ABC-INT-GO categories
Integrator Unit: GO term categories
(Integrator unit: define GO term selection)
Abstract:
This page integrates leraning units on code development and data structures, graph theory and the Gene Ontology.
Deliverables:
- Integrator unit: Deliverables can be submitted for course marks. See below for details.
Prerequisites:
This unit builds on material covered in the following prerequisite units:
Contents
Evaluation
This "Integrator Unit" is not for evaluation. We will work through this unit in class to illustrate the process of translating requirements to tasks, and bringing a project to a defined conclusion, supported by the ABC knowledge network.
- Report option
- Work through the tasks described in the scenario.
- Document your results in a short report on a subpage of your User page on the Student Wiki. Describe your methods (R-code!) in an appendix;
Contents
Scenario
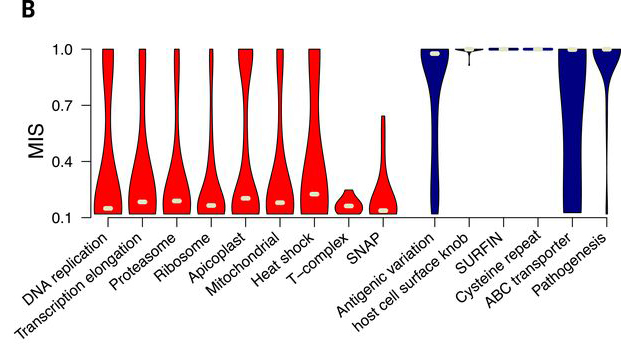
This is panel B' of Figure 3 from Zhang et al.s analysis of essential genes of Plasmodium falciparum by saturation mutagenesis[1]. A typical question of large-scale experiments that discover sets of genes is: what do these genes do? Is there a trend among functional categories? As you see, the data is derived from experiments, but the interpretation is entirely dependent on how these functional categories are defined.
How were these categories defined? Is there a principled way to do so?
The Gene Ontology contains more than 270,000 terms and these are far too many categories to provide a meaningful overview such as the one that Zhang et al. required. GO maintains a number of subsets, but the generic "GO slim" also contains more than 2,200 terms.
Your task is:
- to consider how a meaningful, balanced subset of GO terms can be defined to a resolution that can be specified by a user, say, between 10 and 100 terms;
- to write R code to produce such a subset, paying attention to R coding style, and best practice of software design, data management, and reproducible research;
- to document and evaluate your results.
The code should use go-basic.obo as its input, it can also use the homo sapiens GOA data. It's output should be useful as input to a function that reads GOA data and a list of gene symbols, and associates each gene symbol with a GO term.
- Note
- This task is open in the sense that there are potentiall many suitable solutions: I expect that balancing terms will consider the number of children on the GO graph and/or the number of genes annotated to the various branches in GOA. Presumably your first task is to explore the various concepts around this task, formulate precise requirements, and to write a project plan.
Self-evaluation
Notes
- ↑
Zhang et al. (2018) Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360:. (pmid: 29724925) [ PubMed ] [ DOI ] Severe malaria is caused by the apicomplexan parasite Plasmodium falciparum. Despite decades of research, the distinct biology of these parasites has made it challenging to establish high-throughput genetic approaches to identify and prioritize therapeutic targets. Using transposon mutagenesis of P. falciparum in an approach that exploited its AT-rich genome, we generated more than 38,000 mutants, saturating the genome and defining mutability and fitness costs for over 87% of genes. Of 5399 genes, our study defined 2680 genes as essential for optimal growth of asexual blood stages in vitro. These essential genes are associated with drug resistance, represent leading vaccine candidates, and include approximately 1000 Plasmodium-conserved genes of unknown function. We validated this approach by testing proteasome pathways for individual mutants associated with artemisinin sensitivity.
Further reading, links and resources
If in doubt, ask! If anything about this learning unit is not clear to you, do not proceed blindly but ask for clarification. Post your question on the course mailing list: others are likely to have similar problems. Or send an email to your instructor.
About ...
Author:
- Boris Steipe <boris.steipe@utoronto.ca>
Created:
- 2018-09-18
Modified:
- 2018-09-18
Version:
- 1.0
Version history:
- 1.0 First live version
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.