyn et al. (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29-33. (pmid: 23609542) |
|
[ PubMed ] [ DOI ] The Basic Local Alignment Search Tool (BLAST) website at the National Center for Biotechnology (NCBI) is an important resource for searching and aligning sequences. A new BLAST report allows faster loading of alignments, adds navigation aids, allows easy downloading of subject sequences and reports and has improved usability. Here, we describe these improvements to the BLAST report, discuss design decisions, describe other improvements to the search page and database documentation and outline plans for future development. The NCBI BLAST URL is http://blast.ncbi.nlm.nih.gov. |
Notes
- ↑
Pearson (2013) An introduction to sequence similarity ("homology") searching. Curr Protoc Bioinformatics Chapter 3:Unit3.1. (pmid: 23749753) [ PubMed ] [ DOI ] Sequence similarity searching, typically with BLAST, is the most widely used and most reliable strategy for characterizing newly determined sequences. Sequence similarity searches can identify "homologous" proteins or genes by detecting excess similarity- statistically significant similarity that reflects common ancestry. This unit provides an overview of the inference of homology from significant similarity, and introduces other units in this chapter that provide more details on effective strategies for identifying homologs.
- ↑ Note that RBMs are usually orthologues, but the definition of orthologue and RBM is not the same. Most importantly, many orthologues are not RBMs. We will explore this more when we discuss phylogenetic inference.
- ↑ One such case we encountered involved a protein that has a corrupted annotation for the DNA binding domain. It appears to be the correct orthologue, but it only has the second highest BLAST score.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2017-08-05
Modified:
- 2020-09-24
Version:
- 1.1
Version history:
- 1.1 2020 Maintenance
- 1.0 Live version 2017
- 0.1 First stub
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.