VMD
Jump to navigation
Jump to search
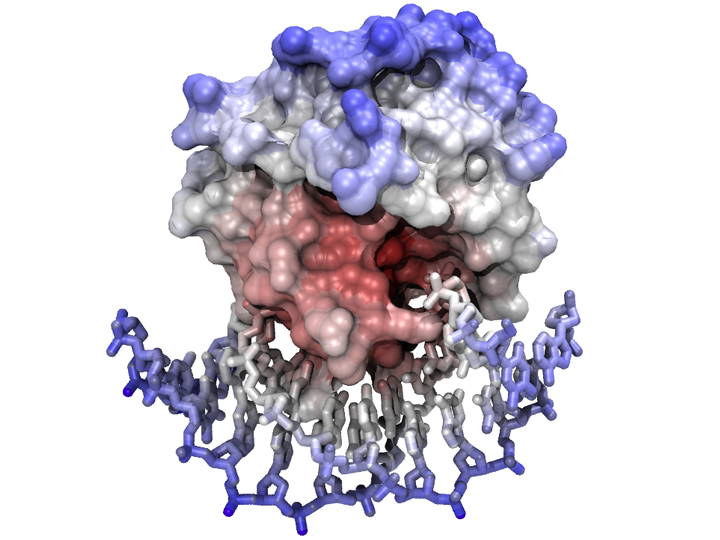

A complex between a model of Mbp1 and a segment of DNA
The model was visualized and positioned in VMD - nucleotides were displayed as "licorice" sticks and the protein was covered by its solvent-accessible surface, calculated with a probe radius of 1.4 Å. Coloring is ramped according to position (i.e. an atom's distance from the complex's centre of mass). The scene was rendered with the tachyon raytracer using ambient occlusion lighting and 8-fold antialiasing.
The model was visualized and positioned in VMD - nucleotides were displayed as "licorice" sticks and the protein was covered by its solvent-accessible surface, calculated with a probe radius of 1.4 Å. Coloring is ramped according to position (i.e. an atom's distance from the complex's centre of mass). The scene was rendered with the tachyon raytracer using ambient occlusion lighting and 8-fold antialiasing.
VMD (viual molecular dynamics) is a powerful, well engineered, molecular graphics package. It is one of a number of freely available tools to visualize and manipulate three-dimensional molecular datasets, but it stands out due to its rich features, the versatility of its use and the ease with which common tasks can be accomplished and the fact that it is being actively developed and maintained.
Installing VMD
- Access the VMD homepage and navigate to the download section.
- Follow the link to the newest version (1.8.6 as of this writing) and review the features.
- Access the Download page and click on the link appropriate for your platform. Follow the instructions to install VMD.
Molecular Models - Basics
This section of the tutorial introduces you to basic molecular rendering. It includes
- The basic function of VMD
- Loading and displaying molecules
- changing display parameters
- drawing styles and multiple representations
- selecting subsets of the molecule
- the Sequence Viewer extension
- saving and reloading your work
- Follow this link for Part 1 of the BCH441 VMD tutorial