PHALY
PHALY: Phagosome/Lysosome Fusion
Contents
- 1 Stage 1: The System seed
- 2 Stage 2: Concepts
- 3 Stage 3: Adding Genes
- 3.1 Components added from literature review
- 3.2 Genes added from direct annotation
- 3.2.1 LC3-II
- 3.2.2 SNAP29-STX17 Qabc-SNARE
- 3.2.3 PHALY
- 3.2.4 Mon1-CCZ1
- 3.2.5 lysosome
- 3.2.6 SNAP29-STX17-VAMP8 SNARE-pin
- 3.2.7 PIK3C3 complex
- 3.2.8 HOPS complex
- 3.2.9 PIKFYVE complex
- 3.2.10 RAB7-RILP-dynein-dynactin complex
- 3.2.11 F-actin network
- 3.2.12 F-actin
- 3.2.13 20s supercomplex
- 3.2.14 NOT IN PHALY
- 3.2.15 New gene facts:
- 3.3 Genes added from network annotation
- 3.4 Genes added from phenotype and behaviour
- 4 Completing the Role Ontology
- 5 System Architecture
- 6 System data
- 7 References
- Name: Boris Steipe
- eMail: boris.steipe@utoronto.ca
Stage 1: The System seed
- Name
- Phagosome/Lysosome Fusion
- Code
- PHALY
- Definition
- Control and process of phagosome-lysosome docking and fusion.
- Description
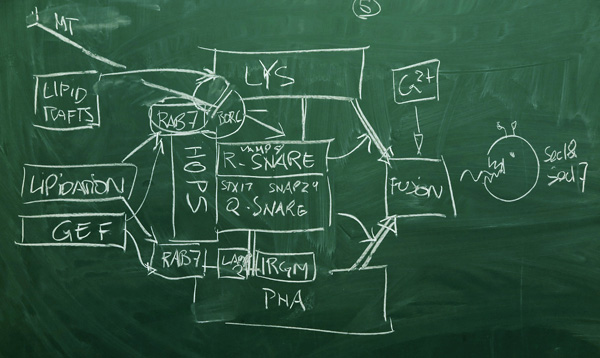
- Phagosome-lysosome fusion is a key step in autophagy, the process in which a lysosome - a package of acidic, lytic enzymes - fuses to a phagosome that has enveloped some cargo, which is targeted for degradation and recycling. The fusion is a highly regulated process, involving the preparation of lipid microdomains, positioning of vesicles by dynein motors that move along the cytoskeleton, membrane anchored SNAREs that provide core docking functions, the facilitating CORVET, HOPS and BORC complexes, and precise regulation of fusion initiation at the protein level, the lipid level, and through the ionic context which involves calcium-selective voltage gated channels.
- Seed term (Biological Process ontology)
-
- GO:0090385 (phagosome-lysosome fusion)
- Relevant related terms(according to GO graph and co-occurring terms)
-
- GO:0001845 phagolysosome assembly (P) (Parent)
- GO:0090384 phagosome-lysosome docking (P) (Sibling)
- GO:0048278 vesicle docking (P) (Parent)
- GO:0031201 SNARE complex (C) (Co-occurring)
- GO:0000149 SNARE binding (F)(Co-occurring)
- GO:0005484 SNAP receptor activity (F) (co-occurring)
- GO:0090383 phagosome acidification (P) (co-occurring)
- Initial associated human genes
Seven additional proteins are annotated to GO:0090385 in GOA, they were excluded from the seed-term table because their evidence codes indicated insufficient curation.
- Seed literature
Corona & Jackson (2018) Finding the Middle Ground for Autophagic Fusion Requirements. Trends Cell Biol 28:869-881. (pmid: 30115558) [ PubMed ] [ DOI ] Autophagosome/amphisome-lysosome fusion is a highly regulated process at the protein, lipid, and biochemical level. Each primary component of fusion, such as the core SNAREs, HOPS complex, or physical positioning by microtubule-associated dynein motors, are regulated at multiple points to ensure optimum conditions for autophagic flux to proceed. With the complexity of the membrane fusion system, it is not difficult to imagine how autophagic flux defect-related disorders, such as Huntington's disease, non-familial Alzheimer's disease, and Vici syndrome develop. Each membrane fusion step is regulated at the protein, lipid, and ion level. This review aims to discuss the recent developments toward understanding the regulation of autophagosome, amphisome, and lysosome fusion requirements for successful autophagic flux.
Zhi et al. (2018) Anatomy of autophagy: from the beginning to the end. Cell Mol Life Sci 75:815-831. (pmid: 28939950) [ PubMed ] [ DOI ] Autophagy is a highly regulated process in eukaryotes to maintain homeostasis and manage stress responses. Understanding the regulatory mechanisms and key players involved in autophagy will provide critical insights into disease-related pathogenesis and potential clinical treatments. In this review, we describe the hallmark events involved in autophagy, from its initiation, to the final destruction of engulfed targets. Furthermore, based on structural and biochemical data, we evaluate the roles of key players in these processes and provide rationale as to how they control autophagic events in a highly ordered manner.
1b: First Sketch
1c: Observables
- Components
- Literature curation
- Annotation to relevant GO terms
- Collaborations
- SMART interactions
- Known complexes
- SNAREs
- HOPS Complex
- BORC complex
- Crystal structures[3]
- Behaviour
- autophagic flux
- Mislocalization of components
- genetic interactions
- Vici syndrome (autosomal recessive EPG5 mutations)
Stage 2: Concepts
Concepts
Phagosome - lysosome fusion is the effector step of phagocytosis and (macro)autophagy.
The context of the PHALY system is defined as follows:
- Phagosome vesicles and lysosome vesicles have been produced and co-localized;
- The PHALY system docks the two vesicles and fuses their membranes;
- As a result, lysosome contents gets mixed into the phagosome and its contents is digested.
Fusion is effected by membrane anchored SNARE proteins (soluble NSF attachment protein receptors) in concert with tether complexes that are activated by Rab proteins[1]. SNAREs are small, membrane-anchored, helical proteins that can assemble into four-helix bundles. Based on sequence motifs, SNARES are subdivided into Q-SNARES and R-SNARES. Since parts of the bundle are be located in a vesicle membrane, and other parts in a target membrane, formation of the bundle brings vesicle and target in close proximity. Thus SNAREs are functionally divided into v-SNARES (incoming vesicle, e.g. lysosome), and t-SNARES (target of the vesicle, e.g. phagosome). On the target side, we usually have three Q-SNARE motifs (Qabc); on the vesicle side we usually have a single R-SNARE.
SNAREs alone cannot support efficient fusion. Additional membrane tethers are provided by the HOPS, CORVET and BORC complexes. These complexes are highly regulated by Rab proteins.
Rab proteins are members of one of the five major families of the Ras superfamily[4] (Ras, Rho, Ran, Rab and Arf). These small, homologous GTPases are molecular switches that are localized to specific membrane microdomains. There, they are turned on through a conformational change due to binding GTP, and turned off by hydrolyzing GTP to GDP[5]. These functions are assisted by auxiliary proteins: GAPs (GTPase Activitating Proteins) activate the GTPase activity, thus leading to conversion of GTP to GDP which brings the protein to the off-state, GEF (Guanine nucleotide exchange factors) promote the realease of GDB, followed by binding of GTP which leads to the on-state. Each of the Ras family members has a distinct profile of roles in the cell. The Rab family proteins are predominantly associated with membrane-trafficking and vesicular transport. In the phagosome-lysosome fusion process, Rab proteins are specifically implicated in SNARE/tether protein assembly, and lipid-domain driven localization[3].
To ensure that the fusion happens at the right time in the right place, the system must include
- functions to synthesize and assemble the required components;
- localization elements that create a targetable environment and localize the required components in the right place;
- contact elements that bring all participants into close proximity;
- on-switches that trigger fusion;
- components that provide energy and/or metabolites to make fusion proceed against the unfavourable geometry of high-curvature membrane domains;
- off-switches that limit the fusion event - if this is not a self-limiting process;
- degradation and recycling functions.
Stage 3: Adding Genes
Components added from literature review
- '''CODE''' is the product of ENST12345. Notes. {{#pmid: <PMID>|LABEL}}.
- '''CODE''' is a component of ''CODE''. Notes. {{#pmid: <PMID>|LABEL}}.
- '''CODE.<Roman numeral>''' is a state defined by: (COMPONENT CODE.STATE) and [OWN STATE VARIABLE REALIZATION]. {{#pmid: <PMID>|LABEL}}.
- '''CODE.<Roman numeral>'''changes to (STATE) when [STATE VARIABLE CHANGE]. {{#pmid: <PMID>|LABEL}}.
It is useful to start by collecting facts that are broken down to relate to one single component each. Summarizing knowledge in structured free-text helps parse the facts to enter them into a common data model. The sentence prototype is:
'''COMPONENT''' [SYMBOL] (NAME) is a component of ''SYSTEM''. NOTES. {{#pmid: <PMID>|LABEL}}.
Make special note of genes that are NOT in your system, even though they are homologous to system components or interact with components.
Preparing the phagosome
- The autophagosome is a component of the PHALY system. Fusion is the process of joining the outer phagosomal membrane of the autophagosome with the membrane of a lysosome. [1] The outer autophagosomal membrane (OAM) and the inner autophagosomal membrane (IAM) form by membrane fission after closure of the phagosomal sac. [6]
- The OAM (outer autophagosomal membrane) is a component of the autophagosome system. The outer autophagosomal membrane (OAM) forms by membrane fission after closure of the phagosomal sac. [6]
- The IAM (inner autophagosomal membrane) is a component of the autophagosome system. The inner autophagosomal membrane (IAM) forms by membrane fission after closure of the phagosomal sac. [6] The IAM is degraded after lysosomal fusion and degradation of the IAM leads to immediate dissociation of STX17 from the cis-SNARE complex. [6]
- LC3-II is a component of the autophagosome. LC3-II (PE lipidated LC3) has been conjugated to the PE-enriched phagosome membrane and thereby serves as phagosome marker. [1] The LC3-II conjugate is produced by the Atg8 conjugation system (Atg7, Atg3, Atg5, Atg12, Atg16L1/2, Atg4A/B/C/D). [3] Priming of the Atg8 conjugation system requires the Atg12 conjugating system (Atg12, Atg5, Atg7, Atg10) [3]
- LC3 [MAP1LC3A] (microtubule associated protein 1 light chain 3 alpha) is a component of LC3-II. LC3 is a member of the Atg8 protein family.[1] LC3 is a cytosolic protein that is conjugated to PE in the PE-enriched phagosome membrane in a stress response.[1] Conjugation of LC3 and PE involves a ubiquitin-like conjugation system. [7]
- PE (phosphatidylethanolamine) is a component of LC3-II. PE is the lipid anchor of LC3 in the phagosome membrane. [1]
- The SNAP29-STX17 Qabc-SNARE is a component of the SNAP29-STX17-VAMP8 SNARE-pin. The SNAP29-STX17 Qabc-SNARE forms through the interaction of SNAP29 and STX17 in the phagosome membrane.[1]
- SNAP29 [SNAP29] (synaptosome associated protein 29) is a component of the SNAP29-STX17 Qabc-SNARE. SNAP29 is a Qa-SNARE (a t-SNARE) that localizes to the membrane in a lipidation independent manner through its STX17 interaction. [1] SNAP29 is held in a non-interacting state by O-GlcNAcylation with GlcNAC. [8]
- GlcNAC (N-acetyl-glucosamine) is a component of the PHALY system. O-GlcNAcylation of SNAP29 in nutrient-sufficient conditions inhibits the association of the SNAP29-STX17 Qabc-SNARE. [1] SNAP29 is O-GlcNAcylated by the promiscuous OGT (UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase) under nutrient-sufficient conditions and thus links the nutrient status sensing system with autophagic flux.[8] SNAP29 has its O-GlcNAcetylation reversed by the N-acetyl-β-glucosaminidase OGA (O-GlcNAcase).[8]
- STX17 [STX17] (Syntaxin 17) is a component of the SNAP29-STX17 Qabc-SNARE. STX17 is a Qa-SNARE which localizes to the phagosome by binding the phagosome membrane marker LC3-II via its LC3-interacting region (LIR).[1] STX17 phosporylation on S2 is a key decision point for fusion to proceed; dephosphorylation relieves an inhibitory interaction with VPS33A and the phosphomimetic S2E mutant cannot form a SNARE bundle. [9] STX17 has two glycine-zipper motif transmembrane domains that are required for membrane insertion. [10]
- LAMP2 [LAMP2] (lysosomal associated membrane protein 2) is a component of the PHALY system. LAMP2 is an integral membrane protein. The LAMP-2A isoform has a lysosomal targeting signature sequence. The presence of LAMP2 is required for the binding of STX17.[11]
- IRGM [IRGM] (immunity related GTPase M) is a component of the PHALY system. IRGM forms a complex (an autophagosome recognition particle, ARP) with STX17 and the Atg8 homologues LC3 and GABARAPL1 that is responsible for phagosomal targeting of STX17. IRGM does not directly bind LC3. IRGM's active conformation is GTP bound. IRGM also has a role in autophagy initiation complexes, binding BECLIN1, ULK1 and ATG16L1, which is indpendent of STX17 binding.[10]
- RAB7 [RAB7A] (RAB7A, member RAS oncogene family) is a component of the PHALY system. RAB7 is a small GTPase that replaces RAB5 in maturing endosomes.[2] RAB7 localization to the membrane requires prenylation. [1] RAB7 is held in an inactive, soluble state by GDI (ARHGDIA), this interaction - and membrane recruitment - is released by the Mon1-CCZ1 complex.[12] RAB7 is activated on the phagosome by the Mon1-CCZ1 GEF complex which localizes to the phagosome by Atg8 homologue protein binding. [1] RAB7 can be localized to the lysosome by interaction with active lysosomal proton pumping vacuolar-type ATPase (V-ATPase). [13] Inits active conformation, RAB7 can bind RILP to promote minus-end directed microtubular transport of the lysosome towards the MTOC. [14]
- Mon1-CCZ1 (Mon1-CCZ1 GEF complex) is a component of the PHALY system. The Mon1-CCZ1 GEF complex is a GEF that activates Rab7 by exchanging bound GDP (inactive form) to GTP (active form).[1] It is found on both phagosome and lysosome membranes. [12]
- MON1A [MON1A] (MON1 homolog A, secretory trafficking associated) is a component of the Mon1-CCZ1 system. MON1A is one of two proteins in the heterooligomeric Mon1-CCZ1 GEF complex. [1]
- CCZ1 [CCZ1] (CCZ1 homolog, vacuolar protein trafficking and biogenesis associated) is a component of the Mon1-CCZ1 system. CCZ1 is one of two proteins in the heterooligomeric Mon1-CCZ1 GEF complex. [1]
- GABARAPL1 [GABARAPL1] (GABA type A receptor associated protein like 1) is a component of the PHALY system. GABARAPL1 is a ubiquitin-like modifier that binds to the Mon1-CCZ1 GEF complex and localizes it to the autophagosome.[1] As a yeast Atg8 family homologue (like LC3), GABARAPL1 is a downstream effector of the mTOR pathway. Lipidation by phosphatidylethanolamine causes it to be enriched in the autophagosome membrane, where it serves as a scaffold to recruit other proteins to the membrane. [15]
- EPG5 [EPG5] (ectopic P-granules autophagy protein 5 homolog) is a component of the PHALY. EPG5 is a RAB7 effector that localizes to both the phagosome and the lysosome by RAB7 binding, and binds to and enhances the SNAP29-STX17 Qabc-SNARE, thus facilitating VAMP8 binding. Binding of EPG5 shifts STX17 affinity from SNAP25 (without EPG5) to SNAP29 (with EPG5). EPG5 mutations cause Vici syndrome. [1]
Preparing the lysosome
- The lysosome is a component of the PHALY system. Fusion is the process of joining the outer phagosomal membrane of the phagosome with the membrane of a lysosome. [1]
- LAMP1 [LAMP1] (lysosomal associated membrane protein 1) is a component of the lysosome. LAMP1 is an integral membrane protein that is a lysosomal marker. [1]
- VAMP8 [VAMP8] (vesicle associated membrane protein) is a component of the SNAP29-STX17-VAMP8 SNARE-pin. VAMP8 is a component of the lysosome membrane. VAMP8 is the single v-SNARE (an R-SNARE) that is recruited into the Qabc-SNARE complex for fusion. This SNARE "pairing" forms a membrane-bridging trans-SNARE complex, the SNAP29-STX17-VAMP8 SNAREpin. Localization of VAMP8 to the lysosome is dependent on active RAB21. [1]RAB21 activation by SBF2 (a guanine nucleotide exchange factor for GTPases) under starvation conditions is an interface to nutrient sensing. [16]
- VAMP7 [VAMP7] (vesicle associated membrane protein 7) is a component of the lysosome. The v-SNARE VAMP7 has been considered to be characteristic of secretory lysosomes, i.e. not PHALY.[17] However, VAMP7 interacts with the VPS33A domain of HOPS complex and has been found to be able to initiate fusion with the SNAP29-STX17 Qabc-SNARE by FRET. [9] VAMP7 is part of the SNARE-pin with STX4 and SNAP23 in secretory lysosomes. [18] VAMP7 has an autoinhibitory longin domain that VAMP8 does not have.[19]
Tether Proteins and Docking
- The SNAP29-STX17-VAMP8 SNARE-pin is a component of the PHALY system. Binding of the SNAP29-STX17 Qabc-SNARE with VAMP8 forms the initial SNAP29-STX17-VAMP8 SNARE-pin which bridges the phagosome and the lysosome. This trans-SNARE complex (membrane bridging) is the fusion initiator which ultimately leads to "zippering" - membrane apposition, lipid mixing, pore-formation, membrane bilayer fusion, which results in the formation of the cis-SNARE complex (inserted into a single membrane), and bringing the cis-SNARE pin into a alpha-SNAP binding competent conformation. [1][20]
- ATG14 [ATG14] (Barkor, beclin 1-associated autophagy-related key regulator) is a component of the PIK3C3 complex. ATG14 stimulates BECN1 phosphorylation.[21] ATG14 regulates the PIK3C3 complex.[22]. ATG14 binds to and stabilizes the SNAP29-STX17 Qabc-SNARE on the membrane. [22] ATG14 homooligomers have a membrane-tethering function via their BATS domains (Barkor autophagosome targeting sequence), which is enhanced in membranes with low curvature and high PI(3)P. [22]
- The HOPS complex (homotypic fusion and protein sorting complex) is a component of the PHALY system. The HOPS complex is a hexa-heterooligomeric membrane tethering complex (MTC complex) that bridges two membranes containing RAB7 molecules in active conformation. It bridges vesicle and target membranes via its Rab7 binding domains and acts as a SNARE chaperone via a SNARE binding domain. It is the major "clamping" factor in SNARE mediated fusion. [2] The chaperoning function is crucial for topologically correct assembly of the trans-SNARE-pin, and prevention of reassembly of a cis-SNARE complex after SNARE disassembly by NSF-SNAP.[23] The BORC complex recruits the HOPS complex to the lysosomal membrane. The BORC complex functions by interacting with kinesins and determining the position of the lysosome by regulating the balance of (+)-end and (-)-end microtubular transport. One of its effectors is Arl8. The BORC complex comprises BLOC1S1, BLOC1S2, SNAPIN, KXD1, BORCS5, BORCS6, BORCS7, and BORCS8. [24]
- VPS16 [VPS16] (VPS16 core subunit of CORVET and HOPS complexes) is a component of the HOPS complex. VPS16 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex. It provides part of the SNARE-pin interaction interface. [2]
- VPS33A [VPS33A] (VPS33A core subunit of CORVET and HOPS complexes) is a component of the HOPS complex. VPS33 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex. It provides part of the SNARE-pin interaction interface. [2] VPS33A is an SM protein (Sec1/Munc18 protein) which stabilizes the nascent SNARE bundle by interacting with both v-SNARES and t-SNARES. [9] VPS33A interacts with a "closed form of STX17. [9]
- VPS18 [VPS18] (VPS18 core subunit of CORVET and HOPS complexes) is a component of the HOPS complex. VPS18 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex. It does not interact with the SNARE-pin. [2]
- VPS11 [VPS11] (VPS18 core subunit of CORVET and HOPS complexes) is a component of the HOPS complex. VPS11 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex. It does not interact with the SNARE-pin. [2]
- VPS41 [VPS41] (VPS41 subunit of HOPS complex) is a component of the HOPS complex. VPS41 (Vacuolar Protein Sorting) is one of the two subunits that are specific to the HOPS complex. It provides one of two Rab7 interaction interfaces. [2]
- VPS39 [VPS39] (VPS39 subunit of HOPS complex) is a component of the HOPS complex. VPS39 (Vacuolar Protein Sorting) is one of the two subunits that are specific to the HOPS complex. It provides one of two Rab7 interaction interfaces. [2]
Lipids
- lipid rafts are a component of the PHALY system. Lipid rafts are required for autophagic flux and play a role in the fusion event. [1]
- cholesterol is a component of lipid rafts. Cholesterol provides specific binding domains and increases membrane thickness and stiffness. [25]
- OSBPL1A [OSBPL1A] (Oxysterol binding protein like 1A) is a component of the PHALY system. OSBPL1A is a RAB7-GTP effector. In active form it interacts with cholesterol in lipd rafts through its ORD domain. This interaction recruits PLEKHM1 and through it the HOPS complex. In the absence of cholesterol, OSBPL1A interacts with VAPA via its FFAT domain.[1] This inhibits PLEKHM1 binding to RAB7, whereupon PLEKHM1 and RILP recruit HOPS complex.[14] The ER-bound VAPA protein can be bound by OSBPL1A via its FFAT domain.This interaction creates contact sites between the ER and the phagosome, which inhibits membrane tethers, microtubular transport, and stalls the fusion process.[1]
- The PIK3C3 complex (RUBCNL-UVRAG–BECN1,2–PIK3C3 complex) is a component of the PHALY system. The PIK3C3 complex creates phosphoinositide-3-phosphate at the phagosome-lysosome fusion site. It integrates a number of general (GPCR) signalling pathways. [25]
- PIK3C3 [PIK3C3] (phosphatidylinositol 3-kinase catalytic subunit type 3) is a component of the PIK3C3 complex. PIK3C3 (also: Vps34) produces phosphatidylinositol-3-phosphate (PI(3)P) from PI.[26]
- UVRAG [UVRAG] (UV radiation resistance associated) is a component of the PIK3C3 complex. UVRAG increases RAB7 concentration in maturing endosomes. It provides an interface to the MTORC1 complex: phosporylation of UVRAG by MTORC1 causes it to sequester with RUBCN, away from the HOPS complex.[1]
- NRBF2 [NRBF2] (nuclear receptor binding factor 2) is a component of the PIK3C3 complex. NRBF2 inhibts PIK3C3 activity and thus reduces PI(3)P levels.[27]
- PIK3R4 [PIK3R4] (phosphoinositide-3-kinase regulatory subunit 4) is a component of the PIK3C3 complex. PK3R4 (also called Vps15) is a protein kinase that regulates PIK3C3.[28]
- RUBCN [RUBCN] (rubicon autophagy regulator) is a component of the PIK3C3 complex. RUBCN sequesters UVRAG away from the HOPS complex.[1]
- RUBCNL [RUBCNL] (rubicon like autophagy enhancer) is a component of the PIK3C3 complex. RUBCNL (Pacer, protein associated with UVRAG as autophagy enhancer) releases UVRAG from RUBCN.[1]. RUBCNL also anchors PI3KC3 as well as HOPS to STX17. Its phosphorylation by mTORC1 integrates the mTOR pathway (active mTOR shuts down RUBCNL enhancement of the system); its acetylation by TIP60 integrates the GSK3-TIP60 pathway. [25]
- BECN1 [BECN1] (Beclin 1) is a component of the PIK3C3 complex. BECN1 (Beclin1) is a core component of the RUBCNL-UVRAG–BECN1,2–PIK3C3 complex.[1] Phosphorylation of BECN1 is stimulated by ATG14.[21]
- PLEKHM1 [PLEKHM1] (pleckstrin homology and RUN domain containing M1) is a component of the PIK3C3 complex. PLEKHM1 (Pleckstrin homology domain-containing family M member 1) is a multivalent adaptor that enhances HOPS complex / LC3 (Atg8) interactions in a RAB7 dependent way.[1]
- PI(3)P (phosphatidylinositol-3-phosphate) is a component of the PIK3C3 complex. PI(3)P interfaces with many signalling pathways. It is produced at the phagosome by PIK3C3 in the PIK3C3 complex, and is further phosphorylated to PI(3,5)P by PIKfyve after dissociation of PIK3C3.[26] Degradation of PI(3)P to PI on phagosomes reduces autophagy.[29]
- INPP5E [INPP5E] (inositol polyphosphate-5-phosphatase E) is a component of the PHALY system. INPP5E decreases lysosomal phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) and increases PI(3)P and is required for the fusion event. Both an excess and a reduction of the PI(3)P to PI(3,5)P2 ratio inhibits fusion.[29]
- The PIKFYVE complex (PIKFYVE complex) is a component of the PHALY system. The PIKFYVE complex controls PI(3,5)P2 levels and consists of PIKFYVE, FIG4, VAC14 (ArPIKfyve), and WIPI1. [30]
- PIKFYVE [PIKFYVE] (phosphoinositide kinase, FYVE-type zinc finger containing) is a component of the PIKFYVE complex. PIKFYVE phosphorylates PI(3)P to PI(3,5)P2. Both an excess and a reduction of the PI(3)P to PI(3,5)P2 ratio inhibits fusion. [29]
- PI(3,5)P2 (phosphatidylinositol-3,5-bisphosphate) is a component of the PIKFYVE complex. (PI(3,5)P2) is produced by the action of PIKFYVE on PI3P. [26] PI(3,5)P2 counteracts cortactin mediated actin filament stabilization on lysosomes; actin on the lysosome surface is required for fusion.[29]
Cytoskeleton
- The RAB7-RILP-dynein-dynactin complex (RAB7-RILP-dynein-dynactin complex) is a component of the PHALY system. The RAB7-RILP-dynein-dynactin complex is responsible for minus-end transport of lysosomes along microtubules towards the MTOC, where most lysosomes are located.[14]
- RILP [RILP] (Rab interacting lysosomal protein) is a component of the RAB7-RILP-dynein-dynactin complex. RILP associates with RAB7-GTP; the complex promotes dynein-dynactin association with the membrane and subsequent transport.[14]
- An F-actin network (actin filament network) is a component of the PHALY system. A cortactin dependent, remodelled, local filamentous actin network between the phagosome and the lysosome promotes fusion. [7]
- CTTN [CTTN] (cortactin) is a component of the F-actin network. CTTN is a branch-stabilizing interactor that remodels the F-actin network, which is a fusion requirement by recruiting the ARP2/3 complex to the fusion site.[7]
- HDAC6 [HDAC6] (histone deacetylase 6) is a component of the F-actin network. HDAC6, a ubiquitin binding deacylase, recruits CTTN to the autophagosome.[7]
- ARP2/3 complex (ARP2/3 complex) is a component of the F-actin network. The actin-nucleator ARP2/3 complex is a seven-subunit complex that can nucleate actin-filament branchpoints to establish a network. It stimulates the local assembly of an F-actin network for efficient fusion. It consists of ARP2, ARP3, ARPC1, ARPC2, ARPC3, ARPC4, and ARPC5. [7]
- F-actin (filamentous actin) is a component of the F-actin network. F-actin is a filament of ACTB (G-actin) monomers which is a scaffold for myosin-motors like the fusion-promoting MYO1C myosin to move on. [7]
- ACTB [ACTB] (actin beta) is a component of F-actin. The cytoplasmic, soluble globular G-actin polymerizes in to a filamentous form: F-actin. [7]
- MYO1C [MYO1C] (myosin 1C) is a component of the F-actin network. MYO1C is a monomeric class I myosin, which associates with cholesterol lipid rafts. It contains a PH domain that binds specifically to PI(4,5)P2. It is a "slow" motor, ideal for translocating heavy cargos, not tethering, and thus is able to move lipid rafts from storage compartments to their site of action. [7]
- PI(4,5)P2 (phosphatidylinositol-4,5-bisphosphate) is a component of the F-actin network. PI(4,5)P2 clusters at lipid rafts in a cholesterol and Ca2+ dependent fashion. These clusters bind the ARP2/3 complex. [31]
Ion regulation
- Ca2+ (calcium ion) is a component of the PHALY system. Ca2+ is required to stabilize the SNAREpin. [1]
- CACNA1A [CACNA1A] (Voltage-dependent P/Q-type calcium channel subunit alpha-1A) is a component of the PHALY system. The lysosome resident population of voltage-gated calcium channel CACNA1A is required for calcium efflux from the lysosome for the fusion event. [1]
Fusion mechanism
- A SNAREpin team is a component of the PHALY system. From three to six SNAP29-STX17-VAMP8 SNARE-pins form a mechanically coupled SNAREpin team in a rigid membrane (cholesterol lipid-raft) which accelerates fusion by four orders of magnitude. [32]
Disassembly and recycling
- The 20s supercomplex is a component of the PHALY system. The 20s supercomplex forms around the cis-SNARE complex, by binding two to four molecules of alpha-SNAP and an NSF homohexamer to the cis-SNARE-pin.[20]
- alpha-SNAP [NAPA] (NSF attachment protein alpha) is a component of the 20s supercomplex. Two to four molecules of cytoplasmic alpha-SNAP wrap around the cis-SNARE-pin to form a SNAP-SNARE subcomplex. [20]
- NSF [NSF] (N-ethylmaleimide sensitive factor) is a component of the 20s supercomplex. NSF is a cytoplasmic AAA+ ATPase that binds to the SNAP-SNARE subcomplex in an ATP-bound state. Subsequent ATP hydrolysis induces major conformational rearrangements of NSF that disassociate the SNARE-pin into individual constituent molecules. Nucleotide exchange presumably disassociates the remaining NSF-SNAP subcomplex, and allows the cycle to restart.[20]
- TBC1D2 [TBC1D2] (TBC1 domain family member 2A) is a component of the PHALY system. TBC1D2 (Armus) is a RAB7 specific GAP that activates hydrolysis of RAB7 bound GTP to GDP and thus catalyzes conversion of RAB7 to its inactive state. [12]
Not in the system
- NOT IN PHALY are genes that are not in the PHALY system. "NOT IN PHALY" genes are related to PHALY system components, but are demonstrably not part of PHALY. Genes that are NOT IN PHALY would need to be present in the cell even if the cell did not have a PHALY system. [33]
- VTI1B [VTI1B] (vesicle transport through interaction with t-SNAREs 1B) is NOT IN PHALY. The t-SNARE VTI1B may work in a pathway that is parallel to STX17, in pathogen containing autophagosomes or recycling endosomes.[1]
- STX6 [STX6] (syntaxin 6) is NOT IN PHALY. The t-SNARE STX6 is the interaction partner of VTI1B. [1]
- VAMP3 [VAMP3] (vesicle associated membrane protein 3) is NOT IN PHALY. The v-SNARE VAMP3 forms a SNAREpin with the VTI1B-STX6 complex.[1]
Since each of the facts above relates to only one component of the system, they can be imported into an Excel spreadsheet that supports the systems data model. Code is in text2tsv.R on the GitHub repository.
- Parallel and/or specialized pathways ...
- YKT6 may work in a pathway that is parallell to STX17.[1]
- Open questions ...
- Which phospatase dephosphorylates the STX17 N-terminal S2 in the VPS33A complex to switch STX17 into a fusion-competent interaction with HOPS complex? This appears to be a key checkpoint. [9]
System hierachy
The Excel spreadsheet has the system and subsystem hierarchy stored in a systemComponent join table. Code to represent this is a tree is in excel2tree.R on the GitHub repository.
--PHALY
|__20s supercomplex
|__alpha-SNAP
|__NSF
|__autophagosome
|__IAM
|__LC3-II
|__LC3
|__PE
|__OAM
|__Ca2+
|__CACNA1A
|__EPG5
|__F-actin network
|__ARP2/3 complex
|__CTTN
|__F-actin
|__ACTB
|__HDAC6
|__MYO1C
|__PI(4,5)P2
|__GABARAPL1
|__GlcNAC
|__HOPS complex
|__VPS11
|__VPS16
|__VPS18
|__VPS33A
|__VPS39
|__VPS41
|__INPP5E
|__IRGM
|__LAMP2
|__lipid rafts
|__cholesterol
|__lysosome
|__LAMP1
|__VAMP7
|__Mon1-CCZ1
|__CCZ1
|__MON1A
|__NOT IN PHALY
|__STX6
|__VAMP3
|__VTI1B
|__OSBPL1A
|__PIK3C3 complex
|__ATG14
|__BECN1
|__NRBF2
|__PI(3)P
|__PIK3C3
|__PIK3R4
|__PLEKHM1
|__RUBCN
|__RUBCNL
|__UVRAG
|__PIKFYVE complex
|__PI(3,5)P2
|__PIKFYVE
|__RAB7
|__RAB7-RILP-dynein-dynactin complex
|__RILP
|__SNAP29-STX17-VAMP8 SNARE-pin
|__SNAP29-STX17 Qabc-SNARE
|__SNAP29
|__STX17
|__VAMP8
|__SNAREpin team
|__TBC1D2
Genes added from direct annotation
It is useful to collect links to gene information via the HGNC resource. Code to generate Wikitext for a table with linked information is in text2annotationLinks.R on the GitHub repository.
This includes genes discovered because they have been annotated with a relationship to the system, in a database such as UniProt, NCBI-Protein or any of the three GO ontologies represented in GOA (GO annotations).
LC3-II
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| MAP1LC3A | microtubule associated protein 1 light chain 3 alpha | Q9H492 | ENSG00000101460 | uc002xaq.3 | 84557 | MAP1LC3A |
SNAP29-STX17 Qabc-SNARE
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| SNAP29 | synaptosome associated protein 29 | O95721 | ENSG00000099940 | uc011ahw.3 | 9342 | SNAP29 |
| STX17 | Syntaxin 17 | P56962 | ENSG00000136874 | uc004bal.5 | 55014 | STX17 |
PHALY
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| LAMP2 | lysosomal associated membrane protein 2 | P13473 | ENSG00000005893 | uc004ess.5 | 3920 | LAMP2 |
| IRGM | immunity related GTPase M | A1A4Y4 | ENSG00000237693 | uc010jhk.3 | 345611 | IRGM |
| RAB7A | RAB7A, member RAS oncogene family | P51149 | ENSG00000075785 | uc003eks.2 | 7879 | RAB7A |
| GABARAPL1 | GABA type A receptor associated protein like 1 | Q9H0R8 | ENSG00000139112 | uc001qxs.4 | 23710 | GABARAPL1 |
| EPG5 | ectopic P-granules autophagy protein 5 homolog | Q9HCE0 | ENSG00000152223 | uc002lbm.4 | 57724 | EPG5 |
| OSBPL1A | Oxysterol binding protein like 1A | Q9BXW6 | ENSG00000141447 | uc002kve.5 | 114876 | OSBPL1A |
| INPP5E | inositol polyphosphate-5-phosphatase E | Q9NRR6 | ENSG00000148384 | uc004cho.4 | 56623 | INPP5E |
| CACNA1A | Voltage-dependent P/Q-type calcium channel subunit alpha-1A | O00555 | ENSG00000141837 | uc002mwy.5 | 773 | CACNA1A |
| TBC1D2 | TBC1 domain family member 2A | Q9BYX2 | ENSG00000095383 | uc011lvb.3 | 55357 | TBC1D2 |
Mon1-CCZ1
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| MON1A | MON1 homolog A, secretory trafficking associated | Q86VX9 | ENSG00000164077 | uc003cxz.4 | 84315 | MON1A |
| CCZ1 | CCZ1 homolog, vacuolar protein trafficking and biogenesis associated | P86791 | ENSG00000122674 | uc003spf.4 | 51622 | CCZ1 |
lysosome
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| LAMP1 | lysosomal associated membrane protein 1 | P11279 | ENSG00000185896 | uc001vtm.2 | 3916 | LAMP1 |
| VAMP7 | vesicle associated membrane protein 7 | P51809 | ENSG00000124333 | uc004fxj.4 | 6845 | VAMP7 |
SNAP29-STX17-VAMP8 SNARE-pin
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| VAMP8 | vesicle associated membrane protein | Q9BV40 | ENSG00000118640 | uc002spt.5 | 8673 | VAMP8 |
PIK3C3 complex
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| ATG14 | Barkor, beclin 1-associated autophagy-related key regulator | Q6ZNE5 | ENSG00000126775 | uc001xbx.3 | 22863 | ATG14 |
| PIK3C3 | phosphatidylinositol 3-kinase catalytic subunit type 3 | Q8NEB9 | ENSG00000078142 | uc002lap.4 | 5289 | PIK3C3 |
| UVRAG | UV radiation resistance associated | Q9P2Y5 | ENSG00000198382 | uc001oxc.4 | 7405 | UVRAG |
| NRBF2 | nuclear receptor binding factor 2 | Q96F24 | ENSG00000148572 | uc001jmj.6 | 29982 | NRBF2 |
| PIK3R4 | phosphoinositide-3-kinase regulatory subunit 4 | Q99570 | ENSG00000196455 | uc003enj.4 | 30849 | PIK3R4 |
| RUBCN | rubicon autophagy regulator | Q92622 | ENSG00000145016 | NA | 9711 | RUBCN |
| RUBCNL | rubicon like autophagy enhancer | Q9H714 | ENSG00000102445 | uc001vbi.6 | 80183 | RUBCNL |
| BECN1 | Beclin 1 | Q14457 | ENSG00000126581 | uc002ibn.3 | 8678 | BECN1 |
| PLEKHM1 | pleckstrin homology and RUN domain containing M1 | Q9Y4G2 | ENSG00000225190 | uc002ija.4 | 9842 | PLEKHM1 |
HOPS complex
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| VPS16 | VPS16 core subunit of CORVET and HOPS complexes | Q9H269 | ENSG00000215305 | uc002whe.5 | 64601 | VPS16 |
| VPS33A | VPS33A core subunit of CORVET and HOPS complexes | Q96AX1 | ENSG00000139719 | uc001ucd.4 | 65082 | VPS33A |
| VPS18 | VPS18 core subunit of CORVET and HOPS complexes | Q9P253 | ENSG00000104142 | uc001zne.3 | 57617 | VPS18 |
| VPS11 | VPS18 core subunit of CORVET and HOPS complexes | Q9H270 | ENSG00000160695 | uc058iep.1 | 55823 | VPS11 |
| VPS41 | VPS41 subunit of HOPS complex | P49754 | ENSG00000006715 | uc003tgy.4 | 27072 | VPS41 |
| VPS39 | VPS39 subunit of HOPS complex | Q96JC1 | ENSG00000166887 | uc001zpc.4 | 23339 | VPS39 |
PIKFYVE complex
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| PIKFYVE | phosphoinositide kinase, FYVE-type zinc finger containing | Q9Y2I7 | ENSG00000115020 | uc002vcz.3 | 200576 | PIKFYVE |
RAB7-RILP-dynein-dynactin complex
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| RILP | Rab interacting lysosomal protein | Q96NA2 | ENSG00000167705 | uc002ftd.4 | 83547 | RILP |
F-actin network
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| CTTN | cortactin | Q14247 | ENSG00000085733 | uc001opw.5 | 2017 | CTTN |
| HDAC6 | histone deacetylase 6 | Q9UBN7 | ENSG00000094631 | uc004dks.2 | 10013 | HDAC6 |
| MYO1C | myosin 1C | O00159 | ENSG00000197879 | uc002fso.4 | 4641 | MYO1C |
F-actin
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| ACTB | actin beta | P60709 | ENSG00000075624 | uc003sot.5 | 60 | ACTB |
20s supercomplex
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| NAPA | NSF attachment protein alpha | P54920 | ENSG00000105402 | uc002pha.3 | 8775 | NAPA |
| NSF | N-ethylmaleimide sensitive factor | P46459 | ENSG00000073969 | uc002iku.4 | 4905 | NSF |
NOT IN PHALY
| Symbol | Name | UniProt | ensembl | UCSC | NCBI gene | AmiGO |
| VTI1B | vesicle transport through interaction with t-SNAREs 1B | Q9UEU0 | ENSG00000100568 | uc001xjt.4 | 10490 | VTI1B |
| STX6 | syntaxin 6 | O43752 | ENSG00000135823 | uc021pfr.3 | 10228 | STX6 |
| VAMP3 | vesicle associated membrane protein 3 | Q15836 | ENSG00000049245 | uc001aol.3 | 9341 | VAMP3 |
New gene facts:
When we collect new gene facts from databases, we need to be able to annotate the source of a fact, independent of whether it has a pmid. I created a template for this purpose: {{DB|<database>|<ID>}}. For now, it only lists the database, but it will be straightforward to have the template create an actual hyperlink to the record - for a select set of databases.text2tsv.R on the GitHub repository has been modified accordingly.
- ATG13 [ATG13] (autophagy related 13) is a component of the ULK1 complex. Mutations in LC3 can destroy ATG13 interactions and reduce autophagosome formation. (UniProt:Q9H492) ATG13 interacts with LC3 via its LIR (LC3-interacting region). [34] ATG13 is a member of the ULK1 complex which consists of ULK1 (formerly Atg1, unc-51 like autophagy activating kinase 1), ATG13, RB1CC1 (formerly Atg17 / FIP200, RB1 inducible coiled-coil 1), and ATG101. The Atg1 complex is activated under stress conditions by TORC1 and PKA. [34]
- The ULK1 complex is a component of the autophagosome. The ULK1 complex integrates a large number of cellular systems: the mTOR pathway, AMPK as an effector of AMP homeostasis, growth factor pathways acting via TIP60, genotoxic stress response via PPM1D, protein biosynthesis, and response attenuation during prolonged starvation via the cullin E3 ligase [35].
- MAP1LC3B [MAP1LC3B] (microtubule associated protein 1 light chain 3 beta) is a component of the LC3-II system. LC3 beta and gamma are functionally equivalent homologues of MAP1LC3A (UCSC:uc002xaq.3).
- MAP1LC3C [MAP1LC3C] (microtubule associated protein 1 light chain 3 gamma) is a component of the LC3-II system. LC3 beta and gamma are functionally equivalent homologues of MAP1LC3A (UCSC:uc002xaq.3).
- SNAP23 [SNAP23] (synaptosome associated protein 23) is NOT IN PHALY. SNAP23 is a t-SNARE for general membrane fusion. (UniProt:O00161)
- SNAP25 [SNAP25] (synaptosome associated protein 25) is NOT IN PHALY. SNAP25 is a t-SNARE involved in neurotransmitter release at the synapse. (UniProt:P61266)
- STX1A [STX1A] (syntaxin 1A) is NOT IN PHALY. STX1A is involved in hormone and neurotransmitter exocytosis. (UniProt:Q16623)
- STX1B [STX1B] (syntaxin 1B) is NOT IN PHALY. STX1B is involved in docking of synaptic vesicles at presynaptic active zones. (UniProt:Q16623)
- STX2 [STX2] (syntaxin 2) is NOT IN PHALY. STX2 is essential for epithelial morphogenesis. (UniProt:P32856)
- STX3 [STX3] (syntaxin 3) is NOT IN PHALY. STX3 is involved in docking of synaptic vesicles at presynaptic active zones. (UniProt:Q13277)
- STX4 [STX4] (syntaxin 4) is NOT IN PHALY. STX3 is a plasma mebrane t-SNARE that is involved in docking of transport vesicles. (UniProt:Q12846)
- STX6 [STX6] (syntaxin 6) is NOT IN PHALY. STX6 is essential involved in vesicle trafficking. (UniProt:O43752)
- STX7 [STX7] (syntaxin 7) is NOT IN PHALY. STX7 is involved in homotypic fusion of endocytic organelles. (UniProt:O15400)
- STX8 [STX8] (syntaxin 8) is NOT IN PHALY. STX8 is involved in early-secretory retrograde transport. (UniProt:Q9UNK0)
- STX10 [STX10] (syntaxin 10) is NOT IN PHALY. STX10 is involved in vesicular transport from the late endosomes to the trans-Golgi network. (UniProt:O60499)
- STX11 [STX11] (syntaxin 11) is NOT IN PHALY. STX11 acts between the late endosome and the trans-Golgi network. (UniProt:O75558)
- STX12 [STX12] (syntaxin 12) is NOT IN PHALY. STX12 acts between the late endosome and the trans-Golgi network. (UniProt:Q86Y82)
- STX16 [STX16] (syntaxin 16) is NOT IN PHALY. STX16 is a t-SNARE localized to the trans-Golgi network. (UniProt:O14662)
- STX18 [STX18] (syntaxin 18) is NOT IN PHALY. STX18 is a SNARE that is involved in Golgi-derived retrograde transport vesicles. (UniProt:Q9P2W9)
- STX19 [STX19] (syntaxin 19) is NOT IN PHALY. STX19 is involved in in endosomal trafficking of the epidermal growth factor receptor (EGFR). (UniProt:Q8N4C7)
Genes added from network annotation
This includes genes discovered in the network neighbourhood of system components, in a database like STRING or IntAct, or in pathways such as KEGG or Reactome.
- New gene facts
- SEC22B [SEC22B] (SEC22 homolog B, vesicle trafficking protein) is NOT IN PHALY. SEC22B is a high-confidence interactor in STRING, but it is a SNARE that is involved in retrograde transport from the Golgi to the ER. (UniProt:O75396)
- ATG4A [ATG4A] (autophagy related 4A cysteine peptidase) is a component of LC3-II. ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly. (KEGG:hsa04140)
- ATG4B [ATG4B] (autophagy related 4B cysteine peptidase) is a component of LC3-II. ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly. (KEGG:hsa04140)
- ATG4C [ATG4C] (autophagy related 4C cysteine peptidase) is a component of LC3-II. ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly. (KEGG:hsa04140)
- ATG4D [ATG4D] (autophagy related 4D cysteine peptidase) is a component of LC3-II. ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly. (KEGG:hsa04140)
Genes added from phenotype and behaviour
This includes genes annotated to a related phenotype in OMIM or the GWAS catalog.
Completing the Role Ontology
...
System Architecture
Sketch a system architecture.
System data
Format the system data for import into a system database. Details TBD.
JSON formatted data ... (click to expand)
{"parameter":[{
"ID":"dame.left-7a1-9ff7-2582-3b5a-b2094984aaa2",
"typeID":"chop.cool-4d3-8407-c9d2-0b38-5b069be72da0",
"value":"2.1.1"}],
"type":[{
"ID":"chop.cool-4d3-8407-c9d2-0b38-5b069be72da0",
"name":"DBversion",
"description":"Version of the systems database schema",
"validation":"^\\\\d+\\\\.\\\\d+\\\\.\\\\d+$"
},
{
"ID":"suit.tang-e32-67a8-4e32-1abb-c9e75ce1f2c1",
"name":"componentType",
"description":"Type of the component: atomic or composed",
"validation":"^(atomic)|(composed)$"
},
{
"ID":"cash.mare-ecf-5799-0f92-19ce-514bce3b5276",
"name":"molType",
"description":"The type of an atomic component",
"validation":"^(protein)|(RNA)|(lipid)|(metabolite)|(concept)|(other)$"
},
{
"ID":"wail.help-74b-baea-9c82-2adb-86e613d00ea2",
"name":"xRefUniProt",
"description":"Cross-reference UniProt KB ID",
"validation":"^UniProt:[0-9A-Z_]+$"
},
{
"ID":"coup.thus-73e-1292-0442-a97f-825167430bb5",
"name":"xRefENST",
"description":"Cross-reference Ensembl Transcript ID",
"validation":"^ENST:\\\\d+$"
},
{
"ID":"when.lute-8f9-b1e1-0ce2-1bd7-e861591869fb",
"name":"xRefENSE",
"description":"Cross-reference Ensembl Exon ID",
"validation":"^ENSE:\\\\d+$"
},
{
"ID":"gems.bird-2b4-0370-b092-da6f-a1ca41435587",
"name":"xRefENSG",
"description":"Cross-reference Ensembl Gene ID",
"validation":"^ENSG:\\\\d+$"
},
{
"ID":"time.barn-768-97b6-7962-684c-6cbd7d366979",
"name":"xRefENSP",
"description":"Cross-reference Ensembl Protein ID",
"validation":"^ENSP:\\\\d+$"
},
{
"ID":"warm.mate-bad-125f-a2a2-4b13-edefc11e1df0",
"name":"xRefPMID",
"description":"PubMed ID",
"validation":"^PMID:\\\\d+$"
},
{
"ID":"ribs.bolt-da3-5f2e-3ae2-5940-0b361a01abff",
"name":"xRefHGNC",
"description":"HGNC symbol",
"validation":"^HGNC:.+$"
},
{
"ID":"know.palm-46d-6143-b892-184d-0d3a420d31db",
"name":"xRefRefSeq",
"description":"RefSeq ID",
"validation":"^RefSeq:.._\\\\d+(\\\\.\\\\d+)?$"
},
{
"ID":"mall.case-f26-4df1-e892-9b70-380b2672a009",
"name":"xRefUCSC",
"description":"UCSC ID",
"validation":"^UCSC:uc[0-9a-z]+(\\\\.\\\\d+)?$"
},
{
"ID":"cork.span-2a0-f809-4182-49e2-2b3ce58c8710",
"name":"xRefNCBIgene",
"description":"NCBI gene ID",
"validation":"^gene:\\\\d+$"
},
{
"ID":"coat.curl-eab-3beb-e8e2-4aae-5e0f2235619c",
"name":"xRefOMIM",
"description":"OMIM ID",
"validation":"^OMIM:\\\\d+$"
},
{
"ID":"logs.nice-b29-62f6-c1d2-29f4-87bdb7e996ab",
"name":"xRefPDB",
"description":"PDB ID with optional chain separated with \".\" or \"_\"",
"validation":"^PDB:[0-9][0-9A-Za-z]{3}([\\\\._][0-9A-Za-z])?$"
},
{
"ID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"name":"genericNote",
"description":"Free-text annotation of an entity",
"validation":".+"
},
{
"ID":"pure.heal-c14-7e5a-65b2-ebbe-d822be29e26c",
"name":"historyNote",
"description":"Information on a superseded entity",
"validation":".+"
},
{
"ID":"seat.fate-150-9459-c452-3a15-98518ebcd683",
"name":"geneType",
"description":"Gene type as defined by HGNC",
"validation":"^(immunoglobulin)|(lncRNA)|(miRNA)|(protein)|(protocadherin)|(RNA, cluster)|(RNA, misc)|(rRNA)|(scRNA)|(snoRNA)|(snRNA)|(TCR)|(tRNA)|(vtRNA)|(Y RNA)$"}],
"system":[{
"ID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"code":"PHALY",
"name":"Phagosome/Lysosome Fusion",
"def":"control and process of phagosome-lysosome docking and fusion.",
"description":"Phagosome-lysosome fusion is a key step in autophagy, the process in which a lysosome - a package of acidic, lytic enzymes - fuses to a phagosome that has enveloped some cargo, which is targeted for degradation and recycling. The fusion is a highly regulated process, involving the preparation of lipid microdomains, positioning of vesicles by dynein motors that move along the cytoskeleton, membrane anchored SNAREs that provide core docking functions, the facilitating CORVET, HOPS and BORC complexes, and precise regulation of fusion initiation at the protein level, the lipid level, and through the ionic context which involves calcium-selective voltage gated channels."
},
{
"ID":"skin.toll-8ab-a5d2-1202-eaf0-807d897969f0",
"code":"autophagosome",
"name":"autophagosome",
"def":"a phagosome that sequesters cell-internal material",
"description":"Autophagosomes digest cell-internal material for general maintenance and under nutrient stress conditions."
},
{
"ID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"code":"LC3-II",
"name":"LC3-II",
"def":"lipidated LC3",
"description":"Prenylated LC3 is membrane resident on phagosomes and lysosomes."
},
{
"ID":"stab.base-4c7-4edd-25d2-4bcc-4086ee82a3a7",
"code":"SNAP29-STX17 Qabc-SNARE",
"name":"SNAP29-STX17 Qabc-SNARE",
"def":"t-snare bundle",
"description":"Contains three of the four required helices on the target side."
},
{
"ID":"verb.wade-b97-7197-f292-bb4c-56a9ab1a5add",
"code":"Mon1-CCZ1",
"name":"Mon1-CCZ1 GEF complex",
"def":"GEF for Rab7",
"description":"Activates Rab7 by catalyzing the GDP-GTP exchange"
},
{
"ID":"pump.trap-67f-eb22-c5d2-4a6e-68673cb5afa3",
"code":"ULK1 complex",
"name":"ULK1 complex",
"def":"heterotrimeric nutrient status sensor complex",
"description":"The ULK1 complex integrates a large number of cellular systems: the mTOR pathway, AMPK as an effector of AMP homeostasis, growth factor pathways acting via TIP60, genotoxic stress response via PPM1D, protein biosynthesis, and response attenuation during prolonged starvation via the cullin E3 ligase."
},
{
"ID":"pant.balm-93f-c169-d342-a8fe-d88028fb890a",
"code":"lysosome",
"name":"lysosome",
"def":"a vesicle that contains acidic, lytic enzymes",
"description":"The cargo of the lysosome is evacuated into the phagosome after fusion and digests the phagosome's contents."
},
{
"ID":"root.boys-359-f424-0572-4bcd-19d6ff64c741",
"code":"SNAP29-STX17-VAMP8 SNARE-pin",
"name":"SNAP29-STX17-VAMP8 SNARE-pin",
"def":"four-helix bundle SNARE",
"description":"The SNARE pin is a trans-SNARE complex as it juxtaposes the membranes to be fused, and turns into a cis-SNARE complex after fusion."
},
{
"ID":"tier.dash-c42-4c22-ad72-6b4b-bfe470e4455f",
"code":"STX17bound",
"name":"Syntaxin 17 (membrane bound)",
"def":"Syntaxin 17 (membrane bound)",
"description":"Membrane bound form of the Q-SNARE syntaxin 17"
},
{
"ID":"deed.mead-d34-551d-1f82-4a65-a5cc8b9e745a",
"code":"RAB7active",
"name":"RAB7 (active)",
"def":"active, membrane bound RAB7",
"description":"The active (GTP bound) form of RAB7, prenylated and membrane bound."
},
{
"ID":"give.need-232-ddd4-b342-aaf0-041e2b87062a",
"code":"HOPS complex",
"name":"homotypic fusion and protein sorting complex",
"def":"a hexa-heterooligomeric membrane tethering complex (MTC complex)",
"description":"A tethering complex that bridges phagosome and lysosome via RAB7 and functions as a SNAREpin chaperone."
},
{
"ID":"stab.belt-ff1-c9fc-dcc2-da67-06c44a8d9005",
"code":"lipid rafts",
"name":"lipid rafts",
"def":"cholesterol enriched membrane subdomains",
"description":"Lipid raft subdomains are cholesterol rich patches in membranes that are foci of signalling activity."
},
{
"ID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"code":"PIK3C3 complex",
"name":"RUBCNL-UVRAG–BECN1,2–PIK3C3 complex",
"def":"PIK3C3 complex",
"description":"The PIK3C3 kinase complex creates phosphoinositide-3-phosphate at the phagosome-lysosome fusion site."
},
{
"ID":"rock.tend-392-4e0a-ae42-5a96-6ffdead1df8f",
"code":"PIKFYVE complex",
"name":"PIKFYVE complex",
"def":"PI(3)P-5 kinase",
"description":"PIKFYVE phosphorylates PI(3)P to PI(3,5)P2. Both an excess and a reduction of the PI(3)P to PI(3,5)P2 ratio inhibits fusion"
},
{
"ID":"fear.lead-daa-76b4-b832-29f8-48cb624ea900",
"code":"RAB7-RILP-dynein-dynactin complex",
"name":"RAB7-RILP-dynein-dynactin complex",
"def":"vesicle-cytoskeleton connector",
"description":"The RAB7-RILP-dynein-dynactin complex is responsible for minus-end transport of lysosomes along microtubules towards the MTOC, where most lysosomes are located."
},
{
"ID":"lull.slab-be7-27e1-df72-2959-b42fa71d83b2",
"code":"F-actin network",
"name":"actin filament network",
"def":"actin network",
"description":"A network of branched actin filaments. The branch points are constituted from the Arp2/3 complex."
},
{
"ID":"doll.fawn-529-099c-6462-fb0b-9ec626eef6a5",
"code":"F-actin",
"name":"filamentous actin",
"def":"actin filament",
"description":"F-actin is a filament that forms by polymerization of actin monomers."
},
{
"ID":"owls.gear-304-36d5-ff92-1913-0de755710d35",
"code":"20s supercomplex",
"name":"20s supercomplex",
"def":"SNARE unfoldase",
"description":"The 20s supercomplex forms around the cis-SNARE complex, by binding two to four molecules of alpha-SNAP and an NSF homohexamer to the cis-SNARE-pin"
},
{
"ID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"code":"NOT IN PHALY",
"name":"NOT IN PHALY",
"def":"External to PHALY",
"description":"Genes that are members of PHALY component protein families and/or interact with PHALY components but are not themselves a part of PHALY."}],
"systemComponent":[{
"ID":"bolt.bell-e18-2ff6-4872-58a9-2a619b9b335f",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"brew.main-b5c-4427-17f2-0a43-015f4f9486d1",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"input",
"notes":"Fusion is the process of joining the outer phagosomal membrane of the autophagosome with the membrane of a lysosome (pmid:30115558). The outer autophagosomal membrane (OAM) and the inner autophagosomal membrane (IAM) form by membrane fission after closure of the phagosomal sac (pmid:27885029)."
},
{
"ID":"rain.jade-116-4fc4-1ef2-aa6e-4d14417ef305",
"systemID":"skin.toll-8ab-a5d2-1202-eaf0-807d897969f0",
"componentID":"mild.spur-d4f-1dd4-3642-7ad1-b93e4b1b6357",
"evidenceType":"TAS",
"evidenceSource":"27885029",
"role":"location",
"notes":"The outer autophagosomal membrane (OAM) forms by membrane fission after closure of the phagosomal sac (pmid:27885029)."
},
{
"ID":"fuel.wool-319-ef94-b862-eb48-2e57de0d2b47",
"systemID":"skin.toll-8ab-a5d2-1202-eaf0-807d897969f0",
"componentID":"wolf.make-780-f3f4-2e42-781c-e985b856e937",
"evidenceType":"TAS",
"evidenceSource":"27885029",
"role":"location",
"notes":"The inner autophagosomal membrane (IAM) forms by membrane fission after closure of the phagosomal sac (pmid:27885029). The IAM is degraded after lysosomal fusion and degradation of the IAM leads to immediate dissociation of STX17 from the cis-SNARE complex (pmid:27885029)."
},
{
"ID":"look.bred-be7-3c6c-4fe2-d9b7-828f6da0b6a4",
"systemID":"skin.toll-8ab-a5d2-1202-eaf0-807d897969f0",
"componentID":"calf.pane-5b5-127b-b3c2-b931-c79d4f9ded09",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"attachment",
"notes":"LC3-II (PE lipidated LC3) has been conjugated to the PE-enriched phagosome membrane and thereby serves as phagosome marker (pmid:30115558). The LC3-II conjugate is produced by the Atg8 conjugation system (Atg7, Atg3, Atg5, Atg12, Atg16L1/2, Atg4A/B/C/D) (pmid:28939950). Priming of the Atg8 conjugation system requires the Atg12 conjugating system (Atg12, Atg5, Atg7, Atg10) (pmid:28939950)."
},
{
"ID":"thin.meal-c1c-b556-3692-484b-e997f431469c",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"isle.gait-a82-1ff7-70b2-e810-fd2dd3c1c465",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"input",
"notes":"LC3 is a cytosolic protein that is conjugated to PE in the PE-enriched phagosome membrane in a stress response (pmid:30115558). Conjugation of LC3 and PE involves a ubiquitin-like conjugation system (pmid:27146966)."
},
{
"ID":"gown.sign-01f-02b0-3ba2-fbbb-e443703bf2a3",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"stag.teas-702-efa2-8822-b9ae-4f7a179fa9c9",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"input",
"notes":"PE is the lipid anchor of LC3 in the phagosome membrane (pmid:30115558)."
},
{
"ID":"mink.foam-636-25ad-11e2-2bcf-c68dec8d56a1",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"gust.mice-439-a46b-71c2-1900-8fc7723c07bb",
"evidenceType":"TAS",
"evidenceSource":"KEGG:hsa04140",
"role":"transform",
"notes":"ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly (KEGG:hsa04140}}"
},
{
"ID":"junk.each-74f-4b53-1612-3ae6-364828533626",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"core.crib-900-34d7-f252-1ae6-8593125d7d3f",
"evidenceType":"TAS",
"evidenceSource":"KEGG:hsa04140",
"role":"transform",
"notes":"ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly (KEGG:hsa04140}}"
},
{
"ID":"silk.sock-773-5629-56f2-6a05-8512e3eb0f10",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"gang.sent-e39-e249-c2d2-8b56-3af299a9b19b",
"evidenceType":"TAS",
"evidenceSource":"KEGG:hsa04140",
"role":"transform",
"notes":"ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly (KEGG:hsa04140}}"
},
{
"ID":"bark.mill-04f-e103-2702-3a18-e97e38c68767",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"bees.lone-c40-a77c-63b2-fafd-8486b1adf5c5",
"evidenceType":"TAS",
"evidenceSource":"KEGG:hsa04140",
"role":"transform",
"notes":"ATG4 proteins play a role in Atg8 protein PTM (priming for lipidation) and in delipidation for phagosome disassembly (KEGG:hsa04140}}"
},
{
"ID":"been.high-d45-3ffa-e232-b81e-afa6836a3187",
"systemID":"root.boys-359-f424-0572-4bcd-19d6ff64c741",
"componentID":"sign.bids-877-06f5-69c2-5a0f-ba6f1df4aca4",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"The SNAP29-STX17 Qabc-SNARE forms through the interaction of SNAP29 and STX17 in the phagosome membrane (pmid:30115558)."
},
{
"ID":"cult.moth-86e-fe5a-6c62-aaf7-8b864dad06ca",
"systemID":"stab.base-4c7-4edd-25d2-4bcc-4086ee82a3a7",
"componentID":"june.seas-24d-971c-f512-5826-33052e1e0ae3",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"SNAP29 is a Qa-SNARE (a t-SNARE) that localizes to the membrane in a lipidation independent manner through its STX17 interaction (pmid:30115558). SNAP29 is held in a non-interacting state by O-GlcNAcylation with GlcNAC (pmid:25419848)."
},
{
"ID":"span.page-579-16b1-ffd2-3a0d-e19723671a57",
"systemID":"stab.base-4c7-4edd-25d2-4bcc-4086ee82a3a7",
"componentID":"sets.lips-81d-d3de-fce2-caf7-5b4393b8ea4b",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"prevent",
"notes":"O-GlcNAcylation of SNAP29 in nutrient-sufficient conditions inhibits the association of the SNAP29-STX17 Qabc-SNARE (pmid:30115558). SNAP29 is O-GlcNAcylated by the promiscuous OGT (UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase) under nutrient-sufficient conditions and thus links the nutrient status sensing system with autophagic flux (pmid:25419848). SNAP29 has its O-GlcNAcetylation reversed by the N-acetyl-β-glucosaminidase OGA (O-GlcNAcase) (pmid:25419848)."
},
{
"ID":"laud.harm-f98-b9c9-11d2-0b43-fffda60215d6",
"systemID":"stab.base-4c7-4edd-25d2-4bcc-4086ee82a3a7",
"componentID":"mess.seal-1e4-158a-0e72-d8d5-ad944aaff957",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"Bound STX17 phosporylation on S2 is a key decision point for fusion to proceed; dephosphorylation relieves an inhibitory interaction with VPS33A and the phosphomimetic S2E mutant cannot form a SNARE bundle (pmid:30655294). STX17 has two glycine-zipper motif transmembrane domains that are required for membrane insertion (pmid:29420192)."
},
{
"ID":"prod.hemp-625-854e-4f92-f977-d044f65e6f66",
"systemID":"tier.dash-c42-4c22-ad72-6b4b-bfe470e4455f",
"componentID":"prod.bins-b67-7850-fca2-5bd0-9e214caf67c0",
"evidenceType":"TAS",
"evidenceSource":"29420192",
"role":"transform",
"notes":"STX17 is a Qa-SNARE which localizes to the phagosome by binding the phagosome membrane marker LC3-II via its LC3-interacting region (LIR) (pmid:30115558)."
},
{
"ID":"laud.chip-973-093d-4d12-b9f3-1248d590702d",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"pond.none-227-a5f6-2cc2-5ab0-f3d38b36d2a4",
"evidenceType":"TAS",
"evidenceSource":"27628032",
"role":"attachment",
"notes":"LAMP2 is an integral membrane protein. The LAMP-2A isoform has a lysosomal targeting signature sequence. The presence of LAMP2 is required for the binding of STX17 (pmid:27628032)."
},
{
"ID":"mink.chin-b99-c186-d312-fa24-f951db653fd4",
"systemID":"tier.dash-c42-4c22-ad72-6b4b-bfe470e4455f",
"componentID":"stir.lids-10a-c2ff-94f2-88fe-cc259492b58b",
"evidenceType":"TAS",
"evidenceSource":"29420192",
"role":"prepare",
"notes":"IRGM forms a complex (an autophagosome recognition particle, ARP) with STX17 and the Atg8 homologues LC3 and GABARAPL1 that is responsible for phagosomal targeting of STX17. IRGM does not directly bind LC3. IRGM's active conformation is GTP bound. IRGM also has a role in autophagy initiation complexes, binding BECLIN1, ULK1 and ATG16L1, which is independent of STX17 binding (pmid:29420192)."
},
{
"ID":"sigh.food-1f7-1717-03b2-3baf-10ea36c3b372",
"systemID":"skin.toll-8ab-a5d2-1202-eaf0-807d897969f0",
"componentID":"reel.gull-3a8-71d1-6212-e9ec-fb252ff13105",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"attachment",
"notes":"RAB7 is a small GTPase that replaces RAB5 in maturing endosomes (pmid:23645161). RAB7 localization to the membrane requires '''prenylation''' (pmid:30115558). RAB7 is held in an inactive, soluble state by GDI (ARHGDIA), this interaction - and membrane recruitment - is released by the Mon1-CCZ1 complex (pmid:30333976). RAB7 is activated on the phagosome by the Mon1-CCZ1 GEF complex which localizes to the phagosome by Atg8 homologue protein binding (pmid:30115558)."
},
{
"ID":"poll.hope-4b2-7d07-4e52-884f-c2340590b760",
"systemID":"pant.balm-93f-c169-d342-a8fe-d88028fb890a",
"componentID":"reel.gull-3a8-71d1-6212-e9ec-fb252ff13105",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"attachment",
"notes":"RAB7 can be localized to the lysosome by interaction with active lysosomal proton pumping vacuolar-type ATPase (V-ATPase) (pmid:30717974). In its active conformation, RAB7 can bind RILP to promote minus-end directed microtubular transport of the lysosome towards the MTOC (pmid:12944476)."
},
{
"ID":"keen.harm-699-f354-7dd2-6bf9-2df5c77c7076",
"systemID":"deed.mead-d34-551d-1f82-4a65-a5cc8b9e745a",
"componentID":"file.mime-4bf-2cfd-6e62-49f6-e5b1aefa698a",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"transform",
"notes":"The Mon1-CCZ1 GEF complex is a GEF that activates Rab7 by exchanging bound GDP (inactive form) to GTP (active form) (pmid:30115558). It is found on both phagosome and lysosome membranes (pmid:30333976)."
},
{
"ID":"knot.dash-94f-dc9b-8252-5978-d3e1cfe76787",
"systemID":"verb.wade-b97-7197-f292-bb4c-56a9ab1a5add",
"componentID":"sage.wood-e9a-5934-c9d2-6aab-dca28ae194ba",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"MON1A is one of two proteins in the heterooligomeric Mon1-CCZ1 GEF complex (pmid:30115558)."
},
{
"ID":"dash.fool-60e-3441-5382-9ab7-f6f6c48c0d1f",
"systemID":"verb.wade-b97-7197-f292-bb4c-56a9ab1a5add",
"componentID":"knit.hiss-047-86fb-e0f2-4ad9-ebafcff9269d",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"CCZ1 is one of two proteins in the heterooligomeric Mon1-CCZ1 GEF complex (pmid:30115558)."
},
{
"ID":"kept.wigs-cef-5af9-38a2-d940-cb36fe33efaa",
"systemID":"verb.wade-b97-7197-f292-bb4c-56a9ab1a5add",
"componentID":"webs.laud-453-0049-6f12-59d2-c55101ab383b",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"localize",
"notes":"GABARAPL1 is a ubiquitin-like modifier that binds to the Mon1-CCZ1 GEF complex and localizes it to the autophagosome (pmid:30115558). As a yeast Atg8 family homologue (like LC3), GABARAPL1 is a downstream effector of the mTOR pathway. Lipidation by phosphatidylethanolamine causes it to be enriched in the autophagosome membrane, where it serves as a scaffold to recruit other proteins to the membrane (pmid:30767700)."
},
{
"ID":"fail.camp-c66-f461-7382-a97d-c55140164ab9",
"systemID":"stab.base-4c7-4edd-25d2-4bcc-4086ee82a3a7",
"componentID":"wipe.bulk-ac5-33c4-a422-992b-15bbadd61344",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"enhance",
"notes":"EPG5 is a RAB7 effector that localizes to both the phagosome and the lysosome by RAB7 binding, and binds to and enhances the SNAP29-STX17 Qabc-SNARE, thus facilitating VAMP8 binding. Binding of EPG5 shifts STX17 affinity from SNAP25 (without EPG5) to SNAP29 (with EPG5). EPG5 mutations cause Vici syndrome (pmid:30115558)."
},
{
"ID":"vent.pane-a87-3e42-c542-b814-e5931d3c7e04",
"systemID":"pump.trap-67f-eb22-c5d2-4a6e-68673cb5afa3",
"componentID":"cage.back-649-14d7-fc22-58c5-21db5c35a36f",
"evidenceType":"TAS",
"evidenceSource":"24290141",
"role":"compose",
"notes":"Mutations in LC3 can destroy ATG13 interactions and reduce autophagosome formation (UniProt:Q9H492}} ATG13 interacts with LC3 via its LIR (LC3-interacting region) (pmid:24290141). ATG13 is a member of the ULK1 complex which consists of ULK1 (formerly Atg1, unc-51 like autophagy activating kinase 1), ATG13, RB1CC1 (formerly Atg17 / FIP200, RB1 inducible coiled-coil 1), and ATG101 (pmid:24290141)."
},
{
"ID":"spin.seam-e1a-ac2b-0332-69d5-3fc0ad54b43b",
"systemID":"skin.toll-8ab-a5d2-1202-eaf0-807d897969f0",
"componentID":"safe.bent-d50-883d-efc2-c947-81c5fc7d0fde",
"evidenceType":"TAS",
"evidenceSource":"24290141",
"role":"transform",
"notes":"The ULK1 complex consists of ULK1 (formerly Atg1, unc-51 like autophagy activating kinase 1), ATG13, RB1CC1 (formerly Atg17 / FIP200, RB1 inducible coiled-coil 1), and ATG101 (pmid:24290141). The ULK1 complex integrates a large number of cellular systems: the mTOR pathway, AMPK as an effector of AMP homeostasis, growth factor pathways acting via TIP60, genotoxic stress response via PPM1D, protein biosynthesis, and response attenuation during prolonged starvation via the cullin E3 ligase (pmid:29233870). The ULK1 complex is activated under stress conditions by TORC1 and PKA, it is thus a sensor of nutrient signals that activates autophagy (pmid:24290141)."
},
{
"ID":"barn.live-4fe-7833-6702-2acb-a43a8dfbd622",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"grog.cold-1e7-3859-a5e2-e917-bc2c0001dad2",
"evidenceType":"TAS",
"evidenceSource":"UCSC:uc002xaq.3",
"role":"input",
"notes":"LC3 beta and gamma are functionally equivalent homologues of MAP1LC3A (UCSC:uc002xaq.3}}."
},
{
"ID":"rich.make-64b-cd9e-97c2-585c-5da155a6e05d",
"systemID":"rail.skid-d47-002b-68c2-0847-551c3015d524",
"componentID":"wasp.deal-c61-5011-f0b2-6b87-b52b5f0c7b44",
"evidenceType":"TAS",
"evidenceSource":"UCSC:uc002xaq.3",
"role":"input",
"notes":"LC3 beta and gamma are functionally equivalent homologues of MAP1LC3A (UCSC:uc002xaq.3}}."
},
{
"ID":"harp.peek-c1b-2c35-54f2-e9a3-3c3330b418d1",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"flip.glad-f50-9299-e452-8978-24620d230bdc",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"input",
"notes":"Fusion is the process of joining the outer phagosomal membrane of the phagosome with the membrane of a lysosome (pmid:30115558)."
},
{
"ID":"fine.urge-c89-006b-e822-fa5a-8510ff8201ba",
"systemID":"pant.balm-93f-c169-d342-a8fe-d88028fb890a",
"componentID":"harp.thaw-f3d-f0e0-0622-0b87-ffc3a9aef0e2",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"attachment",
"notes":"LAMP1 is an integral membrane protein that is a lysosomal marker (pmid:30115558)."
},
{
"ID":"fast.clay-29f-fef8-d7c2-5b33-0b07d578df97",
"systemID":"root.boys-359-f424-0572-4bcd-19d6ff64c741",
"componentID":"toes.rods-c61-ca74-9b72-ca0c-96cfe2a82869",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"VAMP8 is a component of the lysosome membrane. VAMP8 is the single v-SNARE (an R-SNARE) that is recruited into the Qabc-SNARE complex for fusion. This SNARE \"pairing\" forms a membrane-bridging trans-SNARE complex, the SNAP29-STX17-VAMP8 SNAREpin. Localization of VAMP8 to the lysosome is dependent on active RAB21 (pmid:30115558). RAB21 activation by SBF2 (a guanine nucleotide exchange factor for GTPases) under starvation conditions is an interface to nutrient sensing (pmid:25648148)."
},
{
"ID":"roar.golf-fec-1919-dc82-1a9e-f976fa4d8011",
"systemID":"pant.balm-93f-c169-d342-a8fe-d88028fb890a",
"componentID":"cave.gull-aac-32e6-43a2-4975-812d31591dfa",
"evidenceType":"TAS",
"evidenceSource":"30240735",
"role":"compose",
"notes":"The v-SNARE VAMP7 has been considered to be characteristic of secretory lysosomes, i.e. not PHALY (pmid:30240735). However, VAMP7 interacts with the VPS33A domain of HOPS complex and has been found to be able to initiate fusion with the SNAP29-STX17 Qabc-SNARE by FRET (pmid:30655294). VAMP7 is part of the SNARE-pin with STX4 and SNAP23 in secretory lysosomes (pmid:28471021). VAMP7 has an autoinhibitory longin domain that VAMP8 does not have (pmid:26567219)."
},
{
"ID":"bids.mane-e66-dd34-a102-3889-a8b1f9bf3937",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"cone.nail-78b-a797-60a2-1a71-4b6973f63bfa",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"integrate",
"notes":"Binding of the SNAP29-STX17 Qabc-SNARE with VAMP8 forms the initial SNAP29-STX17-VAMP8 SNARE-pin which bridges the phagosome and the lysosome. This trans-SNARE complex (membrane bridging) is the fusion initiator which ultimately leads to \"zippering\" - membrane apposition, lipid mixing, pore-formation, membrane bilayer fusion, which results in the formation of the cis-SNARE complex (inserted into a single membrane), and bringing the cis-SNARE pin into a alpha-SNAP binding competent conformation (pmid:30115558). (pmid:25581794)."
},
{
"ID":"pint.free-64c-6e51-c3f2-69b5-11e5e9bae248",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"coat.dean-37b-7c8c-b692-581e-548b259def46",
"evidenceType":"TAS",
"evidenceSource":"23878393",
"role":"transform",
"notes":"ATG14 stimulates BECN1 phosphorylation (pmid:23878393). ATG14 regulates the PIK3C3 complex (pmid:25686604). ATG14 binds to and stabilizes the SNAP29-STX17 Qabc-SNARE on the membrane (pmid:25686604). ATG14 homooligomers have a membrane-tethering function via their BATS domains (Barkor autophagosome targeting sequence), which is enhanced in membranes with low curvature and high PI(3)P (pmid:25686604)."
},
{
"ID":"mode.ants-6b8-b09c-1b82-19fa-99f37c57d40c",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"hits.kite-345-8536-8682-8aa8-5da04b7a4ff8",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"locate",
"notes":"The HOPS complex is a hexa-heterooligomeric membrane tethering complex (MTC complex) that bridges two membranes containing RAB7 molecules in active conformation. It bridges vesicle and target membranes via its Rab7 binding domains and acts as a SNARE chaperone via a SNARE binding domain. It is the major \"clamping\" factor in SNARE mediated fusion (pmid:23645161). The chaperoning function is crucial for topologically correct assembly of the trans-SNARE-pin, and prevention of reassembly of a cis-SNARE complex after SNARE disassembly by NSF-SNAP (pmid:27301672). The BORC complex recruits the HOPS complex to the lysosomal membrane. The BORC complex functions by interacting with kinesins and determining the position of the lysosome by regulating the balance of (+)-end and (-)-end microtubular transport. One of its effectors is Arl8. The BORC complex comprises BLOC1S1, BLOC1S2, SNAPIN, KXD1, BORCS5, BORCS6, BORCS7, and BORCS8 (pmid:25898167)."
},
{
"ID":"drum.team-041-4d03-84e2-9a2d-35f6006313c8",
"systemID":"give.need-232-ddd4-b342-aaf0-041e2b87062a",
"componentID":"thru.fort-62b-f289-e932-b910-85ed42d6588b",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"compose",
"notes":"VPS16 provides part of the SNARE-pin interaction interface (pmid:23645161)."
},
{
"ID":"vise.docs-2fb-1c10-3bd2-f843-a04f72566d3e",
"systemID":"give.need-232-ddd4-b342-aaf0-041e2b87062a",
"componentID":"meet.high-e7f-67d6-4fa2-8ae0-1a5338569f23",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"compose",
"notes":"VPS33 provides part of the SNARE-pin interaction interface (pmid:23645161). VPS33A is an SM protein (Sec1/Munc18 protein) which stabilizes the nascent SNARE bundle by interacting with both v-SNARES and t-SNARES (pmid:30655294). VPS33A interacts with a \"closed form of STX17 (pmid:30655294)."
},
{
"ID":"knit.boar-c8c-bc2e-1a42-8aa6-6c55154fdf27",
"systemID":"give.need-232-ddd4-b342-aaf0-041e2b87062a",
"componentID":"wave.deem-5c8-d6ef-d192-5920-be2617eaa320",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"compose",
"notes":"VPS18 does not interact with the SNARE-pin (pmid:23645161)."
},
{
"ID":"whey.loop-081-3926-40c2-790c-a564baaaf74a",
"systemID":"give.need-232-ddd4-b342-aaf0-041e2b87062a",
"componentID":"hook.take-193-7c5c-b3b2-ba68-9b26aeb88e4f",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"compose",
"notes":"VPS11 does not interact with the SNARE-pin (pmid:23645161)."
},
{
"ID":"cure.newt-cb8-b736-af42-8b73-e6c3a794928e",
"systemID":"give.need-232-ddd4-b342-aaf0-041e2b87062a",
"componentID":"soak.have-ead-d696-4292-8a1c-c6bd6dab86d8",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"compose",
"notes":"VPS41 provides one of two Rab7 interaction interfaces (pmid:23645161)."
},
{
"ID":"page.thug-7a0-e1af-6542-a95a-588432e9a518",
"systemID":"give.need-232-ddd4-b342-aaf0-041e2b87062a",
"componentID":"leap.code-1e6-a805-9e82-88da-dc3c7a9b0939",
"evidenceType":"TAS",
"evidenceSource":"23645161",
"role":"compose",
"notes":"VPS39 provides one of two Rab7 interaction interfaces (pmid:23645161)."
},
{
"ID":"than.bulk-206-8a9a-52b2-59b7-356f2320f718",
"systemID":"skin.toll-8ab-a5d2-1202-eaf0-807d897969f0",
"componentID":"runs.ploy-bfc-c454-45d2-dbdf-7adcde1b3758",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"location",
"notes":"Lipid rafts are required for autophagic flux and play a role in the fusion event (pmid:30115558)."
},
{
"ID":"vent.soup-82d-42d7-e732-f9e6-90bc94db939e",
"systemID":"pant.balm-93f-c169-d342-a8fe-d88028fb890a",
"componentID":"runs.ploy-bfc-c454-45d2-dbdf-7adcde1b3758",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"location",
"notes":"Lipid rafts are required for autophagic flux and play a role in the fusion event (pmid:30115558)."
},
{
"ID":"says.feud-17f-0711-e972-08cf-861f38cc0ac3",
"systemID":"stab.belt-ff1-c9fc-dcc2-da67-06c44a8d9005",
"componentID":"thin.brew-e9c-fa09-4462-290d-797babf87586",
"evidenceType":"TAS",
"evidenceSource":"30747526",
"role":"compose",
"notes":"Cholesterol provides specific binding domains and increases membrane thickness and stiffness (pmid:30747526)."
},
{
"ID":"tick.rift-bf4-6d24-0f62-ea0c-eaaa877f8500",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"hugs.stub-676-67ee-a7d2-a8a3-cff6ea1a60f0",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"localize",
"notes":"OSBPL1A is a RAB7-GTP effector. In active form it interacts with cholesterol in lipd rafts through its ORD domain. This interaction recruits PLEKHM1 and through it the HOPS complex. In the absence of cholesterol, OSBPL1A interacts with VAPA via its FFAT domain (pmid:30115558). This inhibits PLEKHM1 binding to RAB7, whereupon PLEKHM1 and RILP recruit HOPS complex (pmid:27283760). The ER-bound VAPA protein can be bound by OSBPL1A via its FFAT domain.This interaction creates contact sites between the ER and the phagosome, which inhibits membrane tethers, microtubular transport, and stalls the fusion process (pmid:30115558)."
},
{
"ID":"eats.milk-5b2-be25-3eb2-6810-55b1d69bfd3f",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"pays.leap-4b7-35ce-5aa2-aa05-c93631a533c4",
"evidenceType":"TAS",
"evidenceSource":"30704899",
"role":"integrate",
"notes":"The PIK3C3 complex creates phosphoinositide-3-phosphate at the phagosome-lysosome fusion site. It integrates a number of general (GPCR) signalling pathways (pmid:30704899)."
},
{
"ID":"gene.fled-c70-8829-0302-5b08-b703cf387b8a",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"word.film-fb9-d0d6-4ef2-6982-9e517633e41f",
"evidenceType":"TAS",
"evidenceSource":"29089378",
"role":"compose",
"notes":"PIK3C3 (also: Vps34) produces phosphatidylinositol-3-phosphate (PI(3)P) from PI (pmid:29089378)."
},
{
"ID":"beds.twig-f65-6c43-bff2-5b21-4bdb173c9aeb",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"note.shin-85d-2c29-5bf2-abcd-506232a14b8d",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"UVRAG increases RAB7 concentration in maturing endosomes. It provides an interface to the MTORC1 complex: phosporylation of UVRAG by MTORC1 causes it to sequester with RUBCN, away from the HOPS complex (pmid:30115558)."
},
{
"ID":"same.prey-2cc-fac9-ab82-78e2-ce291c12a486",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"walk.next-cee-da9e-3802-5970-688389626c3b",
"evidenceType":"TAS",
"evidenceSource":"25086043",
"role":"compose",
"notes":"NRBF2 inhibts PIK3C3 activity and thus reduces PI(3)P levels (pmid:25086043)."
},
{
"ID":"runs.odds-b47-3aaa-45a2-dbcd-63b8c85b8f1c",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"pies.slip-a4b-a984-7722-cac5-86ccadd86a1d",
"evidenceType":"TAS",
"evidenceSource":"18326940",
"role":"compose",
"notes":"PK3R4 (also called Vps15) is a protein kinase that regulates PIK3C3 (pmid:18326940)."
},
{
"ID":"debt.robe-aae-1366-13b2-0849-b6435d0d81cc",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"bore.vain-cbb-04d5-bfc2-c8cf-bf964f752fc8",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"RUBCN sequesters UVRAG away from the HOPS complex (pmid:30115558)."
},
{
"ID":"gait.slap-41a-2472-1e42-9ae0-67023bef60fa",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"toll.fell-e8c-8b22-1082-7bb0-8ce383ade0ea",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"RUBCNL (Pacer, protein associated with UVRAG as autophagy enhancer) releases UVRAG from RUBCN (pmid:30115558). . RUBCNL also anchors PI3KC3 as well as HOPS to STX17. Its phosphorylation by mTORC1 integrates the mTOR pathway (active mTOR shuts down RUBCNL enhancement of the system); its acetylation by TIP60 integrates the GSK3-TIP60 pathway (pmid:30704899)."
},
{
"ID":"ties.barn-f4c-7730-3322-5bf9-0aa47eea15cc",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"swim.doom-f60-b92a-d092-4ae6-535916d7ac3a",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"BECN1 (Beclin1) is a core component of the RUBCNL-UVRAG–BECN1,2–PIK3C3 complex (pmid:30115558). Phosphorylation of BECN1 is stimulated by ATG14 (pmid:23878393)."
},
{
"ID":"caps.chat-62a-f55b-a6f2-ab48-ace5fc572d05",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"hens.sang-044-e09f-a102-a9b6-63a33e9f8e2e",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"PLEKHM1 (Pleckstrin homology domain-containing family M member 1) is a multivalent adaptor that enhances HOPS complex / LC3 (Atg8) interactions in a RAB7 dependent way (pmid:30115558)."
},
{
"ID":"oaks.next-f5a-15d1-0e52-7b5b-6c43f86a688b",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"wake.stop-028-e0d1-dbd2-39dc-24af09d1894d",
"evidenceType":"TAS",
"evidenceSource":"29089378",
"role":"output",
"notes":"PI(3)P interfaces with many signalling pathways. It is produced at the phagosome by PIK3C3 in the PIK3C3 complex, and is further phosphorylated to PI(3,5)P by PIKfyve after dissociation of PIK3C3 (pmid:29089378). Degradation of PI(3)P to PI on phagosomes reduces autophagy (pmid:27340123)."
},
{
"ID":"bone.gage-392-a7ea-a602-1834-b69324a5b3ac",
"systemID":"stub.sigh-a47-a0a5-b7c2-c832-1a2bcec5753f",
"componentID":"give.toad-b23-1014-6712-9b1f-d76f1abe9aae",
"evidenceType":"TAS",
"evidenceSource":"29089378",
"role":"input",
"notes":"PI is phosphorylated at the phagosome toPI(3)P by PIK3C3 in the PIK3C3 complex. Degradation of PI(3)P to PI on phagosomes reduces autophagy (pmid:27340123)."
},
{
"ID":"care.myth-84b-8f47-aae2-7840-957ac3c7d9c5",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"mole.heel-580-b3cf-ec42-7b8e-33a0504c5b5b",
"evidenceType":"TAS",
"evidenceSource":"27340123",
"role":"set",
"notes":"INPP5E decreases lysosomal phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) and increases PI(3)P and is required for the fusion event. Both an excess and a reduction of the PI(3)P to PI(3,5)P2 ratio inhibits fusion (pmid:27340123)."
},
{
"ID":"fill.howl-085-d857-3782-c998-bd6d034f3ef6",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"cell.raft-364-a39c-6362-abe2-797a0dcf9276",
"evidenceType":"TAS",
"evidenceSource":"28302928",
"role":"set",
"notes":"The PIKFYVE complex controls PI(3,5)P2 levels and consists of PIKFYVE, FIG4, VAC14 (ArPIKfyve), and WIPI1 (pmid:28302928)."
},
{
"ID":"else.pine-dbf-9c59-a5c2-2a78-86671c651f80",
"systemID":"rock.tend-392-4e0a-ae42-5a96-6ffdead1df8f",
"componentID":"turn.need-3fe-2bc6-1312-7923-3dd66b25371f",
"evidenceType":"TAS",
"evidenceSource":"27340123",
"role":"set",
"notes":"PIKFYVE phosphorylates PI(3)P to PI(3,5)P2. Both an excess and a reduction of the PI(3)P to PI(3,5)P2 ratio inhibits fusion (pmid:27340123)."
},
{
"ID":"lamp.swap-83a-e9dd-fb22-28f8-a7ceefcedae1",
"systemID":"rock.tend-392-4e0a-ae42-5a96-6ffdead1df8f",
"componentID":"team.flex-712-75fe-9e02-a999-9aded07a24cb",
"evidenceType":"TAS",
"evidenceSource":"29089378",
"role":"output",
"notes":"(PI(3,5)P2) is produced by the action of PIKFYVE on PI3P (pmid:29089378). PI(3,5)P2 counteracts cortactin mediated actin filament stabilization on lysosomes; actin on the lysosome surface is required for fusion (pmid:27340123)."
},
{
"ID":"sock.peak-cc3-f027-59d2-4902-d552d6d0337d",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"rang.torn-8e1-1ff3-4bc2-7962-137d0835f74a",
"evidenceType":"TAS",
"evidenceSource":"27283760",
"role":"locate",
"notes":"The RAB7-RILP-dynein-dynactin complex is responsible for minus-end transport of lysosomes along microtubules towards the MTOC, where most lysosomes are located (pmid:27283760)."
},
{
"ID":"damp.snip-5f5-cdcf-4d42-7b0b-61870a6323ec",
"systemID":"fear.lead-daa-76b4-b832-29f8-48cb624ea900",
"componentID":"prep.used-3a0-36b4-c5e2-eb17-df6d634b8277",
"evidenceType":"TAS",
"evidenceSource":"12944476",
"role":"compose",
"notes":"RILP associates with RAB7-GTP; the complex promotes dynein-dynactin association with the membrane and subsequent transport (pmid:12944476)."
},
{
"ID":"trip.mill-1c2-4adf-30d2-db77-b8850e3f60b2",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"head.maps-b51-5222-9e02-9a83-317947b0f03d",
"evidenceType":"TAS",
"evidenceSource":"27146966",
"role":"location",
"notes":"A cortactin dependent, remodelled, local filamentous actin network between the phagosome and the lysosome promotes fusion (pmid:27146966)."
},
{
"ID":"cast.warm-784-8365-e2c2-2866-2ece261021a2",
"systemID":"lull.slab-be7-27e1-df72-2959-b42fa71d83b2",
"componentID":"pair.flaw-425-1875-ed02-dbf9-277bb04e893e",
"evidenceType":"TAS",
"evidenceSource":"27146966",
"role":"compose",
"notes":"CTTN is a branch-stabilizing interactor that remodels the F-actin network, which is a fusion requirement by recruiting the ARP2/3 complex to the fusion site (pmid:27146966)."
},
{
"ID":"cake.shoe-56c-e70f-78b2-0871-9521ca8461a4",
"systemID":"lull.slab-be7-27e1-df72-2959-b42fa71d83b2",
"componentID":"grub.luck-728-4d5c-02c2-d923-9232b842de40",
"evidenceType":"TAS",
"evidenceSource":"27146966",
"role":"locate",
"notes":"HDAC6, a ubiquitin binding deacylase, recruits CTTN to the autophagosome (pmid:27146966)."
},
{
"ID":"gaps.miss-5a3-38cc-f972-6a1a-6c9c7c35cfc8",
"systemID":"lull.slab-be7-27e1-df72-2959-b42fa71d83b2",
"componentID":"mens.lack-87b-d4b4-a3a2-9aa7-752a2b098f4d",
"evidenceType":"TAS",
"evidenceSource":"27146966",
"role":"compose",
"notes":"The actin-nucleator ARP2/3 complex is a seven-subunit complex that can nucleate actin-filament branchpoints to establish a network. It stimulates the local assembly of an F-actin network for efficient fusion. It consists of ARP2, ARP3, ARPC1, ARPC2, ARPC3, ARPC4, and ARPC5 (pmid:27146966)."
},
{
"ID":"ears.wide-f7a-3e36-0262-eae4-41de1c812c2d",
"systemID":"lull.slab-be7-27e1-df72-2959-b42fa71d83b2",
"componentID":"stub.tile-0e5-909f-6942-8bce-c38696a2d4a0",
"evidenceType":"TAS",
"evidenceSource":"27146966",
"role":"compose",
"notes":"F-actin is a filament of ACTB (G-actin) monomers which is a scaffold for myosin-motors like the fusion-promoting MYO1C myosin to move on (pmid:27146966)."
},
{
"ID":"worm.raft-766-5b1e-b712-a894-a0fa20c4fa92",
"systemID":"doll.fawn-529-099c-6462-fb0b-9ec626eef6a5",
"componentID":"dare.bans-b1a-0594-e102-badf-d8b373f431b7",
"evidenceType":"TAS",
"evidenceSource":"27146966",
"role":"compose",
"notes":"The cytoplasmic, soluble globular G-actin polymerizes in to a filamentous form: F-actin (pmid:27146966)."
},
{
"ID":"hunt.sand-38e-9c6d-20c2-2b59-041e13f8d9d7",
"systemID":"lull.slab-be7-27e1-df72-2959-b42fa71d83b2",
"componentID":"thin.pier-e5c-032e-e622-8ae1-e5d46bfe78ac",
"evidenceType":"TAS",
"evidenceSource":"27146966",
"role":"locate",
"notes":"MYO1C is a monomeric class I myosin, which associates with cholesterol lipid rafts. It contains a PH domain that binds specifically to PI(4,5)P2. It is a \"slow\" motor, ideal for translocating heavy cargos, not tethering, and thus is able to move lipid rafts from storage compartments to their site of action (pmid:27146966)."
},
{
"ID":"tell.take-da1-2b0e-1082-09f0-5eaf46754632",
"systemID":"lull.slab-be7-27e1-df72-2959-b42fa71d83b2",
"componentID":"fees.yarn-c6a-179d-4952-4918-5bccc8330297",
"evidenceType":"TAS",
"evidenceSource":"30692198",
"role":"attachment",
"notes":"PI(4,5)P2 clusters at lipid rafts in a cholesterol and Ca2+ dependent fashion. These clusters bind the ARP2/3 complex (pmid:30692198)."
},
{
"ID":"foul.lung-999-21ee-1602-1865-4ee3b84d42d0",
"systemID":"root.boys-359-f424-0572-4bcd-19d6ff64c741",
"componentID":"some.fled-330-0e4c-4362-fa2b-6ca9b43a1075",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"compose",
"notes":"Ca2+ is required to stabilize the SNAREpin (pmid:30115558)."
},
{
"ID":"mail.wren-16e-362d-d2d2-6bf3-61ca2d3517aa",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"bead.hour-c64-88c8-a1f2-993f-723284239b26",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"input",
"notes":"The lysosome resident population of voltage-gated calcium channel CACNA1A is required for calcium efflux from the lysosome for the fusion event (pmid:30115558)."
},
{
"ID":"rust.peat-984-967c-1192-d9b0-b6d090102ed4",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"pour.bugs-bdb-0abf-e892-e885-9c20e949c76a",
"evidenceType":"TAS",
"evidenceSource":"30700546",
"role":"transmit",
"notes":"From three to six SNAP29-STX17-VAMP8 SNARE-pins form a mechanically coupled SNAREpin team in a rigid membrane (cholesterol lipid-raft) which accelerates fusion by four orders of magnitude (pmid:30700546)."
},
{
"ID":"says.lull-fe4-2d88-d912-a98c-57cd3eaedadf",
"systemID":"root.boys-359-f424-0572-4bcd-19d6ff64c741",
"componentID":"vise.hash-35f-ead8-99d2-2947-9ab8f6561c13",
"evidenceType":"TAS",
"evidenceSource":"25581794",
"role":"decompose",
"notes":"The 20s supercomplex forms around the cis-SNARE complex, by binding two to four molecules of alpha-SNAP and an NSF homohexamer to the cis-SNARE-pin (pmid:25581794)."
},
{
"ID":"dove.rust-d29-c0b5-e052-580e-987747841192",
"systemID":"owls.gear-304-36d5-ff92-1913-0de755710d35",
"componentID":"suit.leak-8bf-2889-5ca2-f9ed-9e91f9b1d069",
"evidenceType":"TAS",
"evidenceSource":"25581794",
"role":"compose",
"notes":"Two to four molecules of cytoplasmic alpha-SNAP wrap around the cis-SNARE-pin to form a SNAP-SNARE subcomplex (pmid:25581794)."
},
{
"ID":"duct.arch-552-5563-80a2-a86f-0ba12ac82042",
"systemID":"owls.gear-304-36d5-ff92-1913-0de755710d35",
"componentID":"chip.lend-5df-d630-ef52-dbc7-59aaa7c71494",
"evidenceType":"TAS",
"evidenceSource":"25581794",
"role":"compose",
"notes":"NSF is a cytoplasmic AAA+ ATPase that binds to the SNAP-SNARE subcomplex in an ATP-bound state. Subsequent ATP hydrolysis induces major conformational rearrangements of NSF that disassociate the SNARE-pin into individual constituent molecules. Nucleotide exchange presumably disassociates the remaining NSF-SNAP subcomplex, and allows the cycle to restart (pmid:25581794)."
},
{
"ID":"laud.flop-bab-1dfa-db02-1b60-9da8e3bd25f8",
"systemID":"deed.mead-d34-551d-1f82-4a65-a5cc8b9e745a",
"componentID":"lawn.gong-987-4d18-70e2-692b-749431f5af5b",
"evidenceType":"TAS",
"evidenceSource":"30333976",
"role":"inactivate",
"notes":"TBC1D2 (Armus) is a RAB7 specific GAP that activates hydrolysis of RAB7 bound GTP to GDP and thus catalyzes conversion of RAB7 to its inactive state (pmid:30333976)."
},
{
"ID":"cove.pray-ec1-5534-3992-39a2-c772a8808378",
"systemID":"sled.bets-e87-946c-4ed2-8b81-171d499085d6",
"componentID":"lamb.hull-aec-9fb5-f932-99b1-e85db79ee1b9",
"evidenceType":"TAS",
"evidenceSource":"Unpublished",
"role":"external",
"notes":"NOT IN PHALY genes are related to PHALY system components, but are demonstrably not part of PHALY (Unpublished}}"
},
{
"ID":"lend.cool-80b-a405-3312-6819-cef6da560b74",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"muse.jobs-c24-59c5-a362-f8f0-f6f8e26f964d",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"external",
"notes":"The t-SNARE VTI1B may work in a pathway that is parallel to STX17, in pathogen containing autophagosomes or recycling endosomes (pmid:30115558)."
},
{
"ID":"laud.dent-a0e-307a-dcf2-6a5e-04a14c070e67",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"cubs.rice-546-05aa-1d52-9a4f-09507cdf1d5f",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"external",
"notes":"The t-SNARE STX6 is the interaction partner of VTI1B (pmid:30115558)."
},
{
"ID":"rice.saws-b50-ebc2-5e92-9baf-6c6608493d11",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"drum.fell-823-079f-8062-5a36-452d68d5c393",
"evidenceType":"TAS",
"evidenceSource":"30115558",
"role":"external",
"notes":"The v-SNARE VAMP3 forms a SNAREpin with the VTI1B-STX6 complex (pmid:30115558)."
},
{
"ID":"kick.beam-399-f82c-9642-2b34-6fe7940831a2",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"owls.turn-c44-bab3-d632-28ff-69b1af2d31a3",
"evidenceType":"TAS",
"evidenceSource":"UniProt:O00161",
"role":"external",
"notes":"SNAP23 is a t-SNARE for general membrane fusion (UniProt:O00161}}"
},
{
"ID":"coal.thru-f1c-3841-1f42-49c7-0d6c2c2e2f2b",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"gall.fees-89f-514f-32d2-cb4d-9c7ceadaad1e",
"evidenceType":"TAS",
"evidenceSource":"UniProt:P61266",
"role":"external",
"notes":"SNAP25 is a t-SNARE involved in neurotransmitter release at the synapse (UniProt:P61266}}"
},
{
"ID":"yawn.sits-ab8-02f0-a3c2-8ae0-5ff796b54c97",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"bout.join-dc8-84fe-4c12-fb23-3f7afee2f8d0",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q16623",
"role":"external",
"notes":"STX1A is involved in hormone and neurotransmitter exocytosis (UniProt:Q16623}}"
},
{
"ID":"veil.size-110-f429-1f12-98b8-36a8b28a978a",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"clan.mash-3f2-bd57-f332-2b3b-cf1f03acf736",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q16623",
"role":"external",
"notes":"STX1B is involved in docking of synaptic vesicles at presynaptic active zones (UniProt:Q16623}}"
},
{
"ID":"term.time-fd8-d7d9-3782-0aa4-fc612a22b0ab",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"trim.male-e1d-1978-2b02-eafb-6b5591cb1d79",
"evidenceType":"TAS",
"evidenceSource":"UniProt:P32856",
"role":"external",
"notes":"STX2 is essential for epithelial morphogenesis (UniProt:P32856}}"
},
{
"ID":"flee.file-ee7-fb71-b6e2-b895-5d5234454ca7",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"path.tuck-be6-c1fd-5df2-1b55-0ccae7f59b0e",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q13277",
"role":"external",
"notes":"STX3 is involved in docking of synaptic vesicles at presynaptic active zones (UniProt:Q13277}}"
},
{
"ID":"cook.tend-286-439f-dc22-e8b2-aff44cb2f068",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"frog.nose-1d4-17a3-a1f2-6a2b-a535ba4ba518",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q12846",
"role":"external",
"notes":"STX3 is a plasma mebrane t-SNARE that is involved in docking of transport vesicles (UniProt:Q12846}}"
},
{
"ID":"grab.thus-b6e-c3b5-b612-28c5-3b8bc9482c07",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"cubs.rice-546-05aa-1d52-9a4f-09507cdf1d5f",
"evidenceType":"TAS",
"evidenceSource":"UniProt:O43752",
"role":"external",
"notes":"STX6 is essential involved in vesicle trafficking (UniProt:O43752}}"
},
{
"ID":"junk.ride-9ae-3d32-4002-a87d-3cff81bdedc5",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"rugs.raid-f56-17e3-0dc2-39c8-7acc151dfe7d",
"evidenceType":"TAS",
"evidenceSource":"UniProt:O15400",
"role":"external",
"notes":"STX7 is involved in homotypic fusion of endocytic organelles (UniProt:O15400}}"
},
{
"ID":"tale.wind-340-cbff-20d2-5acb-28412f0d472f",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"mash.yawn-fa3-12cb-9ad2-797f-db540783b7db",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q9UNK0",
"role":"external",
"notes":"STX8 is involved in early-secretory retrograde transport (UniProt:Q9UNK0}}"
},
{
"ID":"kind.math-7ce-0d76-58c2-488f-cbd2da03575e",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"from.snip-78c-4de9-dc02-abcf-c80a48851217",
"evidenceType":"TAS",
"evidenceSource":"UniProt:O60499",
"role":"external",
"notes":"STX10 is involved in vesicular transport from the late endosomes to the trans-Golgi network (UniProt:O60499}}"
},
{
"ID":"fawn.bulk-5aa-78a1-e992-884b-54830177189f",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"dome.calf-07a-cff6-ebc2-4899-6c254e4ab7bb",
"evidenceType":"TAS",
"evidenceSource":"UniProt:O75558",
"role":"external",
"notes":"STX11 acts between the late endosome and the trans-Golgi network (UniProt:O75558}}"
},
{
"ID":"cans.salt-119-4cfe-1292-aad1-7ac77c865d26",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"golf.park-56f-a5db-f132-0b74-34887708dee8",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q86Y82",
"role":"external",
"notes":"STX12 acts between the late endosome and the trans-Golgi network (UniProt:Q86Y82}}"
},
{
"ID":"goat.fact-735-d9db-0bf2-89ea-fa951cf6ae42",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"asks.drop-bfd-7feb-4d92-4b5f-8fffd85deea1",
"evidenceType":"TAS",
"evidenceSource":"UniProt:O14662",
"role":"external",
"notes":"STX16 is a t-SNARE localized to the trans-Golgi network (UniProt:O14662}}"
},
{
"ID":"yard.plus-ae5-f711-3c12-f81d-620053869882",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"cook.mile-c28-9ec7-a532-dba9-e4ea0774f8fd",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q9P2W9",
"role":"external",
"notes":"STX18 is a SNARE that is involved in Golgi-derived retrograde transport vesicles (UniProt:Q9P2W9}}"
},
{
"ID":"cone.tent-73a-bc7f-cf82-984e-30d02923d9ed",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"deep.sift-2e6-95e6-fa32-aa4b-bf00f510f54e",
"evidenceType":"TAS",
"evidenceSource":"UniProt:Q8N4C7",
"role":"external",
"notes":"STX19 is involved in in endosomal trafficking of the epidermal growth factor receptor (EGFR) (UniProt:Q8N4C7}}"
},
{
"ID":"deed.each-bbc-d18b-3d42-1a8f-ec4d4a7c7617",
"systemID":"rash.stir-fe0-1bb3-ae62-28f8-4e6cab1e945b",
"componentID":"watt.bind-8c1-752c-61c2-3ab3-7734965c6621",
"evidenceType":"TAS",
"evidenceSource":"UniProt:O75396",
"role":"external",
"notes":"SEC22B is a high-confidence interactor in STRING, but it is a SNARE that is involved in retrograde transport from the Golgi to the ER (UniProt:O75396}}"}],
"component":[{
"ID":"brew.main-b5c-4427-17f2-0a43-015f4f9486d1",
"code":"autophagosome",
"componentType":"composed"
},
{
"ID":"mild.spur-d4f-1dd4-3642-7ad1-b93e4b1b6357",
"code":"OAM",
"componentType":"atomic"
},
{
"ID":"wolf.make-780-f3f4-2e42-781c-e985b856e937",
"code":"IAM",
"componentType":"atomic"
},
{
"ID":"calf.pane-5b5-127b-b3c2-b931-c79d4f9ded09",
"code":"LC3-II",
"componentType":"composed"
},
{
"ID":"isle.gait-a82-1ff7-70b2-e810-fd2dd3c1c465",
"code":"LC3",
"componentType":"atomic"
},
{
"ID":"stag.teas-702-efa2-8822-b9ae-4f7a179fa9c9",
"code":"PE",
"componentType":"atomic"
},
{
"ID":"sign.bids-877-06f5-69c2-5a0f-ba6f1df4aca4",
"code":"SNAP29-STX17 Qabc-SNARE",
"componentType":"composed"
},
{
"ID":"june.seas-24d-971c-f512-5826-33052e1e0ae3",
"code":"SNAP29",
"componentType":"atomic"
},
{
"ID":"sets.lips-81d-d3de-fce2-caf7-5b4393b8ea4b",
"code":"GlcNAC",
"componentType":"atomic"
},
{
"ID":"mess.seal-1e4-158a-0e72-d8d5-ad944aaff957",
"code":"STX17bound",
"componentType":"composed"
},
{
"ID":"prod.bins-b67-7850-fca2-5bd0-9e214caf67c0",
"code":"STX17free",
"componentType":"atomic"
},
{
"ID":"pond.none-227-a5f6-2cc2-5ab0-f3d38b36d2a4",
"code":"LAMP2",
"componentType":"atomic"
},
{
"ID":"stir.lids-10a-c2ff-94f2-88fe-cc259492b58b",
"code":"IRGM",
"componentType":"atomic"
},
{
"ID":"reel.gull-3a8-71d1-6212-e9ec-fb252ff13105",
"code":"RAB7active",
"componentType":"composed"
},
{
"ID":"doll.club-c20-7172-ace2-5a9c-145dd3ff8e41",
"code":"RAB7",
"componentType":"atomic"
},
{
"ID":"file.mime-4bf-2cfd-6e62-49f6-e5b1aefa698a",
"code":"Mon1-CCZ1",
"componentType":"composed"
},
{
"ID":"sage.wood-e9a-5934-c9d2-6aab-dca28ae194ba",
"code":"MON1A",
"componentType":"atomic"
},
{
"ID":"knit.hiss-047-86fb-e0f2-4ad9-ebafcff9269d",
"code":"CCZ1",
"componentType":"atomic"
},
{
"ID":"webs.laud-453-0049-6f12-59d2-c55101ab383b",
"code":"GABARAPL1",
"componentType":"atomic"
},
{
"ID":"wipe.bulk-ac5-33c4-a422-992b-15bbadd61344",
"code":"EPG5",
"componentType":"atomic"
},
{
"ID":"cage.back-649-14d7-fc22-58c5-21db5c35a36f",
"code":"ATG13",
"componentType":"atomic"
},
{
"ID":"safe.bent-d50-883d-efc2-c947-81c5fc7d0fde",
"code":"ULK1 complex",
"componentType":"composed"
},
{
"ID":"grog.cold-1e7-3859-a5e2-e917-bc2c0001dad2",
"code":"MAP1LC3B",
"componentType":"atomic"
},
{
"ID":"wasp.deal-c61-5011-f0b2-6b87-b52b5f0c7b44",
"code":"MAP1LC3C",
"componentType":"atomic"
},
{
"ID":"flip.glad-f50-9299-e452-8978-24620d230bdc",
"code":"lysosome",
"componentType":"composed"
},
{
"ID":"harp.thaw-f3d-f0e0-0622-0b87-ffc3a9aef0e2",
"code":"LAMP1",
"componentType":"atomic"
},
{
"ID":"toes.rods-c61-ca74-9b72-ca0c-96cfe2a82869",
"code":"VAMP8",
"componentType":"atomic"
},
{
"ID":"cave.gull-aac-32e6-43a2-4975-812d31591dfa",
"code":"VAMP7",
"componentType":"atomic"
},
{
"ID":"cone.nail-78b-a797-60a2-1a71-4b6973f63bfa",
"code":"SNAP29-STX17-VAMP8 SNARE-pin",
"componentType":"composed"
},
{
"ID":"coat.dean-37b-7c8c-b692-581e-548b259def46",
"code":"ATG14",
"componentType":"atomic"
},
{
"ID":"hits.kite-345-8536-8682-8aa8-5da04b7a4ff8",
"code":"HOPS complex",
"componentType":"composed"
},
{
"ID":"thru.fort-62b-f289-e932-b910-85ed42d6588b",
"code":"VPS16",
"componentType":"atomic"
},
{
"ID":"meet.high-e7f-67d6-4fa2-8ae0-1a5338569f23",
"code":"VPS33A",
"componentType":"atomic"
},
{
"ID":"wave.deem-5c8-d6ef-d192-5920-be2617eaa320",
"code":"VPS18",
"componentType":"atomic"
},
{
"ID":"hook.take-193-7c5c-b3b2-ba68-9b26aeb88e4f",
"code":"VPS11",
"componentType":"atomic"
},
{
"ID":"soak.have-ead-d696-4292-8a1c-c6bd6dab86d8",
"code":"VPS41",
"componentType":"atomic"
},
{
"ID":"leap.code-1e6-a805-9e82-88da-dc3c7a9b0939",
"code":"VPS39",
"componentType":"atomic"
},
{
"ID":"runs.ploy-bfc-c454-45d2-dbdf-7adcde1b3758",
"code":"lipid rafts",
"componentType":"composed"
},
{
"ID":"thin.brew-e9c-fa09-4462-290d-797babf87586",
"code":"cholesterol",
"componentType":"atomic"
},
{
"ID":"hugs.stub-676-67ee-a7d2-a8a3-cff6ea1a60f0",
"code":"OSBPL1A",
"componentType":"atomic"
},
{
"ID":"pays.leap-4b7-35ce-5aa2-aa05-c93631a533c4",
"code":"PIK3C3 complex",
"componentType":"composed"
},
{
"ID":"word.film-fb9-d0d6-4ef2-6982-9e517633e41f",
"code":"PIK3C3",
"componentType":"atomic"
},
{
"ID":"note.shin-85d-2c29-5bf2-abcd-506232a14b8d",
"code":"UVRAG",
"componentType":"atomic"
},
{
"ID":"walk.next-cee-da9e-3802-5970-688389626c3b",
"code":"NRBF2",
"componentType":"atomic"
},
{
"ID":"pies.slip-a4b-a984-7722-cac5-86ccadd86a1d",
"code":"PIK3R4",
"componentType":"atomic"
},
{
"ID":"bore.vain-cbb-04d5-bfc2-c8cf-bf964f752fc8",
"code":"RUBCN",
"componentType":"atomic"
},
{
"ID":"toll.fell-e8c-8b22-1082-7bb0-8ce383ade0ea",
"code":"RUBCNL",
"componentType":"atomic"
},
{
"ID":"swim.doom-f60-b92a-d092-4ae6-535916d7ac3a",
"code":"BECN1",
"componentType":"atomic"
},
{
"ID":"hens.sang-044-e09f-a102-a9b6-63a33e9f8e2e",
"code":"PLEKHM1",
"componentType":"atomic"
},
{
"ID":"give.toad-b23-1014-6712-9b1f-d76f1abe9aae",
"code":"PI",
"componentType":"atomic"
},
{
"ID":"wake.stop-028-e0d1-dbd2-39dc-24af09d1894d",
"code":"PI(3)P",
"componentType":"atomic"
},
{
"ID":"team.flex-712-75fe-9e02-a999-9aded07a24cb",
"code":"PI(3,5)P2",
"componentType":"atomic"
},
{
"ID":"mole.heel-580-b3cf-ec42-7b8e-33a0504c5b5b",
"code":"INPP5E",
"componentType":"atomic"
},
{
"ID":"cell.raft-364-a39c-6362-abe2-797a0dcf9276",
"code":"PIKFYVE complex",
"componentType":"composed"
},
{
"ID":"turn.need-3fe-2bc6-1312-7923-3dd66b25371f",
"code":"PIKFYVE",
"componentType":"atomic"
},
{
"ID":"rang.torn-8e1-1ff3-4bc2-7962-137d0835f74a",
"code":"RAB7-RILP-dynein-dynactin complex",
"componentType":"composed"
},
{
"ID":"prep.used-3a0-36b4-c5e2-eb17-df6d634b8277",
"code":"RILP",
"componentType":"atomic"
},
{
"ID":"head.maps-b51-5222-9e02-9a83-317947b0f03d",
"code":"F-actin network",
"componentType":"composed"
},
{
"ID":"pair.flaw-425-1875-ed02-dbf9-277bb04e893e",
"code":"CTTN",
"componentType":"atomic"
},
{
"ID":"grub.luck-728-4d5c-02c2-d923-9232b842de40",
"code":"HDAC6",
"componentType":"atomic"
},
{
"ID":"mens.lack-87b-d4b4-a3a2-9aa7-752a2b098f4d",
"code":"ARP2/3 complex",
"componentType":"composed"
},
{
"ID":"stub.tile-0e5-909f-6942-8bce-c38696a2d4a0",
"code":"F-actin",
"componentType":"composed"
},
{
"ID":"dare.bans-b1a-0594-e102-badf-d8b373f431b7",
"code":"ACTB",
"componentType":"atomic"
},
{
"ID":"thin.pier-e5c-032e-e622-8ae1-e5d46bfe78ac",
"code":"MYO1C",
"componentType":"atomic"
},
{
"ID":"fees.yarn-c6a-179d-4952-4918-5bccc8330297",
"code":"PI(4,5)P2",
"componentType":"atomic"
},
{
"ID":"some.fled-330-0e4c-4362-fa2b-6ca9b43a1075",
"code":"Ca2+",
"componentType":"atomic"
},
{
"ID":"bead.hour-c64-88c8-a1f2-993f-723284239b26",
"code":"CACNA1A",
"componentType":"atomic"
},
{
"ID":"pour.bugs-bdb-0abf-e892-e885-9c20e949c76a",
"code":"SNAREpin team",
"componentType":"atomic"
},
{
"ID":"vise.hash-35f-ead8-99d2-2947-9ab8f6561c13",
"code":"20s supercomplex",
"componentType":"composed"
},
{
"ID":"suit.leak-8bf-2889-5ca2-f9ed-9e91f9b1d069",
"code":"alpha-SNAP",
"componentType":"atomic"
},
{
"ID":"chip.lend-5df-d630-ef52-dbc7-59aaa7c71494",
"code":"NSF",
"componentType":"atomic"
},
{
"ID":"lawn.gong-987-4d18-70e2-692b-749431f5af5b",
"code":"TBC1D2",
"componentType":"atomic"
},
{
"ID":"lamb.hull-aec-9fb5-f932-99b1-e85db79ee1b9",
"code":"NOT IN PHALY",
"componentType":"composed"
},
{
"ID":"muse.jobs-c24-59c5-a362-f8f0-f6f8e26f964d",
"code":"VTI1B",
"componentType":"atomic"
},
{
"ID":"drum.fell-823-079f-8062-5a36-452d68d5c393",
"code":"VAMP3",
"componentType":"atomic"
},
{
"ID":"bout.join-dc8-84fe-4c12-fb23-3f7afee2f8d0",
"code":"STX1A",
"componentType":"atomic"
},
{
"ID":"clan.mash-3f2-bd57-f332-2b3b-cf1f03acf736",
"code":"STX1B",
"componentType":"atomic"
},
{
"ID":"trim.male-e1d-1978-2b02-eafb-6b5591cb1d79",
"code":"STX2",
"componentType":"atomic"
},
{
"ID":"path.tuck-be6-c1fd-5df2-1b55-0ccae7f59b0e",
"code":"STX3",
"componentType":"atomic"
},
{
"ID":"frog.nose-1d4-17a3-a1f2-6a2b-a535ba4ba518",
"code":"STX4",
"componentType":"atomic"
},
{
"ID":"cubs.rice-546-05aa-1d52-9a4f-09507cdf1d5f",
"code":"STX6",
"componentType":"atomic"
},
{
"ID":"rugs.raid-f56-17e3-0dc2-39c8-7acc151dfe7d",
"code":"STX7",
"componentType":"atomic"
},
{
"ID":"mash.yawn-fa3-12cb-9ad2-797f-db540783b7db",
"code":"STX8",
"componentType":"atomic"
},
{
"ID":"from.snip-78c-4de9-dc02-abcf-c80a48851217",
"code":"STX10",
"componentType":"atomic"
},
{
"ID":"dome.calf-07a-cff6-ebc2-4899-6c254e4ab7bb",
"code":"STX11",
"componentType":"atomic"
},
{
"ID":"golf.park-56f-a5db-f132-0b74-34887708dee8",
"code":"STX12",
"componentType":"atomic"
},
{
"ID":"asks.drop-bfd-7feb-4d92-4b5f-8fffd85deea1",
"code":"STX16",
"componentType":"atomic"
},
{
"ID":"cook.mile-c28-9ec7-a532-dba9-e4ea0774f8fd",
"code":"STX18",
"componentType":"atomic"
},
{
"ID":"deep.sift-2e6-95e6-fa32-aa4b-bf00f510f54e",
"code":"STX19",
"componentType":"atomic"
},
{
"ID":"watt.bind-8c1-752c-61c2-3ab3-7734965c6621",
"code":"SEC22B",
"componentType":"atomic"
},
{
"ID":"gust.mice-439-a46b-71c2-1900-8fc7723c07bb",
"code":"ATG4A",
"componentType":"atomic"
},
{
"ID":"core.crib-900-34d7-f252-1ae6-8593125d7d3f",
"code":"ATG4B",
"componentType":"atomic"
},
{
"ID":"gang.sent-e39-e249-c2d2-8b56-3af299a9b19b",
"code":"ATG4C",
"componentType":"atomic"
},
{
"ID":"bees.lone-c40-a77c-63b2-fafd-8486b1adf5c5",
"code":"ATG4D",
"componentType":"atomic"
},
{
"ID":"owls.turn-c44-bab3-d632-28ff-69b1af2d31a3",
"code":"SNAP23",
"componentType":"atomic"
},
{
"ID":"gall.fees-89f-514f-32d2-cb4d-9c7ceadaad1e",
"code":"SNAP25",
"componentType":"atomic"}],
"componentMolecule":[{
"ID":"kept.bird-61b-f929-0002-fb73-b2b2da1717b2",
"componentID":"mild.spur-d4f-1dd4-3642-7ad1-b93e4b1b6357",
"moleculeID":"clue.loft-b60-17e4-7aa2-89d5-ac55f8bfc171"
},
{
"ID":"crow.gave-9e2-b810-bee2-d8a7-00328a00f172",
"componentID":"wolf.make-780-f3f4-2e42-781c-e985b856e937",
"moleculeID":"tubs.full-d6c-5c3e-41e2-795a-de6f234b7427"
},
{
"ID":"boot.lean-873-49ce-e5c2-abb1-bbc15dc5fbf9",
"componentID":"isle.gait-a82-1ff7-70b2-e810-fd2dd3c1c465",
"moleculeID":"burn.sure-463-5a92-b302-487b-8d543ee5ca04"
},
{
"ID":"wins.hurt-e91-828e-ea22-ab5e-45f402cb04c9",
"componentID":"stag.teas-702-efa2-8822-b9ae-4f7a179fa9c9",
"moleculeID":"cell.lynx-b0d-c61f-4442-ca86-6056262af5a0"
},
{
"ID":"pine.comb-191-572c-9042-6bd6-fc121951811c",
"componentID":"june.seas-24d-971c-f512-5826-33052e1e0ae3",
"moleculeID":"ease.same-d07-2587-0d12-da22-eb93b7b06519"
},
{
"ID":"part.sung-bce-68c1-fe42-2a5b-9ee7afc00810",
"componentID":"sets.lips-81d-d3de-fce2-caf7-5b4393b8ea4b",
"moleculeID":"fate.swat-de5-9446-e762-a817-4a170cb5d0e1"
},
{
"ID":"them.deal-1c4-d677-45c2-5ac1-413cb2ffd31d",
"componentID":"prod.bins-b67-7850-fca2-5bd0-9e214caf67c0",
"moleculeID":"jars.sets-770-a690-8312-d97c-37d3db9eb2da"
},
{
"ID":"fang.crow-a0f-6582-6d82-395b-1e031f87f6b0",
"componentID":"pond.none-227-a5f6-2cc2-5ab0-f3d38b36d2a4",
"moleculeID":"hugs.dump-30b-76ba-ef62-890d-91d464502746"
},
{
"ID":"love.robe-c2d-d813-9e52-0a7c-0eeb95c4a007",
"componentID":"stir.lids-10a-c2ff-94f2-88fe-cc259492b58b",
"moleculeID":"cats.eggs-597-296a-edb2-4bd5-b7f7cc29ff4b"
},
{
"ID":"pane.lame-923-be64-d942-482b-d8c2f40095b0",
"componentID":"doll.club-c20-7172-ace2-5a9c-145dd3ff8e41",
"moleculeID":"grip.paid-df1-2830-0552-3839-4ddd6865e612"
},
{
"ID":"mere.mugs-ac9-1e52-e422-2828-a16f0ae847b4",
"componentID":"sage.wood-e9a-5934-c9d2-6aab-dca28ae194ba",
"moleculeID":"grim.folk-88b-3e9e-72a2-c869-e9c2b85c8cf5"
},
{
"ID":"muse.gain-9f3-5f06-25a2-9aed-a7c435d5cf56",
"componentID":"knit.hiss-047-86fb-e0f2-4ad9-ebafcff9269d",
"moleculeID":"mole.hymn-e25-7fe0-6682-0b8f-1bdbae452dae"
},
{
"ID":"weak.seat-219-2e74-93a2-e8bc-705502968d42",
"componentID":"webs.laud-453-0049-6f12-59d2-c55101ab383b",
"moleculeID":"spam.mead-0da-02ca-d3b2-088b-f8c7ad1ab4b6"
},
{
"ID":"nice.flip-bb0-e206-bd32-1bee-52f5de38a6ba",
"componentID":"wipe.bulk-ac5-33c4-a422-992b-15bbadd61344",
"moleculeID":"rule.side-174-a533-f6b2-2be4-202534c93d0f"
},
{
"ID":"tort.quit-072-cf49-5032-8ae3-4c54fbc18223",
"componentID":"cage.back-649-14d7-fc22-58c5-21db5c35a36f",
"moleculeID":"does.mane-3fb-4cff-b792-4921-adc353d5e286"
},
{
"ID":"owls.down-90a-9d1e-2dc2-4a3d-ea90996a13b1",
"componentID":"grog.cold-1e7-3859-a5e2-e917-bc2c0001dad2",
"moleculeID":"pear.calf-f86-1558-b222-bb6d-9e90aeefe6a4"
},
{
"ID":"till.vase-789-7763-27f2-9862-3b3ad07e365b",
"componentID":"wasp.deal-c61-5011-f0b2-6b87-b52b5f0c7b44",
"moleculeID":"bits.bets-cf6-2b15-6112-d832-abb95ee40b8a"
},
{
"ID":"mule.snap-a12-e73c-a562-cbc1-92b47d504f19",
"componentID":"harp.thaw-f3d-f0e0-0622-0b87-ffc3a9aef0e2",
"moleculeID":"arch.fawn-e96-f078-2bd2-38a4-e32997129487"
},
{
"ID":"real.coup-727-90c7-a5c2-4bac-6b14f77ee285",
"componentID":"toes.rods-c61-ca74-9b72-ca0c-96cfe2a82869",
"moleculeID":"join.toes-974-c2d2-23f2-bb8e-949eb40370e3"
},
{
"ID":"room.wool-95e-07f5-0fa2-7950-d4baf81b854a",
"componentID":"cave.gull-aac-32e6-43a2-4975-812d31591dfa",
"moleculeID":"trim.raid-061-0df2-38b2-dac7-5a348436728a"
},
{
"ID":"pick.moss-ddf-8b5f-1b22-5b83-41d4a074ac0e",
"componentID":"coat.dean-37b-7c8c-b692-581e-548b259def46",
"moleculeID":"door.cure-3a0-ed59-c782-0a74-186630532fa4"
},
{
"ID":"fact.bull-74f-7240-8be2-1a8d-9e5f2c3c6ebe",
"componentID":"thru.fort-62b-f289-e932-b910-85ed42d6588b",
"moleculeID":"gems.chin-e26-1247-e7c2-6bf0-78e6b51d7217"
},
{
"ID":"feed.norm-dd9-3afa-ccd2-4867-cfcdf530019f",
"componentID":"meet.high-e7f-67d6-4fa2-8ae0-1a5338569f23",
"moleculeID":"maze.both-289-f20d-c9d2-b82f-6edab5e48a43"
},
{
"ID":"loom.lute-149-879e-16b2-7908-617a2a545b19",
"componentID":"wave.deem-5c8-d6ef-d192-5920-be2617eaa320",
"moleculeID":"what.hits-b3a-ffe2-d672-ba66-20a1a0355ae2"
},
{
"ID":"lose.crab-25e-17a2-a252-0881-c7750e6ef5af",
"componentID":"hook.take-193-7c5c-b3b2-ba68-9b26aeb88e4f",
"moleculeID":"fake.wild-8e8-3a08-cf62-8914-2629716216eb"
},
{
"ID":"byte.frog-994-910c-8a72-aa41-a0819e2c338c",
"componentID":"soak.have-ead-d696-4292-8a1c-c6bd6dab86d8",
"moleculeID":"coil.chat-de0-7f57-6db2-cbeb-125d6e727efc"
},
{
"ID":"thaw.pope-7e4-3d8b-a5b2-199c-d72c8a09a4b4",
"componentID":"leap.code-1e6-a805-9e82-88da-dc3c7a9b0939",
"moleculeID":"wink.past-19d-8100-1172-690b-7c620504b665"
},
{
"ID":"quay.hose-5e2-e045-47e2-19bf-a3a9e3f3355e",
"componentID":"thin.brew-e9c-fa09-4462-290d-797babf87586",
"moleculeID":"last.cane-f0b-458f-b242-aa38-d927f6644295"
},
{
"ID":"gale.huge-1d0-f125-32e2-cae2-c2bb8d3e24a3",
"componentID":"hugs.stub-676-67ee-a7d2-a8a3-cff6ea1a60f0",
"moleculeID":"lids.crow-452-d65b-4ad2-68c2-80fd027bc249"
},
{
"ID":"barb.dust-df8-5e51-4af2-4b50-87f932efc377",
"componentID":"word.film-fb9-d0d6-4ef2-6982-9e517633e41f",
"moleculeID":"meet.hoop-63e-b993-1f82-9b63-86f3d21b30a5"
},
{
"ID":"days.bans-955-da89-ee32-0bda-bc587fe3b459",
"componentID":"note.shin-85d-2c29-5bf2-abcd-506232a14b8d",
"moleculeID":"tree.hall-c25-841f-ccb2-7ab2-1e489ccbb657"
},
{
"ID":"skin.hide-3c3-b40d-89d2-0a8f-6287ea9c8df0",
"componentID":"walk.next-cee-da9e-3802-5970-688389626c3b",
"moleculeID":"lied.nice-438-e683-16a2-8b36-fd044c7968fb"
},
{
"ID":"feet.rush-9fe-a715-53c2-8bf7-76c1a10c4d9b",
"componentID":"pies.slip-a4b-a984-7722-cac5-86ccadd86a1d",
"moleculeID":"dock.post-154-520f-4812-8add-b97f2af54119"
},
{
"ID":"fine.book-119-ea63-0d82-4a3a-3a6242a0bca9",
"componentID":"bore.vain-cbb-04d5-bfc2-c8cf-bf964f752fc8",
"moleculeID":"tape.husk-957-4b33-90d2-a910-7ade9064c21b"
},
{
"ID":"wall.pork-e5c-9ca5-9522-5a86-691e38ee0284",
"componentID":"toll.fell-e8c-8b22-1082-7bb0-8ce383ade0ea",
"moleculeID":"nice.they-dd0-63af-7922-5a98-1dd4614fca2d"
},
{
"ID":"case.bans-795-0cf1-d4e2-ca5d-28e8634b91ec",
"componentID":"swim.doom-f60-b92a-d092-4ae6-535916d7ac3a",
"moleculeID":"teal.soak-43a-2e01-8cd2-b89d-99490350d947"
},
{
"ID":"less.wish-103-1be1-f4b2-b91b-e4342a60469c",
"componentID":"hens.sang-044-e09f-a102-a9b6-63a33e9f8e2e",
"moleculeID":"down.flea-f20-e0f6-3bb2-bb9b-f601b5267488"
},
{
"ID":"pets.lump-a58-eb30-bbb2-ea0e-25171644d428",
"componentID":"give.toad-b23-1014-6712-9b1f-d76f1abe9aae",
"moleculeID":"last.rugs-96c-6e97-1372-2bdc-0a300eafca40"
},
{
"ID":"lake.gems-26f-5dc3-3362-9b2b-f1cef78fd5a9",
"componentID":"wake.stop-028-e0d1-dbd2-39dc-24af09d1894d",
"moleculeID":"loan.torn-8cb-d250-1222-6b7f-3c96ba522e77"
},
{
"ID":"sees.quit-bc8-8f66-6e12-a97b-208ccb4766e2",
"componentID":"team.flex-712-75fe-9e02-a999-9aded07a24cb",
"moleculeID":"mine.sons-d32-af23-e632-8885-0c9b80bd494c"
},
{
"ID":"sets.tree-cfb-d11f-ff52-4830-190d5d2bf7bd",
"componentID":"mole.heel-580-b3cf-ec42-7b8e-33a0504c5b5b",
"moleculeID":"clip.pork-22e-a961-7e92-8803-5bc2d842ad91"
},
{
"ID":"cane.dame-b39-2761-c272-ab86-2abf549d5321",
"componentID":"turn.need-3fe-2bc6-1312-7923-3dd66b25371f",
"moleculeID":"sigh.sure-225-2e81-bd32-b931-0576514e2956"
},
{
"ID":"move.then-e64-4d59-4522-aa0e-107057693531",
"componentID":"prep.used-3a0-36b4-c5e2-eb17-df6d634b8277",
"moleculeID":"lime.kept-e70-2a60-a3a2-79d4-ec6af02ab95e"
},
{
"ID":"vine.roll-6ba-2234-ef72-b94c-d74d9bc9ac41",
"componentID":"pair.flaw-425-1875-ed02-dbf9-277bb04e893e",
"moleculeID":"mist.glow-e5c-5922-8892-39ab-1b1ed77c7a79"
},
{
"ID":"duck.tent-3bc-ccd8-9a82-5a56-ab696dcf1454",
"componentID":"grub.luck-728-4d5c-02c2-d923-9232b842de40",
"moleculeID":"root.dove-736-9b70-7012-c95a-4fbbe6324131"
},
{
"ID":"stab.maze-d3c-e17a-fc02-3ba4-058af6b75d4b",
"componentID":"dare.bans-b1a-0594-e102-badf-d8b373f431b7",
"moleculeID":"bead.call-cf6-8034-c842-2b8b-952fb7479f85"
},
{
"ID":"reap.flag-f97-1143-0352-fa8f-3926d56f1d5d",
"componentID":"thin.pier-e5c-032e-e622-8ae1-e5d46bfe78ac",
"moleculeID":"dues.just-630-d936-51f2-98c5-4507f3f7821b"
},
{
"ID":"star.germ-346-4447-31a2-c9e6-64a0866e81b0",
"componentID":"fees.yarn-c6a-179d-4952-4918-5bccc8330297",
"moleculeID":"tack.gone-c83-649b-26c2-abb7-c8ba82dda5c7"
},
{
"ID":"shim.bets-839-3832-01d2-aa11-800b6c13a851",
"componentID":"some.fled-330-0e4c-4362-fa2b-6ca9b43a1075",
"moleculeID":"cash.clan-056-0d0f-72a2-69ce-7943e4443c98"
},
{
"ID":"shin.grab-61a-5d05-cd82-bb8f-6ce240c874f0",
"componentID":"bead.hour-c64-88c8-a1f2-993f-723284239b26",
"moleculeID":"muse.this-5d2-9cf3-d952-0b61-3f9a1604cbf7"
},
{
"ID":"pest.span-3ad-5683-3f52-5a21-a838e00109df",
"componentID":"pour.bugs-bdb-0abf-e892-e885-9c20e949c76a",
"moleculeID":"five.than-b6f-2776-24a2-0932-12f4b137aff3"
},
{
"ID":"foil.flip-398-c1b9-62a2-8939-f00ee1b8bc48",
"componentID":"suit.leak-8bf-2889-5ca2-f9ed-9e91f9b1d069",
"moleculeID":"tied.chow-c73-caa9-83b2-abb6-019f6098f5d6"
},
{
"ID":"gall.dove-af6-254b-0472-8bf4-c94c8aa04478",
"componentID":"chip.lend-5df-d630-ef52-dbc7-59aaa7c71494",
"moleculeID":"ship.lime-cc6-34d9-dd42-6a36-c8c4137dd468"
},
{
"ID":"wail.nets-2c4-2890-bbf2-2948-36d008d1f061",
"componentID":"lawn.gong-987-4d18-70e2-692b-749431f5af5b",
"moleculeID":"says.skid-a85-b446-6642-59be-5fd80dbb3176"
},
{
"ID":"yell.hear-414-176b-6ed2-5a8f-1504d9e206e1",
"componentID":"muse.jobs-c24-59c5-a362-f8f0-f6f8e26f964d",
"moleculeID":"cult.cake-fe2-4771-58e2-2a58-f672af7dae6a"
},
{
"ID":"text.lots-145-ab52-be92-6aec-1b9046142ef8",
"componentID":"drum.fell-823-079f-8062-5a36-452d68d5c393",
"moleculeID":"soft.rare-8d1-a65a-1302-1a4c-1af05b3aa48c"
},
{
"ID":"span.live-bf3-a4c4-9652-fa8e-fad4b6a362ac",
"componentID":"bout.join-dc8-84fe-4c12-fb23-3f7afee2f8d0",
"moleculeID":"jade.sang-852-4f80-a632-4af7-f471d66dd16c"
},
{
"ID":"sect.paid-938-2808-4f92-cb30-c2fcdc9b0f85",
"componentID":"clan.mash-3f2-bd57-f332-2b3b-cf1f03acf736",
"moleculeID":"bead.tape-6df-48c8-6992-0a7f-16402253f140"
},
{
"ID":"cuff.pain-dbe-3855-1522-0976-4c2d3c573d6f",
"componentID":"trim.male-e1d-1978-2b02-eafb-6b5591cb1d79",
"moleculeID":"dune.life-a65-2c7a-63a2-4bdf-b9fd3d6cd6e1"
},
{
"ID":"camp.wear-add-31b5-4fa2-497a-d1c91fac3945",
"componentID":"path.tuck-be6-c1fd-5df2-1b55-0ccae7f59b0e",
"moleculeID":"coin.glue-6cd-b94a-d902-98eb-a3793388fe6f"
},
{
"ID":"fern.jade-1f0-906a-d462-6814-e94b5a41156a",
"componentID":"frog.nose-1d4-17a3-a1f2-6a2b-a535ba4ba518",
"moleculeID":"yell.cold-7c3-beb8-f662-5b96-740e1bc31d22"
},
{
"ID":"stir.must-ff0-21a0-dbb2-f856-ae4666d398eb",
"componentID":"cubs.rice-546-05aa-1d52-9a4f-09507cdf1d5f",
"moleculeID":"soak.ploy-daf-fcbe-bbb2-99e1-0217c0599262"
},
{
"ID":"gave.name-cce-a2ad-b972-fb74-b42ca58ad46b",
"componentID":"rugs.raid-f56-17e3-0dc2-39c8-7acc151dfe7d",
"moleculeID":"maps.fall-730-fb0d-1722-7957-3691a926e663"
},
{
"ID":"pose.slip-b87-a1c6-5312-d90d-81dab7682ebe",
"componentID":"mash.yawn-fa3-12cb-9ad2-797f-db540783b7db",
"moleculeID":"sell.gram-76f-6a2f-7572-9afe-ffdcf2a3c359"
},
{
"ID":"chip.desk-8a9-206b-98f2-4a4e-7cfb10c41df9",
"componentID":"from.snip-78c-4de9-dc02-abcf-c80a48851217",
"moleculeID":"sift.dirt-fb4-0566-efa2-7bdd-4b142cc42e84"
},
{
"ID":"pins.than-f5e-fbab-f2f2-b8f0-34bb325430c3",
"componentID":"dome.calf-07a-cff6-ebc2-4899-6c254e4ab7bb",
"moleculeID":"hard.fern-446-82ab-bcc2-2b1d-69d039617882"
},
{
"ID":"scar.feel-db9-857c-e222-ab38-2fe8f11f91e3",
"componentID":"golf.park-56f-a5db-f132-0b74-34887708dee8",
"moleculeID":"fail.here-eb8-61b1-8db2-9aeb-b4c201371609"
},
{
"ID":"chin.door-e6a-8ee9-e0e2-6a94-a77ac9ebccfd",
"componentID":"asks.drop-bfd-7feb-4d92-4b5f-8fffd85deea1",
"moleculeID":"pens.jobs-7aa-d0d6-7922-49f9-99477550088c"
},
{
"ID":"high.hard-0ff-28ca-7d02-7bd5-92e64596a153",
"componentID":"cook.mile-c28-9ec7-a532-dba9-e4ea0774f8fd",
"moleculeID":"ends.rear-b81-8d0d-3612-1bfd-087794fca1d2"
},
{
"ID":"each.club-4a7-6a90-0202-c8c7-9886f4a17568",
"componentID":"deep.sift-2e6-95e6-fa32-aa4b-bf00f510f54e",
"moleculeID":"tend.coat-415-4c4f-6fa2-2a31-ff9762d2d08f"
},
{
"ID":"bend.pins-bc9-1006-7fc2-39c1-0abd8cbff682",
"componentID":"watt.bind-8c1-752c-61c2-3ab3-7734965c6621",
"moleculeID":"sane.fins-dd5-3c12-cd82-3b7b-b1e21bcd3849"
},
{
"ID":"hour.haul-837-a359-ed22-7bfd-27ed40403013",
"componentID":"gust.mice-439-a46b-71c2-1900-8fc7723c07bb",
"moleculeID":"knew.verb-202-9830-1dc2-5b88-4669ce346699"
},
{
"ID":"blot.tape-372-a2cd-fc22-09d1-f0430b98cb5b",
"componentID":"core.crib-900-34d7-f252-1ae6-8593125d7d3f",
"moleculeID":"frog.brag-0c1-409b-69b2-a944-18365bce56f7"
},
{
"ID":"will.bill-184-239f-6002-8a7a-a48bb56297b3",
"componentID":"gang.sent-e39-e249-c2d2-8b56-3af299a9b19b",
"moleculeID":"wins.tomb-417-b2e0-6af2-5b83-1bd9eff534a6"
},
{
"ID":"urge.tale-6aa-3eb9-6b32-98cc-738ef19465be",
"componentID":"bees.lone-c40-a77c-63b2-fafd-8486b1adf5c5",
"moleculeID":"guts.slug-006-6241-5832-5a94-c1db346306e2"
},
{
"ID":"they.turn-0ca-1f4c-d762-e870-3913e73914d9",
"componentID":"owls.turn-c44-bab3-d632-28ff-69b1af2d31a3",
"moleculeID":"chat.bare-89c-81e0-e832-1920-4c6a45e55607"
},
{
"ID":"hush.keys-e56-8024-1352-dbe3-67dc93cc2c35",
"componentID":"gall.fees-89f-514f-32d2-cb4d-9c7ceadaad1e",
"moleculeID":"talk.weld-3e8-ba6c-19b2-aa18-3b11c9137a0e"}],
"molecule":[{
"ID":"clue.loft-b60-17e4-7aa2-89d5-ac55f8bfc171",
"name":"outer autophagosomal membrane",
"moleculeType":"concept",
"structure":"TBD"
},
{
"ID":"tubs.full-d6c-5c3e-41e2-795a-de6f234b7427",
"name":"inner autophagosomal membrane",
"moleculeType":"concept",
"structure":"TBD"
},
{
"ID":"burn.sure-463-5a92-b302-487b-8d543ee5ca04",
"name":"microtubule associated protein 1 light chain 3 alpha",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"cell.lynx-b0d-c61f-4442-ca86-6056262af5a0",
"name":"phosphatidylethanolamine",
"moleculeType":"lipid",
"structure":"TBD"
},
{
"ID":"ease.same-d07-2587-0d12-da22-eb93b7b06519",
"name":"synaptosome associated protein 29",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"fate.swat-de5-9446-e762-a817-4a170cb5d0e1",
"name":"N-acetyl-glucosamine",
"moleculeType":"metabolite",
"structure":"TBD"
},
{
"ID":"jars.sets-770-a690-8312-d97c-37d3db9eb2da",
"name":"Syntaxin 17",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"hugs.dump-30b-76ba-ef62-890d-91d464502746",
"name":"lysosomal associated membrane protein 2",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"cats.eggs-597-296a-edb2-4bd5-b7f7cc29ff4b",
"name":"immunity related GTPase M",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"grip.paid-df1-2830-0552-3839-4ddd6865e612",
"name":"RAB7A, member RAS oncogene family",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"grim.folk-88b-3e9e-72a2-c869-e9c2b85c8cf5",
"name":"MON1 homolog A, secretory trafficking associated",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"mole.hymn-e25-7fe0-6682-0b8f-1bdbae452dae",
"name":"CCZ1 homolog, vacuolar protein trafficking and biogenesis associated",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"spam.mead-0da-02ca-d3b2-088b-f8c7ad1ab4b6",
"name":"GABA type A receptor associated protein like 1",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"rule.side-174-a533-f6b2-2be4-202534c93d0f",
"name":"ectopic P-granules autophagy protein 5 homolog",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"does.mane-3fb-4cff-b792-4921-adc353d5e286",
"name":"autophagy related 13",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"pear.calf-f86-1558-b222-bb6d-9e90aeefe6a4",
"name":"microtubule associated protein 1 light chain 3 beta",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"bits.bets-cf6-2b15-6112-d832-abb95ee40b8a",
"name":"microtubule associated protein 1 light chain 3 gamma",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"arch.fawn-e96-f078-2bd2-38a4-e32997129487",
"name":"lysosomal associated membrane protein 1",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"join.toes-974-c2d2-23f2-bb8e-949eb40370e3",
"name":"vesicle associated membrane protein",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"trim.raid-061-0df2-38b2-dac7-5a348436728a",
"name":"vesicle associated membrane protein 7",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"door.cure-3a0-ed59-c782-0a74-186630532fa4",
"name":"Barkor, beclin 1-associated autophagy-related key regulator",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"gems.chin-e26-1247-e7c2-6bf0-78e6b51d7217",
"name":"VPS16 core subunit of CORVET and HOPS complexes",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"maze.both-289-f20d-c9d2-b82f-6edab5e48a43",
"name":"VPS33A core subunit of CORVET and HOPS complexes",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"what.hits-b3a-ffe2-d672-ba66-20a1a0355ae2",
"name":"VPS18 core subunit of CORVET and HOPS complexes",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"fake.wild-8e8-3a08-cf62-8914-2629716216eb",
"name":"VPS18 core subunit of CORVET and HOPS complexes",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"coil.chat-de0-7f57-6db2-cbeb-125d6e727efc",
"name":"VPS41 subunit of HOPS complex",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"wink.past-19d-8100-1172-690b-7c620504b665",
"name":"VPS39 subunit of HOPS complex",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"last.cane-f0b-458f-b242-aa38-d927f6644295",
"name":"cholesterol",
"moleculeType":"lipid",
"structure":"TBD"
},
{
"ID":"lids.crow-452-d65b-4ad2-68c2-80fd027bc249",
"name":"Oxysterol binding protein like 1A",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"meet.hoop-63e-b993-1f82-9b63-86f3d21b30a5",
"name":"phosphatidylinositol 3-kinase catalytic subunit type 3",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"tree.hall-c25-841f-ccb2-7ab2-1e489ccbb657",
"name":"UV radiation resistance associated",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"lied.nice-438-e683-16a2-8b36-fd044c7968fb",
"name":"nuclear receptor binding factor 2",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"dock.post-154-520f-4812-8add-b97f2af54119",
"name":"phosphoinositide-3-kinase regulatory subunit 4",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"tape.husk-957-4b33-90d2-a910-7ade9064c21b",
"name":"rubicon autophagy regulator",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"nice.they-dd0-63af-7922-5a98-1dd4614fca2d",
"name":"rubicon like autophagy enhancer",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"teal.soak-43a-2e01-8cd2-b89d-99490350d947",
"name":"Beclin 1",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"down.flea-f20-e0f6-3bb2-bb9b-f601b5267488",
"name":"pleckstrin homology and RUN domain containing M1",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"last.rugs-96c-6e97-1372-2bdc-0a300eafca40",
"name":"phosphatidylinositol",
"moleculeType":"lipid",
"structure":"TBD"
},
{
"ID":"loan.torn-8cb-d250-1222-6b7f-3c96ba522e77",
"name":"phosphatidylinositol-3-phosphate",
"moleculeType":"lipid",
"structure":"TBD"
},
{
"ID":"mine.sons-d32-af23-e632-8885-0c9b80bd494c",
"name":"phosphatidylinositol-3,5-bisphosphate",
"moleculeType":"lipid",
"structure":"TBD"
},
{
"ID":"clip.pork-22e-a961-7e92-8803-5bc2d842ad91",
"name":"inositol polyphosphate-5-phosphatase E",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"sigh.sure-225-2e81-bd32-b931-0576514e2956",
"name":"phosphoinositide kinase, FYVE-type zinc finger containing",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"lime.kept-e70-2a60-a3a2-79d4-ec6af02ab95e",
"name":"Rab interacting lysosomal protein",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"mist.glow-e5c-5922-8892-39ab-1b1ed77c7a79",
"name":"cortactin",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"root.dove-736-9b70-7012-c95a-4fbbe6324131",
"name":"histone deacetylase 6",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"bead.call-cf6-8034-c842-2b8b-952fb7479f85",
"name":"actin beta",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"dues.just-630-d936-51f2-98c5-4507f3f7821b",
"name":"myosin 1C",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"tack.gone-c83-649b-26c2-abb7-c8ba82dda5c7",
"name":"phosphatidylinositol-4,5-bisphosphate",
"moleculeType":"lipid",
"structure":"TBD"
},
{
"ID":"cash.clan-056-0d0f-72a2-69ce-7943e4443c98",
"name":"calcium ion",
"moleculeType":"metabolite",
"structure":"TBD"
},
{
"ID":"muse.this-5d2-9cf3-d952-0b61-3f9a1604cbf7",
"name":"Voltage-dependent P/Q-type calcium channel subunit alpha-1A",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"five.than-b6f-2776-24a2-0932-12f4b137aff3",
"name":"SNAREpin team",
"moleculeType":"concept",
"structure":"TBD"
},
{
"ID":"tied.chow-c73-caa9-83b2-abb6-019f6098f5d6",
"name":"NSF attachment protein alpha",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"ship.lime-cc6-34d9-dd42-6a36-c8c4137dd468",
"name":"N-ethylmaleimide sensitive factor",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"says.skid-a85-b446-6642-59be-5fd80dbb3176",
"name":"TBC1 domain family member 2A",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"cult.cake-fe2-4771-58e2-2a58-f672af7dae6a",
"name":"vesicle transport through interaction with t-SNAREs 1B",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"soft.rare-8d1-a65a-1302-1a4c-1af05b3aa48c",
"name":"vesicle associated membrane protein 3",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"jade.sang-852-4f80-a632-4af7-f471d66dd16c",
"name":"syntaxin 1A",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"bead.tape-6df-48c8-6992-0a7f-16402253f140",
"name":"syntaxin 1B",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"dune.life-a65-2c7a-63a2-4bdf-b9fd3d6cd6e1",
"name":"syntaxin 2",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"coin.glue-6cd-b94a-d902-98eb-a3793388fe6f",
"name":"syntaxin 3",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"yell.cold-7c3-beb8-f662-5b96-740e1bc31d22",
"name":"syntaxin 4",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"soak.ploy-daf-fcbe-bbb2-99e1-0217c0599262",
"name":"syntaxin 6",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"maps.fall-730-fb0d-1722-7957-3691a926e663",
"name":"syntaxin 7",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"sell.gram-76f-6a2f-7572-9afe-ffdcf2a3c359",
"name":"syntaxin 8",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"sift.dirt-fb4-0566-efa2-7bdd-4b142cc42e84",
"name":"syntaxin 10",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"hard.fern-446-82ab-bcc2-2b1d-69d039617882",
"name":"syntaxin 11",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"fail.here-eb8-61b1-8db2-9aeb-b4c201371609",
"name":"syntaxin 12",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"pens.jobs-7aa-d0d6-7922-49f9-99477550088c",
"name":"syntaxin 16",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"ends.rear-b81-8d0d-3612-1bfd-087794fca1d2",
"name":"syntaxin 18",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"tend.coat-415-4c4f-6fa2-2a31-ff9762d2d08f",
"name":"syntaxin 19",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"sane.fins-dd5-3c12-cd82-3b7b-b1e21bcd3849",
"name":"SEC22 homolog B, vesicle trafficking protein",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"knew.verb-202-9830-1dc2-5b88-4669ce346699",
"name":"autophagy related 4A cysteine peptidase",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"frog.brag-0c1-409b-69b2-a944-18365bce56f7",
"name":"autophagy related 4B cysteine peptidase",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"wins.tomb-417-b2e0-6af2-5b83-1bd9eff534a6",
"name":"autophagy related 4C cysteine peptidase",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"guts.slug-006-6241-5832-5a94-c1db346306e2",
"name":"autophagy related 4D cysteine peptidase",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"chat.bare-89c-81e0-e832-1920-4c6a45e55607",
"name":"synaptosome associated protein 23",
"moleculeType":"protein",
"structure":"TBD"
},
{
"ID":"talk.weld-3e8-ba6c-19b2-aa18-3b11c9137a0e",
"name":"synaptosome associated protein 25",
"moleculeType":"protein",
"structure":"TBD"}],
"geneProduct":[{
"ID":"heed.reap-731-8d1d-58f2-a9bb-a3ea26f6c803",
"geneID":"bars.self-bef-7151-f602-da1c-839ce7803544",
"moleculeID":"burn.sure-463-5a92-b302-487b-8d543ee5ca04"
},
{
"ID":"bout.hike-aa3-3ada-a5d2-4a69-028371e1e43a",
"geneID":"huts.rent-2c1-f032-0a32-096a-1fafb3664ffd",
"moleculeID":"ease.same-d07-2587-0d12-da22-eb93b7b06519"
},
{
"ID":"spur.rows-57f-f860-dd62-8add-9363255e275d",
"geneID":"volt.snow-f6f-3125-6602-caf2-1cd1b9d82fc0",
"moleculeID":"jars.sets-770-a690-8312-d97c-37d3db9eb2da"
},
{
"ID":"type.zinc-fa0-7441-34f2-5886-cc594c1de5ee",
"geneID":"last.bees-95f-acc3-17c2-18a6-3589a860dbca",
"moleculeID":"hugs.dump-30b-76ba-ef62-890d-91d464502746"
},
{
"ID":"mast.ware-73e-0ec8-db32-a9fb-eca1dd3e992a",
"geneID":"pods.edge-798-0955-1722-79c3-051ed4007012",
"moleculeID":"cats.eggs-597-296a-edb2-4bd5-b7f7cc29ff4b"
},
{
"ID":"sage.shed-01e-ccf3-a852-891b-d76420835e96",
"geneID":"feel.prop-381-c6e1-b192-e999-69a9afbef174",
"moleculeID":"grip.paid-df1-2830-0552-3839-4ddd6865e612"
},
{
"ID":"gear.eats-8f4-e629-1bc2-d8a6-ee73137997a3",
"geneID":"next.toes-7a8-bd88-fa12-d89b-6b7c2a3ef998",
"moleculeID":"grim.folk-88b-3e9e-72a2-c869-e9c2b85c8cf5"
},
{
"ID":"used.hits-352-c551-bc72-4aad-ecb444e76a9a",
"geneID":"shim.warp-b73-b606-ddc2-984a-892aa0814d4e",
"moleculeID":"mole.hymn-e25-7fe0-6682-0b8f-1bdbae452dae"
},
{
"ID":"land.dust-403-40ab-c3e2-da19-7f4767189c7b",
"geneID":"sees.bail-a6e-c223-94e2-a851-caedb27e4d2a",
"moleculeID":"spam.mead-0da-02ca-d3b2-088b-f8c7ad1ab4b6"
},
{
"ID":"curl.bays-e83-ef0a-fcd2-faf8-0994fb60517c",
"geneID":"haze.loan-f49-9ba3-6272-a886-ea18acd3decd",
"moleculeID":"rule.side-174-a533-f6b2-2be4-202534c93d0f"
},
{
"ID":"docs.rang-4af-dc0c-b0e2-48ac-d4e6f5eab2f9",
"geneID":"spin.dusk-9d0-db4f-4352-8bfe-12e7b4be1e8a",
"moleculeID":"does.mane-3fb-4cff-b792-4921-adc353d5e286"
},
{
"ID":"bout.wood-468-de20-08e2-68ed-5187b68c3764",
"geneID":"such.came-6fc-0adc-5df2-c885-2b038d7bc039",
"moleculeID":"pear.calf-f86-1558-b222-bb6d-9e90aeefe6a4"
},
{
"ID":"ease.turf-af0-63af-e692-28e2-c573b356c604",
"geneID":"cool.bark-6cf-3c58-a062-6a4f-73d44f264656",
"moleculeID":"bits.bets-cf6-2b15-6112-d832-abb95ee40b8a"
},
{
"ID":"gust.newt-ced-a715-8ec2-1b48-65e6a1549b7a",
"geneID":"paid.call-5c5-dc34-6992-fa9f-ac9a4557069f",
"moleculeID":"arch.fawn-e96-f078-2bd2-38a4-e32997129487"
},
{
"ID":"leak.bent-c3d-8672-7d72-489a-72393e9aebb4",
"geneID":"hook.five-f07-e1a1-4952-ead4-47d2b865ab11",
"moleculeID":"join.toes-974-c2d2-23f2-bb8e-949eb40370e3"
},
{
"ID":"volt.logs-38c-fea1-2602-d906-dbeb229f55fb",
"geneID":"wing.norm-efc-c118-2d32-8a14-f0fa09a7a88c",
"moleculeID":"trim.raid-061-0df2-38b2-dac7-5a348436728a"
},
{
"ID":"shim.dose-dcb-f0cf-8dc2-2b62-430781183452",
"geneID":"leak.loop-89c-b43a-83f2-5b5c-bedb2bae92be",
"moleculeID":"door.cure-3a0-ed59-c782-0a74-186630532fa4"
},
{
"ID":"fled.line-b20-ca1a-5632-b9e7-16ebce5a8a39",
"geneID":"rose.loan-f00-7e86-ab02-6bf5-6c8cdb929a50",
"moleculeID":"gems.chin-e26-1247-e7c2-6bf0-78e6b51d7217"
},
{
"ID":"pick.flax-ba6-c702-bac2-78c2-3d8ce952a67b",
"geneID":"nine.ship-eb1-d5bf-de82-6baf-253a9626174a",
"moleculeID":"maze.both-289-f20d-c9d2-b82f-6edab5e48a43"
},
{
"ID":"path.frog-dec-f97c-0812-0837-26a7316ab58c",
"geneID":"tool.slot-f9d-dd19-1bf2-0918-ad6cdbc09718",
"moleculeID":"what.hits-b3a-ffe2-d672-ba66-20a1a0355ae2"
},
{
"ID":"peek.rate-bfd-a876-8212-59cc-334278e29a82",
"geneID":"clay.take-104-8d9d-8022-4a62-5e44d8de55f2",
"moleculeID":"fake.wild-8e8-3a08-cf62-8914-2629716216eb"
},
{
"ID":"glow.gale-c62-e27e-7602-eacb-50a79eddc247",
"geneID":"pies.tile-463-006c-2cc2-09f7-91a370232006",
"moleculeID":"coil.chat-de0-7f57-6db2-cbeb-125d6e727efc"
},
{
"ID":"tint.bays-042-e494-04c2-c985-265abed728e0",
"geneID":"look.hawk-9b4-1545-01f2-b8f1-5ae6e72fd766",
"moleculeID":"wink.past-19d-8100-1172-690b-7c620504b665"
},
{
"ID":"hail.ears-a1e-7136-5872-a99e-7f7eaf848b35",
"geneID":"docs.toll-2f6-e371-9632-189d-a2fc9ecc4799",
"moleculeID":"lids.crow-452-d65b-4ad2-68c2-80fd027bc249"
},
{
"ID":"mugs.sand-333-fa13-1132-4876-5565317ab0fe",
"geneID":"maps.cars-fb5-79fe-1432-7893-80ce58b2d296",
"moleculeID":"meet.hoop-63e-b993-1f82-9b63-86f3d21b30a5"
},
{
"ID":"room.stay-abc-d711-33e2-1ab0-0d0ea2861dcb",
"geneID":"lump.taps-39a-047d-fe12-78ff-dcb09bb79290",
"moleculeID":"tree.hall-c25-841f-ccb2-7ab2-1e489ccbb657"
},
{
"ID":"came.crew-256-475e-3052-894d-082866668fee",
"geneID":"part.dine-292-d0df-8672-8a1e-4c840101d45f",
"moleculeID":"lied.nice-438-e683-16a2-8b36-fd044c7968fb"
},
{
"ID":"rash.jars-e87-a93e-3882-1b0c-494347f4a614",
"geneID":"card.book-b2a-f1fc-01f2-ba7a-0d418956b997",
"moleculeID":"dock.post-154-520f-4812-8add-b97f2af54119"
},
{
"ID":"herd.hose-da7-7ec2-f132-f80d-33f582a80739",
"geneID":"sled.fist-417-4377-f5e2-d9eb-14650cb8cf97",
"moleculeID":"tape.husk-957-4b33-90d2-a910-7ade9064c21b"
},
{
"ID":"flow.fish-f5d-20eb-0062-d86c-b69d8f503c32",
"geneID":"been.kids-de8-294c-0522-f9e8-068b2465e1b6",
"moleculeID":"nice.they-dd0-63af-7922-5a98-1dd4614fca2d"
},
{
"ID":"nose.fold-05b-9694-2572-6b26-5091361f54e3",
"geneID":"leak.base-13e-9cfe-afd2-4803-cc408fc8daab",
"moleculeID":"teal.soak-43a-2e01-8cd2-b89d-99490350d947"
},
{
"ID":"cane.wing-da9-6cdf-93f2-8989-e44ca6eb6ddc",
"geneID":"pork.lids-e88-bd29-21c2-1895-b0ec8a98cc17",
"moleculeID":"down.flea-f20-e0f6-3bb2-bb9b-f601b5267488"
},
{
"ID":"well.vain-7e1-5b20-f422-dbc5-a30517e0a82a",
"geneID":"mall.yarn-b98-f736-9822-2bbc-37e5732991c4",
"moleculeID":"clip.pork-22e-a961-7e92-8803-5bc2d842ad91"
},
{
"ID":"belt.dome-651-03c6-5442-baad-0b6a209f88ec",
"geneID":"mink.fell-6fa-e535-1562-3b41-dc857ebaeddc",
"moleculeID":"sigh.sure-225-2e81-bd32-b931-0576514e2956"
},
{
"ID":"pile.suns-2ff-14f7-9ca2-ebe0-b8e0cb6147d2",
"geneID":"chip.barb-dbc-d422-2292-fa56-dddd7bea571b",
"moleculeID":"lime.kept-e70-2a60-a3a2-79d4-ec6af02ab95e"
},
{
"ID":"wish.week-579-5ea4-a242-8952-63e339170474",
"geneID":"sort.rate-108-e91a-b632-5899-c874b5ff0ffa",
"moleculeID":"mist.glow-e5c-5922-8892-39ab-1b1ed77c7a79"
},
{
"ID":"cape.flux-62a-d229-ab32-5a35-08b3db8d3a44",
"geneID":"dear.brow-cab-4621-4fd2-fafe-bc0588bfdaad",
"moleculeID":"root.dove-736-9b70-7012-c95a-4fbbe6324131"
},
{
"ID":"gram.door-f5b-5f5c-97f2-0819-74e5a983e1ac",
"geneID":"deal.rows-82b-cbee-0982-eb46-90efade301b7",
"moleculeID":"bead.call-cf6-8034-c842-2b8b-952fb7479f85"
},
{
"ID":"fork.bits-875-1124-ecf2-6b4e-148da5ff9762",
"geneID":"nail.mint-fbe-7b24-a232-fa5b-5db07203a669",
"moleculeID":"dues.just-630-d936-51f2-98c5-4507f3f7821b"
},
{
"ID":"soak.bake-d38-b306-1cf2-0923-85d2dff9c37b",
"geneID":"none.mash-6ee-608b-d9e2-284d-7d2db948d078",
"moleculeID":"muse.this-5d2-9cf3-d952-0b61-3f9a1604cbf7"
},
{
"ID":"mugs.drag-767-6717-6552-7b31-fe472a9bdd7f",
"geneID":"hats.till-6a4-5c72-0112-58af-aac1edd5aa0f",
"moleculeID":"tied.chow-c73-caa9-83b2-abb6-019f6098f5d6"
},
{
"ID":"loaf.game-982-601d-7452-29ff-133bf28cd8c4",
"geneID":"fume.near-e41-91dc-af02-9900-d78a3fa1031a",
"moleculeID":"ship.lime-cc6-34d9-dd42-6a36-c8c4137dd468"
},
{
"ID":"weak.rare-71b-e93c-fd52-2a71-3b8015c25872",
"geneID":"paid.vise-e5b-deb2-7e72-7960-378b5a1d27c0",
"moleculeID":"says.skid-a85-b446-6642-59be-5fd80dbb3176"
},
{
"ID":"keys.crop-4f2-9175-f1f2-6a3d-6c1150321319",
"geneID":"urge.hike-76f-b330-0a52-2adb-6b17f3724db5",
"moleculeID":"cult.cake-fe2-4771-58e2-2a58-f672af7dae6a"
},
{
"ID":"nose.name-b4f-e47d-1202-8986-140b5fb27706",
"geneID":"bead.rice-032-5852-29d2-8a90-89c727a7da2b",
"moleculeID":"soft.rare-8d1-a65a-1302-1a4c-1af05b3aa48c"
},
{
"ID":"sets.news-ff9-e61d-d742-39d9-18931fffdc24",
"geneID":"look.send-576-285b-dae2-2af6-40d043c5a96f",
"moleculeID":"jade.sang-852-4f80-a632-4af7-f471d66dd16c"
},
{
"ID":"belt.bore-f9b-854b-2502-2a64-77a36e319de5",
"geneID":"four.puff-843-0bfd-73c2-8983-a82a022965a8",
"moleculeID":"bead.tape-6df-48c8-6992-0a7f-16402253f140"
},
{
"ID":"gang.duke-4ee-6261-2422-18ca-c894295fdff3",
"geneID":"pour.slug-9cc-a75b-8602-79cb-451eb9dab471",
"moleculeID":"dune.life-a65-2c7a-63a2-4bdf-b9fd3d6cd6e1"
},
{
"ID":"pope.club-d70-0346-f302-3876-1f9e0fe032f0",
"geneID":"your.lead-464-1a33-6842-1bce-a2565bf4ab3c",
"moleculeID":"coin.glue-6cd-b94a-d902-98eb-a3793388fe6f"
},
{
"ID":"neat.gale-e9c-82ae-c712-eb49-60d58e58cd8e",
"geneID":"sway.left-e9d-3ad2-7f02-0be0-a9cc0c1c57d4",
"moleculeID":"yell.cold-7c3-beb8-f662-5b96-740e1bc31d22"
},
{
"ID":"fame.gene-68c-b3b8-78d2-2881-1252715529a5",
"geneID":"scar.gate-3e2-1c71-5db2-dbe4-b3ccb66e4704",
"moleculeID":"soak.ploy-daf-fcbe-bbb2-99e1-0217c0599262"
},
{
"ID":"good.mugs-cbd-db0e-e092-3ab3-64e4e44a4903",
"geneID":"lynx.type-3ff-cb0b-0eb2-18e8-1720b1fbe852",
"moleculeID":"maps.fall-730-fb0d-1722-7957-3691a926e663"
},
{
"ID":"bead.coup-293-6f1d-cb82-4a3b-37f6cb35db05",
"geneID":"spun.lack-12f-ffd7-f832-0be3-0ecf7bf431af",
"moleculeID":"sell.gram-76f-6a2f-7572-9afe-ffdcf2a3c359"
},
{
"ID":"twig.land-210-729d-5c62-5af3-7beffd748b3f",
"geneID":"lark.dark-38b-44aa-41d2-989b-8cf19a1ddeb0",
"moleculeID":"sift.dirt-fb4-0566-efa2-7bdd-4b142cc42e84"
},
{
"ID":"tint.boil-296-95fe-d622-6835-26522b4eb00a",
"geneID":"your.half-d39-064a-d3f2-89b7-47300eca98d1",
"moleculeID":"hard.fern-446-82ab-bcc2-2b1d-69d039617882"
},
{
"ID":"cups.cold-201-7155-dbb2-09b3-f506e7ec37bc",
"geneID":"mite.gems-7d8-2798-dd02-581a-b202e2d6c657",
"moleculeID":"fail.here-eb8-61b1-8db2-9aeb-b4c201371609"
},
{
"ID":"lush.rail-e33-1fb2-b2f2-9b8a-8b2e14055697",
"geneID":"snip.hoop-bf2-a3c8-d2c2-6b24-f492eebca97e",
"moleculeID":"pens.jobs-7aa-d0d6-7922-49f9-99477550088c"
},
{
"ID":"reef.spun-128-208a-98f2-182a-7f56bafffbd5",
"geneID":"dash.arms-612-6244-dc22-8a41-e6020da03bb7",
"moleculeID":"ends.rear-b81-8d0d-3612-1bfd-087794fca1d2"
},
{
"ID":"both.cold-bcc-d738-2592-3865-7990ef16083a",
"geneID":"ends.pump-b58-a995-ca62-3afa-d8eabcccf566",
"moleculeID":"tend.coat-415-4c4f-6fa2-2a31-ff9762d2d08f"
},
{
"ID":"toys.moat-61b-70a3-9772-fb32-7ed08780b89a",
"geneID":"rise.rail-b97-a9b1-d852-39d4-ba1e66a68310",
"moleculeID":"sane.fins-dd5-3c12-cd82-3b7b-b1e21bcd3849"
},
{
"ID":"fade.drug-615-2bb2-70f2-5b8b-3fe10f087fa4",
"geneID":"melt.span-184-b3c0-c902-a8cf-6579894682a2",
"moleculeID":"knew.verb-202-9830-1dc2-5b88-4669ce346699"
},
{
"ID":"door.room-294-a3b7-e0f2-7be1-10848afa9a0c",
"geneID":"june.neat-93c-537f-8202-5b27-873449a7ce28",
"moleculeID":"frog.brag-0c1-409b-69b2-a944-18365bce56f7"
},
{
"ID":"earn.pool-984-b09b-8872-9907-838f20aec955",
"geneID":"rail.rail-357-d5a2-0dd2-aaab-ec0c435af6fa",
"moleculeID":"wins.tomb-417-b2e0-6af2-5b83-1bd9eff534a6"
},
{
"ID":"claw.tubs-2cf-6f18-0542-0bf5-f16034330e23",
"geneID":"wise.hive-648-cfb0-f042-281c-27c30eea6a21",
"moleculeID":"guts.slug-006-6241-5832-5a94-c1db346306e2"
},
{
"ID":"fool.vent-19f-ac04-cc52-9abe-4181e1a0c8ee",
"geneID":"hash.cute-394-0fe5-5102-2bd5-c3c87ec06919",
"moleculeID":"chat.bare-89c-81e0-e832-1920-4c6a45e55607"
},
{
"ID":"hugs.roar-cf4-099d-36f2-7a35-4eed3ae53fc1",
"geneID":"foil.tune-b62-896e-7112-5995-bd865236a222",
"moleculeID":"talk.weld-3e8-ba6c-19b2-aa18-3b11c9137a0e"}],
"gene":[{
"ID":"bars.self-bef-7151-f602-da1c-839ce7803544",
"symbol":"MAP1LC3A",
"name":"microtubule associated protein 1 light chain 3 alpha"
},
{
"ID":"huts.rent-2c1-f032-0a32-096a-1fafb3664ffd",
"symbol":"SNAP29",
"name":"synaptosome associated protein 29"
},
{
"ID":"volt.snow-f6f-3125-6602-caf2-1cd1b9d82fc0",
"symbol":"STX17",
"name":"Syntaxin 17"
},
{
"ID":"last.bees-95f-acc3-17c2-18a6-3589a860dbca",
"symbol":"LAMP2",
"name":"lysosomal associated membrane protein 2"
},
{
"ID":"pods.edge-798-0955-1722-79c3-051ed4007012",
"symbol":"IRGM",
"name":"immunity related GTPase M"
},
{
"ID":"feel.prop-381-c6e1-b192-e999-69a9afbef174",
"symbol":"RAB7A",
"name":"RAB7A, member RAS oncogene family"
},
{
"ID":"next.toes-7a8-bd88-fa12-d89b-6b7c2a3ef998",
"symbol":"MON1A",
"name":"MON1 homolog A, secretory trafficking associated"
},
{
"ID":"shim.warp-b73-b606-ddc2-984a-892aa0814d4e",
"symbol":"CCZ1",
"name":"CCZ1 homolog, vacuolar protein trafficking and biogenesis associated"
},
{
"ID":"sees.bail-a6e-c223-94e2-a851-caedb27e4d2a",
"symbol":"GABARAPL1",
"name":"GABA type A receptor associated protein like 1"
},
{
"ID":"haze.loan-f49-9ba3-6272-a886-ea18acd3decd",
"symbol":"EPG5",
"name":"ectopic P-granules autophagy protein 5 homolog"
},
{
"ID":"spin.dusk-9d0-db4f-4352-8bfe-12e7b4be1e8a",
"symbol":"ATG13",
"name":"autophagy related 13"
},
{
"ID":"such.came-6fc-0adc-5df2-c885-2b038d7bc039",
"symbol":"MAP1LC3B",
"name":"microtubule associated protein 1 light chain 3 beta"
},
{
"ID":"cool.bark-6cf-3c58-a062-6a4f-73d44f264656",
"symbol":"MAP1LC3C",
"name":"microtubule associated protein 1 light chain 3 gamma"
},
{
"ID":"paid.call-5c5-dc34-6992-fa9f-ac9a4557069f",
"symbol":"LAMP1",
"name":"lysosomal associated membrane protein 1"
},
{
"ID":"hook.five-f07-e1a1-4952-ead4-47d2b865ab11",
"symbol":"VAMP8",
"name":"vesicle associated membrane protein"
},
{
"ID":"wing.norm-efc-c118-2d32-8a14-f0fa09a7a88c",
"symbol":"VAMP7",
"name":"vesicle associated membrane protein 7"
},
{
"ID":"leak.loop-89c-b43a-83f2-5b5c-bedb2bae92be",
"symbol":"ATG14",
"name":"Barkor, beclin 1-associated autophagy-related key regulator"
},
{
"ID":"rose.loan-f00-7e86-ab02-6bf5-6c8cdb929a50",
"symbol":"VPS16",
"name":"VPS16 core subunit of CORVET and HOPS complexes"
},
{
"ID":"nine.ship-eb1-d5bf-de82-6baf-253a9626174a",
"symbol":"VPS33A",
"name":"VPS33A core subunit of CORVET and HOPS complexes"
},
{
"ID":"tool.slot-f9d-dd19-1bf2-0918-ad6cdbc09718",
"symbol":"VPS18",
"name":"VPS18 core subunit of CORVET and HOPS complexes"
},
{
"ID":"clay.take-104-8d9d-8022-4a62-5e44d8de55f2",
"symbol":"VPS11",
"name":"VPS18 core subunit of CORVET and HOPS complexes"
},
{
"ID":"pies.tile-463-006c-2cc2-09f7-91a370232006",
"symbol":"VPS41",
"name":"VPS41 subunit of HOPS complex"
},
{
"ID":"look.hawk-9b4-1545-01f2-b8f1-5ae6e72fd766",
"symbol":"VPS39",
"name":"VPS39 subunit of HOPS complex"
},
{
"ID":"docs.toll-2f6-e371-9632-189d-a2fc9ecc4799",
"symbol":"OSBPL1A",
"name":"Oxysterol binding protein like 1A"
},
{
"ID":"maps.cars-fb5-79fe-1432-7893-80ce58b2d296",
"symbol":"PIK3C3",
"name":"phosphatidylinositol 3-kinase catalytic subunit type 3"
},
{
"ID":"lump.taps-39a-047d-fe12-78ff-dcb09bb79290",
"symbol":"UVRAG",
"name":"UV radiation resistance associated"
},
{
"ID":"part.dine-292-d0df-8672-8a1e-4c840101d45f",
"symbol":"NRBF2",
"name":"nuclear receptor binding factor 2"
},
{
"ID":"card.book-b2a-f1fc-01f2-ba7a-0d418956b997",
"symbol":"PIK3R4",
"name":"phosphoinositide-3-kinase regulatory subunit 4"
},
{
"ID":"sled.fist-417-4377-f5e2-d9eb-14650cb8cf97",
"symbol":"RUBCN",
"name":"rubicon autophagy regulator"
},
{
"ID":"been.kids-de8-294c-0522-f9e8-068b2465e1b6",
"symbol":"RUBCNL",
"name":"rubicon like autophagy enhancer"
},
{
"ID":"leak.base-13e-9cfe-afd2-4803-cc408fc8daab",
"symbol":"BECN1",
"name":"Beclin 1"
},
{
"ID":"pork.lids-e88-bd29-21c2-1895-b0ec8a98cc17",
"symbol":"PLEKHM1",
"name":"pleckstrin homology and RUN domain containing M1"
},
{
"ID":"mall.yarn-b98-f736-9822-2bbc-37e5732991c4",
"symbol":"INPP5E",
"name":"inositol polyphosphate-5-phosphatase E"
},
{
"ID":"mink.fell-6fa-e535-1562-3b41-dc857ebaeddc",
"symbol":"PIKFYVE",
"name":"phosphoinositide kinase, FYVE-type zinc finger containing"
},
{
"ID":"chip.barb-dbc-d422-2292-fa56-dddd7bea571b",
"symbol":"RILP",
"name":"Rab interacting lysosomal protein"
},
{
"ID":"sort.rate-108-e91a-b632-5899-c874b5ff0ffa",
"symbol":"CTTN",
"name":"cortactin"
},
{
"ID":"dear.brow-cab-4621-4fd2-fafe-bc0588bfdaad",
"symbol":"HDAC6",
"name":"histone deacetylase 6"
},
{
"ID":"deal.rows-82b-cbee-0982-eb46-90efade301b7",
"symbol":"ACTB",
"name":"actin beta"
},
{
"ID":"nail.mint-fbe-7b24-a232-fa5b-5db07203a669",
"symbol":"MYO1C",
"name":"myosin 1C"
},
{
"ID":"none.mash-6ee-608b-d9e2-284d-7d2db948d078",
"symbol":"CACNA1A",
"name":"Voltage-dependent P/Q-type calcium channel subunit alpha-1A"
},
{
"ID":"hats.till-6a4-5c72-0112-58af-aac1edd5aa0f",
"symbol":"NAPA",
"name":"NSF attachment protein alpha"
},
{
"ID":"fume.near-e41-91dc-af02-9900-d78a3fa1031a",
"symbol":"NSF",
"name":"N-ethylmaleimide sensitive factor"
},
{
"ID":"paid.vise-e5b-deb2-7e72-7960-378b5a1d27c0",
"symbol":"TBC1D2",
"name":"TBC1 domain family member 2A"
},
{
"ID":"urge.hike-76f-b330-0a52-2adb-6b17f3724db5",
"symbol":"VTI1B",
"name":"vesicle transport through interaction with t-SNAREs 1B"
},
{
"ID":"bead.rice-032-5852-29d2-8a90-89c727a7da2b",
"symbol":"VAMP3",
"name":"vesicle associated membrane protein 3"
},
{
"ID":"look.send-576-285b-dae2-2af6-40d043c5a96f",
"symbol":"STX1A",
"name":"syntaxin 1A"
},
{
"ID":"four.puff-843-0bfd-73c2-8983-a82a022965a8",
"symbol":"STX1B",
"name":"syntaxin 1B"
},
{
"ID":"pour.slug-9cc-a75b-8602-79cb-451eb9dab471",
"symbol":"STX2",
"name":"syntaxin 2"
},
{
"ID":"your.lead-464-1a33-6842-1bce-a2565bf4ab3c",
"symbol":"STX3",
"name":"syntaxin 3"
},
{
"ID":"sway.left-e9d-3ad2-7f02-0be0-a9cc0c1c57d4",
"symbol":"STX4",
"name":"syntaxin 4"
},
{
"ID":"scar.gate-3e2-1c71-5db2-dbe4-b3ccb66e4704",
"symbol":"STX6",
"name":"syntaxin 6"
},
{
"ID":"lynx.type-3ff-cb0b-0eb2-18e8-1720b1fbe852",
"symbol":"STX7",
"name":"syntaxin 7"
},
{
"ID":"spun.lack-12f-ffd7-f832-0be3-0ecf7bf431af",
"symbol":"STX8",
"name":"syntaxin 8"
},
{
"ID":"lark.dark-38b-44aa-41d2-989b-8cf19a1ddeb0",
"symbol":"STX10",
"name":"syntaxin 10"
},
{
"ID":"your.half-d39-064a-d3f2-89b7-47300eca98d1",
"symbol":"STX11",
"name":"syntaxin 11"
},
{
"ID":"mite.gems-7d8-2798-dd02-581a-b202e2d6c657",
"symbol":"STX12",
"name":"syntaxin 12"
},
{
"ID":"snip.hoop-bf2-a3c8-d2c2-6b24-f492eebca97e",
"symbol":"STX16",
"name":"syntaxin 16"
},
{
"ID":"dash.arms-612-6244-dc22-8a41-e6020da03bb7",
"symbol":"STX18",
"name":"syntaxin 18"
},
{
"ID":"ends.pump-b58-a995-ca62-3afa-d8eabcccf566",
"symbol":"STX19",
"name":"syntaxin 19"
},
{
"ID":"rise.rail-b97-a9b1-d852-39d4-ba1e66a68310",
"symbol":"SEC22B",
"name":"SEC22 homolog B, vesicle trafficking protein"
},
{
"ID":"melt.span-184-b3c0-c902-a8cf-6579894682a2",
"symbol":"ATG4A",
"name":"autophagy related 4A cysteine peptidase"
},
{
"ID":"june.neat-93c-537f-8202-5b27-873449a7ce28",
"symbol":"ATG4B",
"name":"autophagy related 4B cysteine peptidase"
},
{
"ID":"rail.rail-357-d5a2-0dd2-aaab-ec0c435af6fa",
"symbol":"ATG4C",
"name":"autophagy related 4C cysteine peptidase"
},
{
"ID":"wise.hive-648-cfb0-f042-281c-27c30eea6a21",
"symbol":"ATG4D",
"name":"autophagy related 4D cysteine peptidase"
},
{
"ID":"hash.cute-394-0fe5-5102-2bd5-c3c87ec06919",
"symbol":"SNAP23",
"name":"synaptosome associated protein 23"
},
{
"ID":"foil.tune-b62-896e-7112-5995-bd865236a222",
"symbol":"SNAP25",
"name":"synaptosome associated protein 25"}],
"note":[{
"ID":"hoop.soul-75e-c6bb-f7a2-2a71-c78d21456ce1",
"targetID":"isle.gait-a82-1ff7-70b2-e810-fd2dd3c1c465",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"LC3 is a member of the Atg8 protein family (pmid:30115558)."
},
{
"ID":"whey.gang-44f-deb3-ce72-3b35-ab092c467c34",
"targetID":"webs.laud-453-0049-6f12-59d2-c55101ab383b",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"GABARAPL1 is a member of the Atg8 protein family (pmid:30115558)."
},
{
"ID":"shot.beds-878-03b0-1b22-fa9f-5cdfa307a978",
"targetID":"thru.fort-62b-f289-e932-b910-85ed42d6588b",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"VPS16 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex (pmid:23645161)."
},
{
"ID":"used.your-5a8-1c69-6722-0946-a9f99f70dc0d",
"targetID":"meet.high-e7f-67d6-4fa2-8ae0-1a5338569f23",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"VPS33 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex (pmid:23645161)."
},
{
"ID":"boat.lots-2b1-6f69-8ca2-783d-a49dec5a64a0",
"targetID":"wave.deem-5c8-d6ef-d192-5920-be2617eaa320",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"VPS18 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex (pmid:23645161)."
},
{
"ID":"fans.trap-e6e-982b-1092-7802-d8d06fd4cd6c",
"targetID":"hook.take-193-7c5c-b3b2-ba68-9b26aeb88e4f",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"VPS11 (Vacuolar Protein Sorting) is one of the four subunits that are common to the HOPS complex and the CORVET complex (pmid:23645161)."
},
{
"ID":"fate.tell-bc9-74e6-c1e2-6b50-789076344e05",
"targetID":"soak.have-ead-d696-4292-8a1c-c6bd6dab86d8",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"VPS41 (Vacuolar Protein Sorting) is one of the two subunits that are specific to the HOPS complex (pmid:23645161)."
},
{
"ID":"saws.care-2f8-34f8-b092-2a17-fedc96ecf116",
"targetID":"leap.code-1e6-a805-9e82-88da-dc3c7a9b0939",
"typeID":"snow.gram-8a2-cb70-96e2-daba-61933e196c9b",
"note":"VPS39 (Vacuolar Protein Sorting) is one of the two subunits that are specific to the HOPS complex (pmid:23645161)."}]}
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 1.25 1.26 1.27 1.28 1.29 1.30 1.31 1.32 1.33 1.34 1.35
Corona & Jackson (2018) Finding the Middle Ground for Autophagic Fusion Requirements. Trends Cell Biol 28:869-881. (pmid: 30115558) [ PubMed ] [ DOI ] Autophagosome/amphisome-lysosome fusion is a highly regulated process at the protein, lipid, and biochemical level. Each primary component of fusion, such as the core SNAREs, HOPS complex, or physical positioning by microtubule-associated dynein motors, are regulated at multiple points to ensure optimum conditions for autophagic flux to proceed. With the complexity of the membrane fusion system, it is not difficult to imagine how autophagic flux defect-related disorders, such as Huntington's disease, non-familial Alzheimer's disease, and Vici syndrome develop. Each membrane fusion step is regulated at the protein, lipid, and ion level. This review aims to discuss the recent developments toward understanding the regulation of autophagosome, amphisome, and lysosome fusion requirements for successful autophagic flux.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8
Balderhaar & Ungermann (2013) CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci 126:1307-16. (pmid: 23645161) [ PubMed ] [ DOI ] Protein and lipid transport along the endolysosomal system of eukaryotic cells depends on multiple fusion and fission events. Over the past few years, the molecular constituents of both fission and fusion machineries have been identified. Here, we focus on the mechanism of membrane fusion at endosomes, vacuoles and lysosomes, and in particular on the role of the two homologous tethering complexes called CORVET and HOPS. Both complexes are heterohexamers; they share four subunits, interact with Rab GTPases and soluble NSF attachment protein receptors (SNAREs) and can tether membranes. Owing to the presence of specific subunits, CORVET is a Rab5 effector complex, whereas HOPS can bind efficiently to late endosomes and lysosomes through Rab7. Based on the recently described overall structure of the HOPS complex and a number of in vivo and in vitro analyses, important insights into their function have been obtained. Here, we discuss the general function of both complexes in yeast and in metazoan cells in the context of endosomal biogenesis and maturation.
- ↑ 3.0 3.1 3.2 3.3
Zhi et al. (2018) Anatomy of autophagy: from the beginning to the end. Cell Mol Life Sci 75:815-831. (pmid: 28939950) [ PubMed ] [ DOI ] Autophagy is a highly regulated process in eukaryotes to maintain homeostasis and manage stress responses. Understanding the regulatory mechanisms and key players involved in autophagy will provide critical insights into disease-related pathogenesis and potential clinical treatments. In this review, we describe the hallmark events involved in autophagy, from its initiation, to the final destruction of engulfed targets. Furthermore, based on structural and biochemical data, we evaluate the roles of key players in these processes and provide rationale as to how they control autophagic events in a highly ordered manner.
- ↑
Wennerberg et al. (2005) The Ras superfamily at a glance. J Cell Sci 118:843-6. (pmid: 15731001) - ↑
Goitre et al. (2014) The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol Biol 1120:1-18. (pmid: 24470015) [ PubMed ] [ DOI ] The Ras superfamily of small GTPases is composed of more than 150 members, which share a conserved structure and biochemical properties, acting as binary molecular switches turned on by binding GTP and off by hydrolyzing GTP to GDP. However, despite considerable structural and biochemical similarities, these proteins play multiple and divergent roles, being versatile and key regulators of virtually all fundamental cellular processes. Conversely, their dysfunction plays a crucial role in the pathogenesis of serious human diseases, including cancer and developmental syndromes. Fuelled by the original identification in 1982 of mutationally activated and transforming human Ras genes in human cancer cell lines, a variety of powerful experimental techniques have been intensively focused on discovering and studying structure, biochemistry, and biology of Ras and Ras-related small GTPases, leading to fundamental research breakthroughs into identification and structural and functional characterization of a huge number of Ras superfamily members, as well as of their multiple regulators and effectors. In this review we provide a general overview of the major milestones that eventually allowed to unlock the secret treasure chest of this large and important superfamily of proteins.
- ↑ 6.0 6.1 6.2 6.3
Tsuboyama et al. (2016) The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354:1036-1041. (pmid: 27885029) [ PubMed ] [ DOI ] In macroautophagy, cytoplasmic contents are sequestered into the double-membrane autophagosome, which fuses with the lysosome to become the autolysosome. It has been thought that the autophagy-related (ATG) conjugation systems are required for autophagosome formation. Here, we found that autophagosomal soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) syntaxin 17-positive autophagosome-like structures could be generated even in the absence of the ATG conjugation systems, although at a reduced rate. These syntaxin 17-positive structures could further fuse with lysosomes, but degradation of the inner autophagosomal membrane was significantly delayed. Accordingly, autophagic activity in ATG conjugation-deficient cells was strongly suppressed. We suggest that the ATG conjugation systems, which are likely required for the closure (i.e., fission) of the autophagosomal edge, are not absolutely essential for autolysosome formation but are important for efficient degradation of the inner autophagosomal membrane.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7
Kruppa et al. (2016) Myosins, Actin and Autophagy. Traffic 17:878-90. (pmid: 27146966) [ PubMed ] [ DOI ] Myosin motor proteins working together with the actin cytoskeleton drive a wide range of cellular processes. In this review, we focus on their roles in autophagy - the pathway the cell uses to ensure homeostasis by targeting pathogens, misfolded proteins and damaged organelles for degradation. The actin cytoskeleton regulated by a host of nucleating, anchoring and stabilizing proteins provides the filament network for the delivery of essential membrane vesicles from different cellular compartments to the autophagosome. Actin networks have also been implicated in structurally supporting the expanding phagophore, moving autophagosomes and enabling efficient fusion with the lysosome. Only a few myosins have so far been shown to play a role in autophagy. Non-muscle myosin IIA functions in the early stages delivering membrane for the initial formation of the autophagosome, whereas myosin IC and myosin VI are involved in the final stages providing specific membranes for autophagosome maturation and its fusion with the lysosome.
- ↑ 8.0 8.1 8.2
Guo et al. (2014) O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat Cell Biol 16:1215-26. (pmid: 25419848) [ PubMed ] [ DOI ] The mechanism by which nutrient status regulates the fusion of autophagosomes with endosomes/lysosomes is poorly understood. Here, we report that O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) mediates O-GlcNAcylation of the SNARE protein SNAP-29 and regulates autophagy in a nutrient-dependent manner. In mammalian cells, OGT knockdown, or mutating the O-GlcNAc sites in SNAP-29, promotes the formation of a SNAP-29-containing SNARE complex, increases fusion between autophagosomes and endosomes/lysosomes, and promotes autophagic flux. In Caenorhabditis elegans, depletion of ogt-1 has a similar effect on autophagy; moreover, expression of an O-GlcNAc-defective SNAP-29 mutant facilitates autophagic degradation of protein aggregates. O-GlcNAcylated SNAP-29 levels are reduced during starvation in mammalian cells and in C. elegans. Our study reveals a mechanism by which O-GlcNAc-modification integrates nutrient status with autophagosome maturation.
- ↑ 9.0 9.1 9.2 9.3 9.4
Saleeb et al. (2019) A VPS33A-binding motif on syntaxin 17 controls autophagy completion in mammalian cells. J Biol Chem 294:4188-4201. (pmid: 30655294) [ PubMed ] [ DOI ] Autophagy is an intracellular degradation pathway that transports cytoplasmic material to the lysosome for hydrolysis. It is completed by SNARE-mediated fusion of the autophagosome and endolysosome membranes. This process must be carefully regulated to maintain the organization of the membrane system and prevent mistargeted degradation. As yet, models of autophagosomal fusion have not been verified within a cellular context because of difficulties with assessing protein interactions in situ Here, we used high-resolution fluorescence lifetime imaging (FLIM)-FRET of HeLa cells to identify protein interactions within the spatiotemporal framework of the cell. We show that autophagosomal syntaxin 17 (Stx17) heterotrimerizes with synaptosome-associated protein 29 (SNAP29) and vesicle-associated membrane protein 7 (VAMP7) in situ, highlighting a functional role for VAMP7 in autophagosome clearance that has previously been sidelined in favor of a role for VAMP8. Additionally, we identified multimodal regulation of SNARE assembly by the Sec1/Munc18 (SM) protein VPS33A, mirroring other syntaxin-SM interactions and therefore suggesting a unified model of SM regulation. Contrary to current theoretical models, we found that the Stx17 N-peptide appears to interact in a positionally conserved, but mechanistically divergent manner with VPS33A, providing a late "go, no-go" step for autophagic fusion via a phosphoserine master-switch. Our findings suggest that Stx17 fusion competency is regulated by a phosphosite in its N-peptide, representing a previously unknown regulatory step in mammalian autophagy.
- ↑ 10.0 10.1
Kumar et al. (2018) Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol 217:997-1013. (pmid: 29420192) [ PubMed ] [ DOI ] Autophagy is a conserved eukaryotic process with metabolic, immune, and general homeostatic functions in mammalian cells. Mammalian autophagosomes fuse with lysosomes in a SNARE-driven process that includes syntaxin 17 (Stx17). How Stx17 translocates to autophagosomes is unknown. In this study, we show that the mechanism of Stx17 recruitment to autophagosomes in human cells entails the small guanosine triphosphatase IRGM. Stx17 directly interacts with IRGM, and efficient Stx17 recruitment to autophagosomes requires IRGM. Both IRGM and Stx17 directly interact with mammalian Atg8 proteins, thus being guided to autophagosomes. We also show that Stx17 is significant in defense against infectious agents and that Stx17-IRGM interaction is targeted by an HIV virulence factor Nef.
- ↑
Hubert et al. (2016) LAMP-2 is required for incorporating syntaxin-17 into autophagosomes and for their fusion with lysosomes. Biol Open 5:1516-1529. (pmid: 27628032) [ PubMed ] [ DOI ] Autophagy is an evolutionarily conserved process used for removing surplus and damaged proteins and organelles from the cytoplasm. The unwanted material is incorporated into autophagosomes that eventually fuse with lysosomes, leading to the degradation of their cargo. The fusion event is mediated by the interaction between the Qa-SNARE syntaxin-17 (STX17) on autophagosomes and the R-SNARE VAMP8 on lysosomes. Cells deficient in lysosome membrane-associated protein-2 (LAMP-2) have increased numbers of autophagosomes but the underlying mechanism is poorly understood. By transfecting LAMP-2-deficient and LAMP-1/2--double-deficient mouse embryonic fibroblasts (MEFs) with a tandem fluorescent-tagged LC3 we observed a failure of fusion between the autophagosomes and the lysosomes that could be rescued by complementation with LAMP-2A. Although we observed no change in expression and localization of VAMP8, its interacting partner STX17 was absent from autophagosomes of LAMP-2-deficient cells. Thus, LAMP-2 is essential for STX17 expression by the autophagosomes and this absence is sufficient to explain their failure to fuse with lysosomes. The results have clear implications for situations associated with a reduction of LAMP-2 expression.
- ↑ 12.0 12.1 12.2
Stroupe (2018) This Is the End: Regulation of Rab7 Nucleotide Binding in Endolysosomal Trafficking and Autophagy. Front Cell Dev Biol 6:129. (pmid: 30333976) [ PubMed ] [ DOI ] Rab7 - or in yeast, Ypt7p - governs membrane trafficking in the late endocytic and autophagic pathways. Rab7 also regulates mitochondrion-lysosome contacts, the sites of mitochondrial fission. Like all Rab GTPases, Rab7 cycles between an "active" GTP-bound form that binds downstream effectors - e.g., the HOPS and retromer complexes and the dynactin-binding Rab-interacting lysosomal protein (RILP) - and an "inactive" GDP-bound form that cannot bind effectors. Accessory proteins regulate the nucleotide binding state of Rab7: guanine nucleotide exchange factors (GEFs) stimulate exchange of bound GDP for GTP, resulting in Rab7 activation, whereas GTPase activating proteins (GAPs) boost Rab7's GTP hydrolysis activity, thereby inactivating Rab7. This review will discuss the GEF and GAPs that control Rab7 nucleotide binding, and thus regulate Rab7's activity in endolysosomal trafficking and autophagy. It will also consider how bacterial pathogens manipulate Rab7 nucleotide binding to support intracellular invasion and immune evasion.
- ↑
Matsumoto & Nakanishi-Matsui (2019) Proton pumping V-ATPase inhibitor bafilomycin A1 affects Rab7 lysosomal localization and abolishes anterograde trafficking of osteoclast secretory lysosomes. Biochem Biophys Res Commun 510:421-426. (pmid: 30717974) [ PubMed ] [ DOI ] Osteoclast lysosomes secrete lytic enzymes into bone resorption lacunae, and sort the lysosomal proton pumping vacuolar-type ATPase (V-ATPase) to the plasma membrane to form the acidic environment required for bone digestion. The a3 isoform of V-ATPase is essential for outward trafficking of the secretory lysosomes and interacts physically with Rab7, a small GTPase that regulates trafficking of late endosomes and lysosomes, to recruit it to lysosomes. However, it is unclear whether organelle acidification by V-ATPase is required for the lysosome trafficking. Here, we showed that incubation of osteoclasts with the V-ATPase inhibitor bafilomycin A1 abolished the osteoclast-characteristic peripheral localization of secretory lysosomes, Rab7, and α-tubulin. Although bafilomycin A1 had little or no effect on Rab7 activation and its interaction with a3, treatment with the inhibitor significantly reduced the lysosomal localization of Rab7. Even constitutively active Rab7 did not localize to lysosomes in the presence of the inhibitor. These results suggest that organelle acidification by V-ATPase is required for localization of activated Rab7 to lysosomes.
- ↑ 14.0 14.1 14.2 14.3
Cite error: InvalidHarrison et al. (2003) Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol 23:6494-506. (pmid: 12944476) [ PubMed ] [ DOI ] Nascent phagosomes must undergo a series of fusion and fission reactions to acquire the microbicidal properties required for the innate immune response. Here we demonstrate that this maturation process involves the GTPase Rab7. Rab7 recruitment to phagosomes was found to precede and to be essential for their fusion with late endosomes and/or lysosomes. Active Rab7 on the phagosomal membrane associates with the effector protein RILP (Rab7-interacting lysosomal protein), which in turn bridges phagosomes with dynein-dynactin, a microtubule-associated motor complex. The motors not only displace phagosomes in the centripetal direction but, strikingly, promote the extension of phagosomal tubules toward late endocytic compartments. Fusion of tubules with these organelles was documented by fluorescence and electron microscopy. Tubule extension and fusion with late endosomes and/or lysosomes were prevented by expression of a truncated form of RILP lacking the dynein-dynactin-recruiting domain. We conclude that full maturation of phagosomes requires the retrograde emission of tubular extensions, which are generated by activation of Rab7, recruitment of RILP, and consequent association of phagosomes with microtubule-associated motors.
<ref>tag; name "Wijdeven2016" defined multiple times with different content Cite error: Invalid<ref>tag; name "Wijdeven2016" defined multiple times with different content Cite error: Invalid<ref>tag; name "Wijdeven2016" defined multiple times with different content - ↑
Birgisdottir et al. (2019) Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy 15:1333-1355. (pmid: 30767700) [ PubMed ] [ DOI ] Autophagosome formation depends on a carefully orchestrated interplay between membrane-associated protein complexes. Initiation of macroautophagy/autophagy is mediated by the ULK1 (unc-51 like autophagy activating kinase 1) protein kinase complex and the autophagy-specific class III phosphatidylinositol 3-kinase complex I (PtdIns3K-C1). The latter contains PIK3C3/VPS34, PIK3R4/VPS15, BECN1/Beclin 1 and ATG14 and phosphorylates phosphatidylinositol to generate phosphatidylinositol 3-phosphate (PtdIns3P). Here, we show that PIK3C3, BECN1 and ATG14 contain functional LIR motifs and interact with the Atg8-family proteins with a preference for GABARAP and GABARAPL1. High resolution crystal structures of the functional LIR motifs of these core components of PtdIns3K-C1were obtained. Variation in hydrophobic pocket 2 (HP2) may explain the specificity for the GABARAP family. Mutation of the LIR motif in ATG14 did not prevent formation of the PtdIns3K-C1 complex, but blocked colocalization with MAP1LC3B/LC3B and impaired mitophagy. The ULK-mediated phosphorylation of S29 in ATG14 was strongly dependent on a functional LIR motif in ATG14. GABARAP-preferring LIR motifs in PIK3C3, BECN1 and ATG14 may, via coincidence detection, contribute to scaffolding of PtdIns3K-C1 on membranes for efficient autophagosome formation. Abbreviations: ATG: autophagy-related; BafA1: bafilomycin A1; GABARAP: GABA type A receptor-associated protein; GABARAPL1: GABA type A receptor associated protein like 1; GFP: enhanced green fluorescent protein; KO: knockout; LDS: LIR docking site; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; PIK3C3: phosphatidylinositol 3-kinase catalytic subunit type 3; PIK3R4: phosphoinositide-3-kinase regulatory subunit 4; PtdIns3K: phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol-3-phosphate; SQSTM1/p62: sequestosome 1; VPS: Vacuolar protein sorting; ULK: unc-51 like autophagy activating kinase.
- ↑
Jean et al. (2015) Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep 16:297-311. (pmid: 25648148) [ PubMed ] [ DOI ] Autophagy, the process for recycling cytoplasm in the lysosome, depends on membrane trafficking. We previously identified Drosophila Sbf as a Rab21 guanine nucleotide exchange factor (GEF) that acts with Rab21 in endosomal trafficking. Here, we show that Sbf/MTMR13 and Rab21 have conserved functions required for starvation-induced autophagy. Depletion of Sbf/MTMR13 or Rab21 blocked endolysosomal trafficking of VAMP8, a SNARE required for autophagosome-lysosome fusion. We show that starvation induces Sbf/MTMR13 GEF and RAB21 activity, as well as their induced binding to VAMP8 (or closest Drosophila homolog, Vamp7). MTMR13 is required for RAB21 activation, VAMP8 interaction and VAMP8 endolysosomal trafficking, defining a novel GEF-Rab-effector pathway. These results identify starvation-responsive endosomal regulators and trafficking that tunes membrane demands with changing autophagy status.
- ↑
Wang et al. (2018) Biomechanical Control of Lysosomal Secretion Via the VAMP7 Hub: A Tug-of-War between VARP and LRRK1. iScience 4:127-143. (pmid: 30240735) [ PubMed ] [ DOI ] The rigidity of the cell environment can vary tremendously between tissues and in pathological conditions. How this property may affect intracellular membrane dynamics is still largely unknown. Here, using atomic force microscopy, we show that cells deficient in the secretory lysosome v-SNARE VAMP7 are impaired in adaptation to substrate rigidity. Conversely, VAMP7-mediated secretion is stimulated by more rigid substrate and this regulation depends on the Longin domain of VAMP7. We further find that the Longin domain binds the kinase and retrograde trafficking adaptor LRRK1 and that LRRK1 negatively regulates VAMP7-mediated exocytosis. Conversely, VARP, a VAMP7- and kinesin 1-interacting protein, further controls the availability for secretion of peripheral VAMP7 vesicles and response of cells to mechanical constraints. LRRK1 and VARP interact with VAMP7 in a competitive manner. We propose a mechanism whereby biomechanical constraints regulate VAMP7-dependent lysosomal secretion via LRRK1 and VARP tug-of-war control of the peripheral pool of secretory lysosomes.
- ↑
Spessott et al. (2017) Syntaxin 4 mediates endosome recycling for lytic granule exocytosis in cytotoxic T-lymphocytes. Traffic 18:442-452. (pmid: 28471021) [ PubMed ] [ DOI ] Adaptive and innate immunity utilize the perforin-killing pathway to eliminate virus-infected or cancer cells. Cytotoxic T-lymphocytes (CTLs) and natural killer cells mediate this process by releasing toxic proteins at the contact area with target cells known as immunological synapse (IS). Formation of a stable IS and exocytosis of toxic proteins requires persistent fusion of Rab11a recycling endosomes with the plasma membrane (PM) that may assure the delivery of key effector proteins. Despite the importance of the recycling endosomal compartment, the membrane fusion proteins that control this process at the IS remain elusive. Here, by performing knockdown experiments we found that syntaxin 4 (STX4) is necessary for cytotoxic activity and CD107a degranulation against target cells in a similar fashion to syntaxin 11, which is involved in lytic granule (LG) exocytosis and immunodeficiency when it is mutated. Using total internal reflection fluorescent microscopy we identified that STX4 mediates fusion of EGFP-Rab11a vesicles at the IS. Immunoprecipitation experiments in lysates of activated CTLs indicate that endogenous STX4 may drive this fusion step by interacting with cognate proteins: Munc18-3/SNAP23/VAMP7 and/or VAMP8. These results reveal the role of STX4 in mediating fusion of Rab11a endosomes upstream of lytic granules (LGs) exocytosis and further demonstrate the importance of this pathway in controlling CTL-mediated cytotoxicity.
- ↑
Daste et al. (2015) Structure and function of longin SNAREs. J Cell Sci 128:4263-72. (pmid: 26567219) [ PubMed ] [ DOI ] Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins constitute the core membrane fusion machinery of intracellular transport and intercellular communication. A little more than ten years ago, it was proposed that the long N-terminal domain of a subset of SNAREs, henceforth called the longin domain, could be a crucial regulator with multiple functions in membrane trafficking. Structural, biochemical and cell biology studies have now produced a large set of data that support this hypothesis and indicate a role for the longin domain in regulating the sorting and activity of SNAREs. Here, we review the first decade of structure-function data on the three prototypical longin SNAREs: Ykt6, VAMP7 and Sec22b. We will, in particular, highlight the conserved molecular mechanisms that allow longin domains to fold back onto the fusion-inducing SNARE coiled-coil domain, thereby inhibiting membrane fusion, and describe the interactions of longin SNAREs with proteins that regulate their intracellular sorting. This dual function of the longin domain in regulating both the membrane localization and membrane fusion activity of SNAREs points to its role as a key regulatory module of intracellular trafficking.
- ↑ 20.0 20.1 20.2 20.3
Zhao et al. (2015) Mechanistic insights into the recycling machine of the SNARE complex. Nature 518:61-7. (pmid: 25581794) [ PubMed ] [ DOI ] Evolutionarily conserved SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptors) proteins form a complex that drives membrane fusion in eukaryotes. The ATPase NSF (N-ethylmaleimide sensitive factor), together with SNAPs (soluble NSF attachment protein), disassembles the SNARE complex into its protein components, making individual SNAREs available for subsequent rounds of fusion. Here we report structures of ATP- and ADP-bound NSF, and the NSF/SNAP/SNARE (20S) supercomplex determined by single-particle electron cryomicroscopy at near-atomic to sub-nanometre resolution without imposing symmetry. Large, potentially force-generating, conformational differences exist between ATP- and ADP-bound NSF. The 20S supercomplex exhibits broken symmetry, transitioning from six-fold symmetry of the NSF ATPase domains to pseudo four-fold symmetry of the SNARE complex. SNAPs interact with the SNARE complex with an opposite structural twist, suggesting an unwinding mechanism. The interfaces between NSF, SNAPs, and SNAREs exhibit characteristic electrostatic patterns, suggesting how one NSF/SNAP species can act on many different SNARE complexes.
- ↑ 21.0 21.1
Fogel et al. (2013) Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol 33:3675-88. (pmid: 23878393) [ PubMed ] [ DOI ] During autophagy, a double membrane envelops cellular material for trafficking to the lysosome. Human beclin-1 and its yeast homologue, Atg6/Vps30, are scaffold proteins bound in a lipid kinase complex with multiple cellular functions, including autophagy. Several different Atg6 complexes exist, with an autophagy-specific form containing Atg14. However, the roles of Atg14 and beclin-1 in the activation of this complex remain unclear. We here addressed the mechanism of beclin-1 complex activation and reveal two critical steps in this pathway. First, we identified a unique domain in beclin-1, conserved in the yeast homologue Atg6, which is involved in membrane association and, unexpectedly, controls autophagosome size and number in yeast. Second, we demonstrated that human Atg14 is critical in controlling an autophagy-dependent phosphorylation of beclin-1. We map these novel phosphorylation sites to serines 90 and 93 and demonstrate that phosphorylation at these sites is necessary for maximal autophagy. These results help clarify the mechanism of beclin-1 and Atg14 during autophagy.
- ↑ 22.0 22.1 22.2
Diao et al. (2015) ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520:563-6. (pmid: 25686604) [ PubMed ] [ DOI ] Autophagy, an important catabolic pathway implicated in a broad spectrum of human diseases, begins by forming double membrane autophagosomes that engulf cytosolic cargo and ends by fusing autophagosomes with lysosomes for degradation. Membrane fusion activity is required for early biogenesis of autophagosomes and late degradation in lysosomes. However, the key regulatory mechanisms of autophagic membrane tethering and fusion remain largely unknown. Here we report that ATG14 (also known as beclin-1-associated autophagy-related key regulator (Barkor) or ATG14L), an essential autophagy-specific regulator of the class III phosphatidylinositol 3-kinase complex, promotes membrane tethering of protein-free liposomes, and enhances hemifusion and full fusion of proteoliposomes reconstituted with the target (t)-SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) syntaxin 17 (STX17) and SNAP29, and the vesicle (v)-SNARE VAMP8 (vesicle-associated membrane protein 8). ATG14 binds to the SNARE core domain of STX17 through its coiled-coil domain, and stabilizes the STX17-SNAP29 binary t-SNARE complex on autophagosomes. The STX17 binding, membrane tethering and fusion-enhancing activities of ATG14 require its homo-oligomerization by cysteine repeats. In ATG14 homo-oligomerization-defective cells, autophagosomes still efficiently form but their fusion with endolysosomes is blocked. Recombinant ATG14 homo-oligomerization mutants also completely lose their ability to promote membrane tethering and to enhance SNARE-mediated fusion in vitro. Taken together, our data suggest an autophagy-specific membrane fusion mechanism in which oligomeric ATG14 directly binds to STX17-SNAP29 binary t-SNARE complex on autophagosomes and primes it for VAMP8 interaction to promote autophagosome-endolysosome fusion.
- ↑
Baker & Hughson (2016) Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol 17:465-79. (pmid: 27301672) [ PubMed ] [ DOI ] Intracellular membrane fusion is mediated in most cases by membrane-bridging complexes of soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). However, the assembly of such complexes in vitro is inefficient, and their uncatalysed disassembly is undetectably slow. Here, we focus on the cellular machinery that orchestrates assembly and disassembly of SNARE complexes, thereby regulating processes ranging from vesicle trafficking to organelle fusion to neurotransmitter release. Rapid progress is being made on many fronts, including the development of more realistic cell-free reconstitutions, the application of single-molecule biophysics, and the elucidation of X-ray and high-resolution electron microscopy structures of the SNARE assembly and disassembly machineries 'in action'.
- ↑
Pu et al. (2015) BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell 33:176-88. (pmid: 25898167) [ PubMed ] [ DOI ] The positioning of lysosomes within the cytoplasm is emerging as a critical determinant of many lysosomal functions. Here we report the identification of a multisubunit complex named BORC that regulates lysosome positioning. BORC comprises eight subunits, some of which are shared with the BLOC-1 complex involved in the biogenesis of lysosome-related organelles, and the others of which are products of previously uncharacterized open reading frames. BORC associates peripherally with the lysosomal membrane, where it functions to recruit the small GTPase Arl8. This initiates a chain of interactions that promotes the kinesin-dependent movement of lysosomes toward the plus ends of microtubules in the peripheral cytoplasm. Interference with BORC or other components of this pathway results in collapse of the lysosomal population into the pericentriolar region. In turn, this causes reduced cell spreading and migration, highlighting the importance of BORC-dependent centrifugal transport for non-degradative functions of lysosomes.
- ↑ 25.0 25.1 25.2
Cite error: InvalidCheng & Smith (2019) Biological Membrane Organization and Cellular Signaling. Chem Rev 119:5849-5880. (pmid: 30747526) [ PubMed ] [ DOI ] To execute their many vital functions, cell membranes are highly organized. Here, we review how membrane structure shapes signal transduction across membranes. Recent experimental and computational advances have shed significant light on mechanisms linking the function of membrane signaling proteins to the composition and physical properties of the membrane lateral structures in which they are embedded. We provide an overview of the structural characteristics of membranes containing heterogeneous mixtures of lipids and other molecules and summarize work on "raft" domains in model and cell membranes, as determined by microscopy, spectroscopy, neutron scattering, and computer simulations. We discuss the principles of partitioning of proteins into membranes and how the structure, dynamics, and function of membrane-embedded and peripheral proteins can be modulated by specific membrane components and physical properties of membranes and raft domains. Finally, we discuss challenges and future directions toward a molecular-level understanding of how membrane organization gives rise to various context-dependent cellular signaling.
<ref>tag; name "Cheng2019" defined multiple times with different content Cite error: Invalid<ref>tag; name "Cheng2019" defined multiple times with different content - ↑ 26.0 26.1 26.2
Naufer et al. (2018) pH of endophagosomes controls association of their membranes with Vps34 and PtdIns(3)P levels. J Cell Biol 217:329-346. (pmid: 29089378) [ PubMed ] [ DOI ] Phagocytosis of filamentous bacteria occurs through tubular phagocytic cups (tPCs) and takes many minutes to engulf these filaments into phagosomes. Contravening the canonical phagocytic pathway, tPCs mature by fusing with endosomes. Using this model, we observed the sequential recruitment of early and late endolysosomal markers to the elongating tPCs. Surprisingly, the regulatory early endosomal lipid phosphatidylinositol-3-phosphate (PtdIns(3)P) persists on tPCs as long as their luminal pH remains neutral. Interestingly, by manipulating cellular pH, we determined that PtdIns(3)P behaves similarly in canonical phagosomes as well as endosomes. We found that this is the product of a pH-based mechanism that induces the dissociation of the Vps34 class III phosphatidylinositol-3-kinase from these organelles as they acidify. The detachment of Vps34 stops the production of PtdIns(3)P, allowing for the turnover of this lipid by PIKfyve. Given that PtdIns(3)P-dependent signaling is important for multiple cellular pathways, this mechanism for pH-dependent regulation of Vps34 could be at the center of many PtdIns(3)P-dependent cellular processes.
- ↑
Zhong et al. (2014) Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J Biol Chem 289:26021-37. (pmid: 25086043) [ PubMed ] [ DOI ] Autophagy is a tightly regulated lysosomal degradation pathway for maintaining cellular homeostasis and responding to stresses. Beclin 1 and its interacting proteins, including the class III phosphatidylinositol-3 kinase Vps34, play crucial roles in autophagy regulation in mammals. We identified nuclear receptor binding factor 2 (Nrbf2) as a Beclin 1-interacting protein from Becn1(-/-);Becn1-EGFP/+ mouse liver and brain. We also found that Nrbf2-Beclin 1 interaction required the N terminus of Nrbf2. We next used the human retinal pigment epithelial cell line RPE-1 as a model system and showed that transiently knocking down Nrbf2 by siRNA increased autophagic flux under both nutrient-rich and starvation conditions. To investigate the mechanism by which Nrbf2 regulates autophagy, we demonstrated that Nrbf2 interacted and colocalized with Atg14L, suggesting that Nrbf2 is a component of the Atg14L-containing Beclin 1-Vps34 complex. Moreover, ectopically expressed Nrbf2 formed cytosolic puncta that were positive for isolation membrane markers. These results suggest that Nrbf2 is involved in autophagosome biogenesis. Furthermore, we showed that Nrbf2 deficiency led to increased intracellular phosphatidylinositol-3 phosphate levels and diminished Atg14L-Vps34/Vps15 interactions, suggesting that Nrbf2-mediated Atg14L-Vps34/Vps15 interactions likely inhibit Vps34 activity. Therefore, we propose that Nrbf2 may interact with the Atg14L-containing Beclin 1-Vps34 protein complex to modulate protein-protein interactions within the complex, leading to suppression of Vps34 activity, autophagosome biogenesis, and autophagic flux. This work reveals a novel aspect of the intricate mechanism for the Beclin 1-Vps34 protein-protein interaction network to achieve precise control of autophagy.
- ↑
Lindmo et al. (2008) The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy 4:500-6. (pmid: 18326940) [ PubMed ] [ DOI ] Autophagy is involved in cellular clearance of aggregate-prone proteins, thereby having a cytoprotective function. Studies in yeast have shown that the PI 3-kinase Vps34 and its regulatory protein kinase Vps15 are important for autophagy, but the possible involvement of these proteins in autophagy in a multicellular animal has not been addressed genetically. Here, we have created a Drosophila deletion mutant of vps15 and studied its role in autophagy and aggregate clearance. Homozygous Deltavps15 Drosophila died at the early L3 larval stage. Using GFP-Atg8a as an autophagic marker, we employed fluorescence microscopy to demonstrate that fat bodies of wild type Drosophila larvae accumulated autophagic structures upon starvation whereas vps15 fat bodies showed no such response. Likewise, electron microscopy revealed starvation-induced autophagy in gut cells from wild type but not Deltavps15 larvae. Fluorescence microscopy showed that Deltavps15 mutant tissues accumulated profiles that were positive for ubiquitin and Ref(2)P, the Drosophila homolog of the sequestosome marker SQSTM1/p62. Biochemical fractionation and Western blotting showed that these structures were partially detergent insoluble, and immuno-electron microscopy further demonstrated the presence of Ref(2)P positive membrane free protein aggregates. These results provide the first genetic evidence for a function of Vps15 in autophagy in multicellular organisms and suggest that the Vps15-containing PI 3-kinase complex may play an important role in clearance of protein aggregates.
- ↑ 29.0 29.1 29.2 29.3
Hasegawa et al. (2016) Autophagosome-lysosome fusion in neurons requires INPP5E, a protein associated with Joubert syndrome. EMBO J 35:1853-67. (pmid: 27340123) [ PubMed ] [ DOI ] Autophagy is a multistep membrane traffic pathway. In contrast to autophagosome formation, the mechanisms underlying autophagosome-lysosome fusion remain largely unknown. Here, we describe a novel autophagy regulator, inositol polyphosphate-5-phosphatase E (INPP5E), involved in autophagosome-lysosome fusion process. In neuronal cells, INPP5E knockdown strongly inhibited autophagy by impairing the fusion step. A fraction of INPP5E is localized to lysosomes, and its membrane anchoring and enzymatic activity are necessary for autophagy. INPP5E decreases lysosomal phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2), one of the substrates of the phosphatase, that counteracts cortactin-mediated actin filament stabilization on lysosomes. Lysosomes require actin filaments on their surface for fusing with autophagosomes. INPP5E is one of the genes responsible for Joubert syndrome, a rare brain abnormality, and mutations found in patients with this disease caused defects in autophagy. Taken together, our data reveal a novel role of phosphoinositide on lysosomes and an association between autophagy and neuronal disease.
- ↑
Hasegawa et al. (2017) PI5P and PI(3,5)P2: Minor, but Essential Phosphoinositides. Cell Struct Funct 42:49-60. (pmid: 28302928) [ PubMed ] [ DOI ] In most eukaryotes, phosphoinositides (PIs) have crucial roles in multiple cellular functions. Although the cellular levels of phosphatidylinositol 5-phosphate (PI5P) and phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) are extremely low relative to some other PIs, emerging evidence demonstrates that both lipids are crucial for the endocytic pathway, intracellular signaling, and adaptation to stress. Mutations that causes defects in the biosynthesis of PI5P and PI(3,5)P2 are linked to human diseases including neurodegenerative disorders. Here, we review recent findings on cellular roles of PI5P and PI(3,5)P2, as well as the pathophysiological importance of these lipids.Key words: Phosphoinositides, Membrane trafficking, Endocytosis, Vacuoles/Lysosomes, Fab1/PIKfyve.
- ↑
Bucki et al. (2019) Lateral distribution of phosphatidylinositol 4,5-bisphosphate in membranes regulates formin- and ARP2/3-mediated actin nucleation. J Biol Chem 294:4704-4722. (pmid: 30692198) [ PubMed ] [ DOI ] Spatial and temporal control of actin polymerization is fundamental for many cellular processes, including cell migration, division, vesicle trafficking, and response to agonists. Many actin-regulatory proteins interact with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) and are either activated or inactivated by local PI(4,5)P2 concentrations that form transiently at the cytoplasmic face of cell membranes. The molecular mechanisms of these interactions and how the dozens of PI(4,5)P2-sensitive actin-binding proteins are selectively recruited to membrane PI(4,5)P2 pools remains undefined. Using a combination of biochemical, imaging, and cell biologic studies, combined with molecular dynamics and analytical theory, we test the hypothesis that the lateral distribution of PI(4,5)P2 within lipid membranes and native plasma membranes alters the capacity of PI(4,5)P2 to nucleate actin assembly in brain and neutrophil extracts and show that activities of formins and the Arp2/3 complex respond to PI(4,5)P2 lateral distribution. Simulations and analytical theory show that cholesterol promotes the cooperative interaction of formins with multiple PI(4,5)P2 headgroups in the membrane to initiate actin nucleation. Masking PI(4,5)P2 with neomycin or disrupting PI(4,5)P2 domains in the plasma membrane by removing cholesterol decreases the ability of these membranes to nucleate actin assembly in cytoplasmic extracts.
- ↑
Manca et al. (2019) SNARE machinery is optimized for ultrafast fusion. Proc Natl Acad Sci U.S.A 116:2435-2442. (pmid: 30700546) [ PubMed ] [ DOI ] SNARE proteins zipper to form complexes (SNAREpins) that power vesicle fusion with target membranes in a variety of biological processes. A single SNAREpin takes about 1 s to fuse two bilayers, yet a handful can ensure release of neurotransmitters from synaptic vesicles much faster: in a 10th of a millisecond. We propose that, similar to the case of muscle myosins, the ultrafast fusion results from cooperative action of many SNAREpins. The coupling originates from mechanical interactions induced by confining scaffolds. Each SNAREpin is known to have enough energy to overcome the fusion barrier of 25-[Formula: see text]; however, the fusion barrier only becomes relevant when the SNAREpins are nearly completely zippered, and from this state, each SNAREpin can deliver only a small fraction of this energy as mechanical work. Therefore, they have to act cooperatively, and we show that at least three of them are needed to ensure fusion in less than a millisecond. However, to reach the prefusion state collectively, starting from the experimentally observed half-zippered metastable state, the SNAREpins have to mechanically synchronize, which takes more time as the number of SNAREpins increases. Incorporating this somewhat counterintuitive idea in a simple coarse-grained model results in the prediction that there should be an optimum number of SNAREpins for submillisecond fusion: three to six over a wide range of parameters. Interestingly, in situ cryoelectron microscope tomography has very recently shown that exactly six SNAREpins participate in the fusion of each synaptic vesicle. This number is in the range predicted by our theory.
- ↑ Unpublished.
- ↑ 34.0 34.1
Suzuki et al. (2014) Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure 22:47-58. (pmid: 24290141) [ PubMed ] [ DOI ] Autophagy is a bulk degradation pathway that removes cytosolic materials to maintain cellular homeostasis. The autophagy-related gene 13 (Atg13) and microtubule associate protein 1 light chain 3 (LC3) proteins are required for autophagosome formation. We demonstrate that each of the human LC3 isoforms (LC3A, LC3B, and LC3C) interacts with Atg13 via the LC3 interacting region (LIR) of Atg13. Using X-ray crystallography, we solved the macromolecular structures of LC3A and LC3C, along with the complex structures of the LC3 isoforms with the Atg13 LIR. Together, our structural and binding analyses reveal that the side-chain of Lys49 of LC3 acts as a gatekeeper to regulate binding of the LIR. We verified this observation by mutation of Lys49 in LC3A, which significantly reduces LC3A positive puncta formation in cultured cells. Our results suggest that specific affinity of the LC3 isoforms to the Atg13 LIR is required for proper autophagosome formation.
- ↑
Zachari & Ganley (2017) The mammalian ULK1 complex and autophagy initiation. Essays Biochem 61:585-596. (pmid: 29233870) [ PubMed ] [ DOI ] Autophagy is a vital lysosomal degradation pathway that serves as a quality control mechanism. It rids the cell of damaged, toxic or excess cellular components, which if left to persist could be detrimental to the cell. It also serves as a recycling pathway to maintain protein synthesis under starvation conditions. A key initial event in autophagy is formation of the autophagosome, a unique double-membrane organelle that engulfs the cytosolic cargo destined for degradation. This step is mediated by the serine/threonine protein kinase ULK1 (unc-51-like kinase 1), which functions in a complex with at least three protein partners: FIP200 (focal adhesion kinase family interacting protein of 200 kDa), ATG (autophagy-related protein) 13 (ATG13), and ATG101. In this artcile, we focus on the regulation of the ULK1 complex during autophagy initiation. The complex pattern of upstream pathways that converge on ULK1 suggests that this complex acts as a node, converting multiple signals into autophagosome formation. Here, we review our current understanding of this regulation and in turn discuss what happens downstream, once the ULK1 complex becomes activated.
![]() This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.
This copyrighted material is licensed under a Creative Commons Attribution 4.0 International License. Follow the link to learn more.