BIO Assignment Week 12
Old materials repository
No more ...
Note: this is not an assignment but material left over from previous iterations of the course. </div-->
Contents
- 1 Some material that is left-over from the major rearrangement
- 2 R code: load alignment and compute information scores
- 3 Calculating conservation scores
- 4 Stereo vision
- 5 Programming R code
- 6 The PDB
- 7 Introduction
- 8 Coloring by conservation
- 9 Modelling the Ankyrin Domain Section
- 10 From Homology Modeling - Modeling alternative binding modes
- 11 Interpretation
- 12 EC
- 13 GO
- 14 Introduction
- 15 GO
- 16 Links and resources
- 17 Footnotes and references
- 18 Ask, if things don't work for you!
Some material that is left-over from the major rearrangement
Computing alignments
try two MSA's algorithms and load them in Jalview. Locally: which one do you prefer? Modify the consensus. Annotate domains.
The EBI has a very convenient page to access a number of MSA algorithms. This is especially convenient when you want to compare, e.g. T-Coffee and Muscle and MAFFT results to see which regions of your alignment are robust. You could use any of these tools, just paste your sequences into a Webform, download the results and load into Jalview. Easy.
But even easier is to calculate the alignments directly from Jalview. available. (Not today. Bummer.)
No. Claculate an external alignment and import.
- Calculate a MAFFT alignment using the Jalview Web service option
Task:
- In Jalview, select Web Service → Alignment → MAFFT with defaults.... The alignment is calculated in a few minutes and displayed in a new window.
- Calculate a MAFFT alignment when the Jalview Web service is NOT available
Task:
- In Jalview, select File → Output to Textbox → FASTA
- Copy the sequences.
- Navigate to the MAFFT Input form at the EBI.
- Paste your sequences into the form.
- Click on Submit.
- Close the Jalview sequence window and either save your MAFFT alignment to file and load in Jalview, or simply 'File → Input Alignment → from Textbox, paste and click New Window.
In any case, you should now have an alignment.
Task:
- Choose Colour → Hydrophobicity and → by Conservation. Then adjust the slider left or right to see which columns are highly conserved. You will notice that the Swi6 sequence that was supposed to align only to the ankyrin domains was in fact aligned to other parts of the sequence as well. This is one part of the MSA that we will have to correct manually and a common problem when aligning sequences of different lengths.
R code: load alignment and compute information scores
As discussed in the lecture, Shannon information is calculated as the difference between expected and observed entropy, where entropy is the negative sum over probabilities times the log of those probabilities:
*a review of regex range characters +?*{min,max}, and greedy.
*build an AT-hook motif matcher https://en.wikipedia.org/wiki/AT-hook
Here we compute Shannon information scores for aligned positions of the APSES domain, and plot the values in R. You can try this with any part of your alignment, but I have used only the aligned residues for the APSES domain for my example. This is a good choice for a first try, since there are (almost) no gaps.
Task:
- Export only the sequences of the aligned APSES domains to a file on your computer, in FASTA format as explained below. You could call this:
Mbp1_All_APSES.fa.- Use your mouse and clik and drag to select the aligned APSES domains in the alignment window.
- Copy your selection to the clipboard.
- Use the main menu (not the menu of your alignment window) and select File → Input alignment → from Textbox; paste the selection into the textbox and click New Window.
- Use File → save as to save the aligned siequences in multi-FASTA format under the filename you want in your R project directory.
- Explore the R-code below. Be sure that you understand it correctly. Note that this code does not implement any sampling bias correction, so positions with large numbers of gaps will receive artificially high scores (the alignment looks like the gap charecter were a conserved character).
# CalculateInformation.R
# Calculate Shannon information for positions in a multiple sequence alignment.
# Requires: an MSA in multi FASTA format
# It is good practice to set variables you might want to change
# in a header block so you don't need to hunt all over the code
# for strings you need to update.
#
setwd("/your/R/working/directory")
mfa <- "MBP1_All_APSES.fa"
# ================================================
# Read sequence alignment fasta file
# ================================================
# read MFA datafile using seqinr function read.fasta()
library(seqinr)
tmp <- read.alignment(mfa, format="fasta")
MSA <- as.matrix(tmp) # convert the list into a characterwise matrix
# with appropriate row and column names using
# the seqinr function as.matrix.alignment()
# You could have a look under the hood of this
# function to understand beter how to convert a
# list into something else ... simply type
# "as.matrix.alignment" - without the parentheses
# to retrieve the function source code (as for any
# function btw).
### Explore contents of and access to the matrix of sequences
MSA
MSA[1,]
MSA[,1]
length(MSA[,1])
# ================================================
# define function to calculate entropy
# ================================================
entropy <- function(v) { # calculate shannon entropy for the aa vector v
# Note: we are not correcting for small sample sizes
# here. Thus if there are a large number of gaps in
# the alignment, this will look like small entropy
# since only a few amino acids are present. In the
# extreme case: if a position is only present in
# one sequence, that one amino acid will be treated
# as 100% conserved - zero entropy. Sampling error
# corrections are discussed eg. in Schneider et al.
# (1986) JMB 188:414
l <- length(v)
a <- rep(0, 21) # initialize a vector with 21 elements (20 aa plus gap)
# the set the name of each row to the one letter
# code. Through this, we can access a row by its
# one letter code.
names(a) <- unlist(strsplit("acdefghiklmnpqrstvwy-", ""))
for (i in 1:l) { # for the whole vector of amino acids
c <- v[i] # retrieve the character
a[c] <- a[c] + 1 # increment its count by one
} # note: we could also have used the table() function for this
tot <- sum(a) - a["-"] # calculate number of observed amino acids
# i.e. subtract gaps
a <- a/tot # frequency is observations of one amino acid
# divided by all observations. We assume that
# frequency equals probability.
a["-"] <- 0
for (i in 1:length(a)) {
if (a[i] != 0) { # if a[i] is not zero, otherwise leave as is.
# By definition, 0*log(0) = 0 but R calculates
# this in parts and returns NaN for log(0).
a[i] <- a[i] * (log(a[i])/log(2)) # replace a[i] with
# p(i) log_2(p(i))
}
}
return(-sum(a)) # return Shannon entropy
}
# ================================================
# calculate entropy for reference distribution
# (from UniProt, c.f. Assignment 2)
# ================================================
refData <- c(
"A"=8.26,
"Q"=3.93,
"L"=9.66,
"S"=6.56,
"R"=5.53,
"E"=6.75,
"K"=5.84,
"T"=5.34,
"N"=4.06,
"G"=7.08,
"M"=2.42,
"W"=1.08,
"D"=5.45,
"H"=2.27,
"F"=3.86,
"Y"=2.92,
"C"=1.37,
"I"=5.96,
"P"=4.70,
"V"=6.87
)
### Calculate the entropy of this distribution
H.ref <- 0
for (i in 1:length(refData)) {
p <- refData[i]/sum(refData) # convert % to probabilities
H.ref <- H.ref - (p * (log(p)/log(2)))
}
# ================================================
# calculate information for each position of
# multiple sequence alignment
# ================================================
lAli <- dim(MSA)[2] # length of row in matrix is second element of dim(<matrix>).
I <- rep(0, lAli) # initialize result vector
for (i in 1:lAli) {
I[i] = H.ref - entropy(MSA[,i]) # I = H_ref - H_obs
}
### evaluate I
I
quantile(I)
hist(I)
plot(I)
# you can see that we have quite a large number of columns with the same,
# high value ... what are these?
which(I > 4)
MSA[,which(I > 4)]
# And what is in the columns with low values?
MSA[,which(I < 1.5)]
# ===================================================
# plot the information
# (c.f. Assignment 5, see there for explanations)
# ===================================================
IP <- (I-min(I))/(max(I) - min(I) + 0.0001)
nCol <- 15
IP <- floor(IP * nCol) + 1
spect <- colorRampPalette(c("#DD0033", "#00BB66", "#3300DD"), bias=0.6)(nCol)
# lets set the information scores from single informations to grey. We
# change the highest level of the spectrum to grey.
#spect[nCol] <- "#CCCCCC"
Icol <- vector()
for (i in 1:length(I)) {
Icol[i] <- spect[ IP[i] ]
}
plot(1,1, xlim=c(0, lAli), ylim=c(-0.5, 5) ,
type="n", bty="n", xlab="position in alignment", ylab="Information (bits)")
# plot as rectangles: height is information and color is coded to information
for (i in 1:lAli) {
rect(i, 0, i+1, I[i], border=NA, col=Icol[i])
}
# As you can see, some of the columns reach very high values, but they are not
# contiguous in sequence. Are they contiguous in structure? We will find out in
# a later assignment, when we map computed values to structure.
Calculating conservation scores
Task:
- Study this code carefully, execute it, section by section and make sure you understand all of it. Ask on the list if anything is not clear.
# BiostringsExample.R
# Short tutorial on sequence alignment with the Biostrings package.
# Boris Steipe for BCH441, 2013 - 2014
#
setwd("~/path/to/your/R_files/")
setwd("~/Documents/07.TEACHING/37-BCH441 Bioinformatics 2014/05-Materials/Assignment_5 data")
# Biostrings is a package within the bioconductor project.
# bioconducter packages have their own installation system,
# they are normally not installed via CRAN.
# First, you load the BioConductor installer...
source("http://bioconductor.org/biocLite.R")
# Then you can install the Biostrings package and all of its dependencies.
biocLite("Biostrings")
# ... and load the library.
library(Biostrings)
# Some basic (technical) information is available ...
library(help=Biostrings)
# ... but for more in depth documentation, use the
# so called "vignettes" that are provided with every R package.
browseVignettes("Biostrings")
# In this code, we mostly use functions that are discussed in the
# pairwise alignement vignette.
# Read in two fasta files - you will need to edit this for YFO
sacce <- readAAStringSet("mbp1-sacce.fa", format="fasta")
# "USTMA" is used only as an example here - modify for YFO :-)
ustma <- readAAStringSet("mbp1-ustma.fa", format="fasta")
sacce
names(sacce)
names(sacce) <- "Mbp1 SACCE"
names(ustma) <- "Mbp1 USTMA" # Example only ... modify for YFO
width(sacce)
as.character(sacce)
# Biostrings takes a sophisticated approach to sequence alignment ...
?pairwiseAlignment
# ... but the use in practice is quite simple:
ali <- pairwiseAlignment(sacce, ustma, substitutionMatrix = "BLOSUM50")
ali
pattern(ali)
subject(ali)
writePairwiseAlignments(ali)
p <- aligned(pattern(ali))
names(p) <- "Mbp1 SACCE aligned"
s <- aligned(subject(ali))
names(s) <- "Mbp1 USTMA aligned"
# don't overwrite your EMBOSS .fal files
writeXStringSet(p, "mbp1-sacce.R.fal", append=FALSE, format="fasta")
writeXStringSet(s, "mbp1-ustma.R.fal", append=FALSE, format="fasta")
# Done.- Compare the alignments you received from the EMBOSS server, and that you computed using R. Are they approximately the same? Exactly? You did use different matrices and gap parameters, so minor differences are to be expected. But by and large you should get the same alignments.
We will now use the aligned sequences to compute a graphical display of alignment quality.
Task:
- Study this code carefully, execute it, section by section and make sure you understand all of it. Ask on the list if anything is not clear.
# aliScore.R
# Evaluating an alignment with a sliding window score
# Boris Steipe, October 2012. Update October 2013
setwd("~/path/to/your/R_files/")
# Scoring matrices can be found at the NCBI.
# ftp://ftp.ncbi.nih.gov/blast/matrices/BLOSUM62
# It is good practice to set variables you might want to change
# in a header block so you don't need to hunt all over the code
# for strings you need to update.
#
fa1 <- "mbp1-sacce.R.fal"
fa2 <- "mbp1-ustma.R.fal"
code1 <- "SACCE"
code2 <- "USTMA"
mdmFile <- "BLOSUM62.mdm"
window <- 9 # window-size (should be an odd integer)
# ================================================
# Read data files
# ================================================
# read fasta datafiles using seqinr function read.fasta()
install.packages("seqinr")
library(seqinr)
tmp <- unlist(read.fasta(fa1, seqtype="AA", as.string=FALSE, seqonly=TRUE))
seq1 <- unlist(strsplit(as.character(tmp), split=""))
tmp <- unlist(read.fasta(fa2, seqtype="AA", as.string=FALSE, seqonly=TRUE))
seq2 <- unlist(strsplit(as.character(tmp), split=""))
if (length(seq1) != length(seq2)) {
print("Error: Sequences have unequal length!")
}
lSeq <- length(seq1)
# ================================================
# Read scoring matrix
# ================================================
MDM <- read.table(mdmFile, skip=6)
# This is a dataframe. Study how it can be accessed:
MDM
MDM[1,]
MDM[,1]
MDM[5,5] # Cys-Cys
MDM[20,20] # Val-Val
MDM[,"W"] # the tryptophan column
MDM["R","W"] # Arg-Trp pairscore
MDM["W","R"] # Trp-Arg pairscore: pairscores are symmetric
colnames(MDM) # names of columns
rownames(MDM) # names of rows
colnames(MDM)[3] # third column
rownames(MDM)[12] # twelfth row
# change the two "*" names to "-" so we can use them to score
# indels of the alignment. This is a bit of a hack, since this
# does not reflect the actual indel penalties (which is, as you)
# remember from your lectures, calculated as a gap opening
# + gap extension penalty; it can't be calculated in a pairwise
# manner) EMBOSS defaults for BLODSUM62 are opening -10 and
# extension -0.5 i.e. a gap of size 3 (-11.5) has approximately
# the same penalty as a 3-character score of "-" matches (-12)
# so a pairscore of -4 is not entirely unreasonable.
colnames(MDM)[24]
rownames(MDM)[24]
colnames(MDM)[24] <- "-"
rownames(MDM)[24] <- "-"
colnames(MDM)[24]
rownames(MDM)[24]
MDM["Q", "-"]
MDM["-", "D"]
# so far so good.
# ================================================
# Tabulate pairscores for alignment
# ================================================
# It is trivial to create a pairscore vector along the
# length of the aligned sequences.
PS <- vector()
for (i in 1:lSeq) {
aa1 <- seq1[i]
aa2 <- seq2[i]
PS[i] = MDM[aa1, aa2]
}
PS
# The same vector could be created - albeit perhaps not so
# easy to understand - with the expression ...
MDM[cbind(seq1,seq2)]
# ================================================
# Calculate moving averages
# ================================================
# In order to evaluate the alignment, we will calculate a
# sliding window average over the pairscores. Somewhat surprisingly
# R doesn't (yet) have a native function for moving averages: options
# that are quoted are:
# - rollmean() in the "zoo" package http://rss.acs.unt.edu/Rdoc/library/zoo/html/rollmean.html
# - MovingAverages() in "TTR" http://rss.acs.unt.edu/Rdoc/library/TTR/html/MovingAverages.html
# - ma() in "forecast" http://robjhyndman.com/software/forecast/
# But since this is easy to code, we shall implement it ourselves.
PSma <- vector() # will hold the averages
winS <- floor(window/2) # span of elements above/below the centre
winC <- winS+1 # centre of the window
# extend the vector PS with zeros (virtual observations) above and below
PS <- c(rep(0, winS), PS , rep(0, winS))
# initialize the window score for the first position
winScore <- sum(PS[1:window])
# write the first score to PSma
PSma[1] <- winScore
# Slide the window along the sequence, and recalculate sum()
# Loop from the next position, to the last position that does not exceed the vector...
for (i in (winC + 1):(lSeq + winS)) {
# subtract the value that has just dropped out of the window
winScore <- winScore - PS[(i-winS-1)]
# add the value that has just entered the window
winScore <- winScore + PS[(i+winS)]
# put score into PSma
PSma[i-winS] <- winScore
}
# convert the sums to averages
PSma <- PSma / window
# have a quick look at the score distributions
boxplot(PSma)
hist(PSma)
# ================================================
# Plot the alignment scores
# ================================================
# normalize the scores
PSma <- (PSma-min(PSma))/(max(PSma) - min(PSma) + 0.0001)
# spread the normalized values to a desired range, n
nCol <- 10
PSma <- floor(PSma * nCol) + 1
# Assign a colorspectrum to a vector (with a bit of colormagic,
# don't worry about that for now). Dark colors are poor scores,
# "hot" colors are high scores
spect <- colorRampPalette(c("black", "red", "yellow", "white"), bias=0.4)(nCol)
# Color is an often abused aspect of plotting. One can use color to label
# *quantities* or *qualities*. For the most part, our pairscores measure amino
# acid similarity. That is a quantity and with the spectrum that we just defined
# we associte the measured quantities with the color of a glowing piece
# of metal: we start with black #000000, then first we ramp up the red
# (i.e. low-energy) part of the visible spectrum to red #FF0000, then we
# add and ramp up the green spectrum giving us yellow #FFFF00 and finally we
# add blue, giving us white #FFFFFF. Let's have a look at the spectrum:
s <- rep(1, nCol)
barplot(s, col=spect, axes=F, main="Color spectrum")
# But one aspect of our data is not quantitatively different: indels.
# We valued indels with pairscores of -4. But indels are not simply poor alignment,
# rather they are non-alignment. This means stretches of -4 values are really
# *qualitatively* different. Let's color them differently by changing the lowest
# level of the spectrum to grey.
spect[1] <- "#CCCCCC"
barplot(s, col=spect, axes=F, main="Color spectrum")
# Now we can display our alignment score vector with colored rectangles.
# Convert the integers in PSma to color values from spect
PScol <- vector()
for (i in 1:length(PSma)) {
PScol[i] <- spect[ PSma[i] ] # this is how a value from PSma is used as an index of spect
}
# Plot the scores. The code is similar to the last assignment.
# Create an empty plot window of appropriate size
plot(1,1, xlim=c(-100, lSeq), ylim=c(0, 2) , type="n", yaxt="n", bty="n", xlab="position in alignment", ylab="")
# Add a label to the left
text (-30, 1, adj=1, labels=c(paste("Mbp1:\n", code1, "\nvs.\n", code2)), cex=0.9 )
# Loop over the vector and draw boxes without border, filled with color.
for (i in 1:lSeq) {
rect(i, 0.9, i+1, 1.1, border=NA, col=PScol[i])
}
# Note that the numbers along the X-axis are not sequence numbers, but numbers
# of the alignment, i.e. sequence number + indel length. That is important to
# realize: if you would like to add the annotations from the last assignment
# which I will leave as an exercise, you need to map your sequence numbering
# into alignment numbering. Let me know in case you try that but need some help.
Stereo vision
Task:
Continue with your stereo practice.
Practice at least ...
- two times daily,
- for 3-5 minutes each session.
- Measure your interocular distance and your fusion distance as explained here on the Student Wiki and add it to the table.
Keep up your practice throughout the course. Once again: do not go through your practice sessions mechanically. If you are not making constant progress in your practice sessions, contact me so we can help you on the right track.
Programming R code
First, we will cover essentials of R programming: the fundamental statements that are needed to write programs–conditional expressions and loops, and how to define functions that allow us to use programming code. But let's start with a few more data types of R so we can use the concepts later on: matrices, lists and data frames.
Task:
Please begin by working through the short R - tutorial: matrices section and the following sections on "Lists" and "Data frames".
Note that what we have done here is just the bare minimum on vectors, matrices and lists. The concepts are very generally useful, and there are many useful ways to extract subsets of values. We'll come across these in various parts of R sample code. But unless you read the provided code examples line by line, make sure you understand every single statement and ask if you are not clear about the syntax, this will all be quickly forgotten. Use it or lose it!
R is a full featured programming language and in order to be that, it needs the ability to manipulate Control Flow, i.e. the order in which instructions are executed. By far the most important control flow statement is a conditional branch i.e. the instruction to execute code at alternative points of the program, depending on the presence or absence of a condition. Using such conditional branch instructions, two main types of programming constructs can be realized: conditional expressions and loops.
Conditional expressions
The template for conditional expression in R is:
if( <expression 1> ) {
<statement 1>
}
else if ( <expresssion 2> ) {
<statement 2>
}
else {
<statement 3>
}...where both the else if (...) { ... } and the else (...) block are optional. We have encountered this construct previously, when we assigned the appropriate colors for amino acids in the frequency plot:
if (names(logRatio[i]) == "F") { barColors[i] <- hydrophobic }
else if (names(logRatio[i]) == "G") { barColors[i] <- plain }
[... etc ...]
else { barColors[i] <- plain }Logical expressions
We have to consider the <expression> in a bit more detail: anything that is, or produces, or can be interpreted as, a Boolean TRUE or FALSE value can serve as an expression in a conditional statement.
Task:
Here are some examples. Copy the code to an R script, predict what will happen on in each line and try it out:
# A boolean constant is interpreted as is:
if (TRUE) {print("true")} else {print("false")}
if (FALSE) {print("true")} else {print("false")}
# Strings of "true" and "false" are coerced to their
# Boolean equivalent, but contrary to other programming languages
# arbitrary, non-empty or empty strings are not interpreted.
if ("true") {print("true")} else {print("false")}
if ("false") {print("true")} else {print("false")}
if ("widdershins") {print("true")} else {print("false")}
if ("") {print("true")} else {print("false")}
# All non-zero, defined numbers are TRUE
if (1) {print("true")} else {print("false")}
if (0) {print("true")} else {print("false")}
if (-1) {print("true")} else {print("false")}
if (pi) {print("true")} else {print("false")}
if (NULL) {print("true")} else {print("false")}
if (NA) {print("true")} else {print("false")}
if (NaN) {print("true")} else {print("false")}
if (Inf) {print("true")} else {print("false")}
# functions can return Boolean values
affirm <- function() { return(TRUE) }
deny <- function() { return(FALSE) }
if (affirm()) {print("true")} else {print("false")}
if (deny()) {print("true")} else {print("false")}
# N.B. coercion of Booleans into numbers can be done as well
and is sometimes useful: consider ...
a <- c(TRUE, TRUE, FALSE, TRUE, FALSE)
a
as.numeric(a)
sum(a)
# ... or coercing the other way ...
as.logical(-1:1)Logical operators
To actually write a conditional statement, we have to be able to test a condition and this is what logical operators do. Is something equal to something else? Is it less? Does something exist? Is it a number?
Task:
Here are some examples. Again, predict what will happen ...
TRUE # Just a statement.
# unary operator
! TRUE # NOT ...
# binary operators
FALSE > TRUE # GREATER THAN ...
FALSE < TRUE # ... these are coerced to numbers
FALSE < -1
0 == FALSE # Careful! == compares, = assigns!!!
"x" == "u" # using lexical sort order ...
"x" >= "u"
"x" <= "u"
"x" != "u"
"aa" > "u" # ... not just length, if different.
"abc" < "u"
TRUE | FALSE # OR: TRUE if either is true
TRUE & FALSE # AND: TRUE if both are TRUE
# equality and identity
?identical
a <- c(TRUE)
b <- c(TRUE)
a; b
a == b
identical(a, b)
b <- 1
a; b
a == b
identical(a, b) # Aha: equal, but not identical
# some other useful tests for conditional expressions
?all
?any
?duplicated
?exists
?is.character
?is.factor
?is.integer
?is.null
?is.numeric
?is.unsorted
?is.vector
Loops
Loops allow you to repeat tasks many times over. The template is:
for (<name> in <vector>) {
<statement>
}Task:
Consider the following: Again, copy the code to a script, study it, predict what will happen and then run it.
# simple for loop
for (i in 1:10) {
print(c(i, i^2, i^3))
}
# Compare excution times: one million square roots from a vector ...
n <- 1000000
x <- 1:n
y <- sqrt(x)
# ... or done explicitly in a for-loop
for (i in 1:n) {
y[i] <- sqrt (x[i])
}If you can achieve your result with an R vector expression, it will be faster than using a loop. But sometimes you need to do things explicitly, for example if you need to access intermediate results.
Here is an example to play with loops: a password generator. Passwords are a pain. We need them everywhere, they are frustrating to type, to remember and since the cracking programs are getting smarter they become more and more likely to be broken. Here is a simple password generator that creates random strings with consonant/vowel alterations. These are melodic and easy to memorize, but actually as strong as an 8-character, fully random password that uses all characters of the keyboard such as )He.{2jJ or #h$bB2X^ (which is pretty much unmemorizable). The former is taken from 207 * 77 1015 possibilities, the latter is from 948 ~ 6*1015 possibilities. HIgh-end GPU supported password crackers can test about 109 passwords a second, the passwords generated by this little algorithm would thus take on the order of 106 seconds or eleven days to crack[1]. This is probably good enough to deter a casual attack.
Task:
Copy, study and run ...
# Suggest memorizable passwords
# Below we use the functions:
?nchar
?sample
?substr
?paste
?print
#define a string of consonants ...
con <- "bcdfghjklmnpqrstvwxz"
# ... and a string of of vowels
vow <- "aeiouy"
for (i in 1:10) { # ten sample passwords to choose from ...
pass = rep("", 14) # make an empty character vector
for (j in 1:7) { # seven consonant/vowel pairs to be created ...
k <- sample(1:nchar(con), 1) # pick a random index for consonants ...
ch <- substr(con,k,k) # ... get the corresponding character ...
idx <- (2*j)-1 # ... compute the position (index) of where to put the consonant ...
pass[idx] <- ch # ... and put it in the right spot
# same thing for the vowel, but coded with fewer intermediate assignments
# of results to variables
k <- sample(1:nchar(vow), 1)
pass[(2*j)] <- substr(vow,k,k)
}
print( paste(pass, collapse="") ) # collapse the vector in to a string and print
}Functions
Finally: functions. Functions look very much like the statements we have seen above. the template looks like:
<name> <- function (<parameters>) {
<statements>
}In this statement, the function is assigned to the name - any valid name in R. Once it is assigned, it the function can be invoked with name(). The parameter list (the values we write into the parentheses followin the function name) can be empty, or hold a list of variable names. If variable names are present, you need to enter the corresponding parameters when you execute the function. These assigned variables are available inside the function, and can be used for computations. This is called "passing the variable into the function".
You have encountered a function to choose YFO names. In this function, your Student ID was the parameter. Here is another example to play with: a function that calculates how old you are. In days. This is neat - you can celebrate your 10,000 birthday - or so.
Task:
Copy, explore and run ...
- Define the function ...
# A lifedays calculator function
myLifeDays <- function(date = NULL) { # give "date" a default value so we can test whether it has been set
if (is.null(date)) {
print ("Enter your birthday as a string in \"YYYY-MM-DD\" format.")
return()
}
x <- strptime(date, "%Y-%m-%d") # convert string to time
y <- format(Sys.time(), "%Y-%m-%d") # convert "now" to time
diff <- round(as.numeric(difftime(y, x, unit="days")))
print(paste("This date was ", diff, " days ago."))
}- Use the function (example)
myLifeDays("1932-09-25") # Glenn Gould's birthdayHere is a good opportunity to play and practice programming: modify this function to accept a second argument. When a second argument is present (e.g. 10000) the function should print the calendar date on which the input date will be that number of days ago. Then you could use it to know when to celebrate your 10,000th lifeDay, or your 777th anniversary day or whatever.
Enjoy.
The PDB
- Search for GO and EC numbers at PDB...
The search options in the PDB structure database are as sophisticated as those at the NCBI. For now, we will try a simple keyword search to get us started.
Task:
- Visit the RCSB PDB website at http://www.pdb.org/
- Briefly orient yourself regarding the database contents and its information offerings and services.
- Enter
Mbp1into the search field. - In your journal, note down the PDB IDs for the three Saccharomyces cerevisiae Mbp1 transcription factor structures your search has retrieved.
- Click on one of the entries and explore the information and services linked from that page.
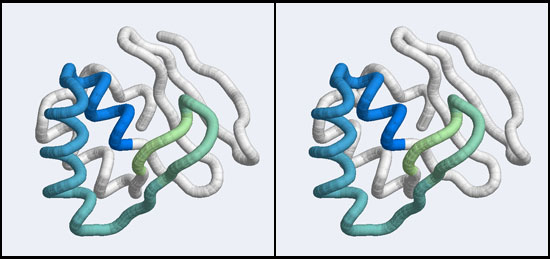
Introduction
Integrating evolutionary information with structural information allows us to establish which residues are invariant in a family–these are presumably structurally important sites–and which residues are functionally important, since they are invariant within, but changeable between subfamilies.
To visualize these relationships, we will load an MSA of APSES domains with VMD and color it by conservation.
The DNA binding site
Now, that you know how YFO Mbp1 aligns with yeast Mbp1, you can evaluate functional conservation in these homologous proteins. You probably already downloaded the two Biochemistry papers by Taylor et al. (2000) and by Deleeuw et al. (2008) that we encountered in Assignment 2. These discuss the residues involved in DNA binding[2]. In particular the residues between 50-74 have been proposed to comprise the DNA recognition domain.
Task:
- Using the APSES domain alignment you have just constructed, find the YFO Mbp1 residues that correspond to the range 50-74 in yeast.
- Note whether the sequences are especially highly conserved in this region.
- Using Chimera, look at the region. Use the sequence window to make sure that the sequence numbering between the paper and the PDB file are the same (they are often not identical!). Then select the residues - the proposed recognition domain - and color them differently for emphasis. Study this in stereo to get a sense of the spatial relationships. Check where the conserved residues are.
- A good representation is stick - but other representations that include sidechains will also serve well.
- Calculate a solvent accessible surface of the protein in a separate representation and make it transparent.
- You could combine three representations: (1) the backbone (in ribbon view), (2) the sidechains of residues that presumably contact DNA, distinctly colored, and (3) a transparent surface of the entire protein. This image should show whether residues annotated as DNA binding form a contiguous binding interface.
DNA binding interfaces are expected to comprise a number of positively charged amino acids, that might form salt-bridges with the phosphate backbone.
Task:
- Study and consider whether this is the case here and which residues might be included.
APSES domains in Chimera (from A4)
What precisely constitutes an APSES domain however is a matter of definition, as you can explore in the following (optional) task.
- Display the protein in ribbon style, e.g. with the Interactive 1 preset.
- Access the Interpro information page for Mbp1 at the EBI: http://www.ebi.ac.uk/interpro/protein/P39678
- In the section Domains and repeats, mouse over the red annotations and note down the residue numbers for the annotated domains. Also follow the links to the respective Interpro domain definition pages.
At this point we have definitions for the following regions on the Mbp1 protein ...
- The KilA-N (pfam 04383) domain definition as applied to the Mbp1 protein sequence by CDD;
- The InterPro KilA, N-terminal/APSES-type HTH, DNA-binding (IPR018004) definition annotated on the Mbp1 sequence;
- The InterPro Transcription regulator HTH, APSES-type DNA-binding domain (IPR003163) definition annotated on the Mbp1 sequence;
- (... in addition – without following the source here – the UniProt record for Mbp1 annotates a "HTH APSES-type" domain from residues 5-111)
... each with its distinct and partially overlapping sequence range. Back to Chimera:
- In the sequence window, select the sequence corresponding to the Interpro KilA-N annotation and colour this fragment red. Remember that you can get the sequence numbers of a residue in the sequence window when you hover the pointer over it - but do confirm that the sequence numbering that Chimera displays matches the numbering of the Interpro domain definition.
- Then select the residue range(s) by which the CDD KilA-N definition is larger, and colour that fragment orange.
- Then select the residue range(s) by which the InterPro APSES domain definition is larger, and colour that fragment yellow.
- If the structure contains residues outside these ranges, colour these white.
- Study this in a side-by-side stereo view and get a sense for how the extra sequence beyond the Kil-A N domain(s) is part of the structure, and how the integrity of the folded structure would be affected if these fragments were missing.
- Display Hydrogen bonds, to get a sense of interactions between residues from the differently colored parts. First show the protein as a stick model, with sticks that are thicker than the default to give a better sense of sidechain packing:
- (i) Select → Select all
- (ii) Actions → Ribbon → hide
- (iii) Select → Structure → protein
- (iv) Actions → Atoms/Bonds → show
- (v) Actions → Atoms/Bonds → stick
- (vi) click on the looking glass icon at the bottom right of the graphics window to bring up the inspector window and choose Inspect ... Bond. Change the radius to 0.4.
- (i) Select → Select all
- Then calculate and display the hydrogen bonds:
- (vii) Tools → Surface/Binding Analysis → FindHbond
- (viii) Set the Line width to 3.0, leave all other parameters with their default values an click Apply
- Clear the selection.
- (vii) Tools → Surface/Binding Analysis → FindHbond
- Let's now simplify the scene a bit and focus on backbone/backbone H-bonds:
- (ix) Select → Structure → Backbone → full
- (x) Actions → Atoms/Bonds → show only
- Clear the selection.
- (ix) Select → Structure → Backbone → full
- Save the resulting image as a jpeg no larger than 600px across and upload it to your Lab notebook on the Wiki.
- When you are done, congratulate yourself on having earned a bonus of 10% on the next quiz.
There is a rather important lesson in this: domain definitions may be fluid, and their boundaries may be computationally derived from sequence comparisons across many families, and do not necessarily correspond to individual structures. Make sure you understand this well.
}}
Given this, it seems appropriate to search the sequence database with the sequence of an Mbp1 structure–this being a structured, stable, subdomain of the whole that presumably contains the protein's most unique and specific function. Let us retrieve this sequence. All PDB structures have their sequences stored in the NCBI protein database. They can be accessed simply via the PDB-ID, which serves as an identifier both for the NCBI and the PDB databases. However there is a small catch (isn't there always?). PDB files can contain more than one protein, e.g. if the crystal structure contains a complex[3]. Each of the individual proteins gets a so-called chain ID–a one letter identifier– to identify them uniquely. To find their unique sequence in the database, you need to know the PDB ID as well as the chain ID. If the file contains only a single protein (as in our case), the chain ID is always A[4]. make sure you understand the concept of protein chains, and chain IDs.
Chimera "sequence": implicit or explicit ?
We discussed the distinction between implicit and explicit sequence. But which one does the Chimera sequence window display? Let's find out.
Task:
- Open Chimera and load the 1BM8 structure from the PDB.
- Save the ccordinate file with File → Save PDB ..., use a filename of
test.pdb. - Open this file in a plain text editor: notepad, TextEdit, nano or the like, but not MS Word! Make sure you view the file in a fixed-width font, not proportionally spaced, i.e. Courier, not Arial. Otherwise the columns in the file won't line up.
- Find the records that begin with
SEQRES ...and confirm that the first amino acid isGLN. - Now scroll down to the
ATOMsection of the file. Identify the records for the first residue in the structure. Delete all lines for side-chain atoms except for theCBatom. This changes the coordinates for glutamine to those of alanine. - Replace the
GLNresidue name withALA(uppercase). This relabels the residue as Alanine in the coordinate section. Therefore you have changed the implicit sequence. Implicit and explicit sequence are now different. The second atom record should now look like this:
ATOM 2 CA ALA A 4 -0.575 5.127 16.398 1.00 51.22 C
- Save the file and load it in Chimera.
- Open the sequence window: does it display
QorAas the first reside?
Therefore, does Chimera use the implicit or explicit sequence in the sequence window?
Coloring by conservation
With VMD, you can import a sequence alignment into the MultiSeq extension and color residues by conservation. The protocol below assumes that an MSA exists - you could have produced it in many different ways, for convenience, I have precalculated one for you. This may not contain the sequences from YFO, if you are curious about these you are welcome to add them and realign.
Task:
- Load the Mbp1 APSES alignment into MultiSeq.
- Access the set of MUSCLE aligned and edited fungal APSES domains.
- Copy the alignment and save it into a convenient directory on your computer as a plain text file. Give it the extension
.aln. - Open VMD and load the
1BM8structure. - As usual, turn the axes off and display your structure in side-by-side stereo.
- Visualize the structure as New Cartoon with Index coloring to re-orient yourself. Identify the recognition helix and the "wing".
- Open Extensions → Analysis → Multiseq.
- You can answer No to download metadata databases, we won't need them here.
- In the MultiSeq Window, navigate to File → Import Data...; Choose "From Files" and Browse to the location of the alignment you have saved. The File navigation window gives you options which files to enable: choose to Enable
ALNfiles (these are CLUSTAL formatted multiple sequence alignments). - Open the alignment file, click on Ok to import the data. If the data can't be loaded, the file may have the wrong extension: .aln is required.
- find the
Mbp1_SACCEsequence in the list, click on it and move it to the top of the Sequences list with your mouse (the list is not static, you can re-order the sequences in any way you like).
You will see that the 1BM8 sequence and the Mbp1_SACCA APSES domain sequence do not match: at the N-terminus the sequence that corresponds to the PDB structure has extra residues, and in the middle the APSES sequences may have gaps inserted.
Task:
- Bring the 1MB1 sequence in register with the APSES alignment.
- MultiSeq supports typical text-editor selection mechanisms. Clicking on a residue selects it, clicking on a row selects the whole sequence. Dragging with the mouse selects several residues, shift-clicking selects ranges, and option-clicking toggles the selection on or off for individual residues. Using the mouse and/or the shift key as required, select the entire first column of the Sequences you have imported. Note: don't include the 1BM8 sequence - this is just for the aligned sequences.
- Select Edit → Enable Editing... → Gaps only to allow changing indels.
- Pressing the spacebar once should insert a gap character before the selected column in all sequences. Insert as many gaps as you need to align the beginning of sequences with the corresponding residues of 1BM8:
S I M .... Note: Have patience - the program's response can be a bit sluggish. - Now insert as many gaps as you need into the
1BM8structure sequence, to align it completely with theMbp1_SACCEAPSES domain sequence. (Simply select residues in the sequence and use the space bar to insert gaps. (Note: I have noticed a bug that sometimes prevents slider or keyboard input to the MultiSeq window; it fails to regain focus after operations in a different window. I don't know whether this is a Mac related problem or a more general bug in MultiSeq. When this happens I quit VMD and restore a saved session. It is a bit annoying but not mission-critical. But to be able to do that, you might want to save your session every now and then.) - When you are done, it may be prudent to save the state of your alignment. Use File → Save Session...
Task:
- Color by similarity
- Use the View → Coloring → Sequence similarity → BLOSUM30 option to color the residues in the alignment and structure. This clearly shows you where conserved and variable residues are located and allows to analyze their structural context.
- Navigate to the Representations window and create a Tube representation of the structure's backbone. Use User coloring to color it according to the conservation score that the Multiseq extension has calculated.
- Create a new representation, choose Licorice as the drawing method, User as the coloring method and select
(sidechain or name CA) and not element H(note:CA, the C-alpha atom must be capitalized.) - Double-click on the NewCartoon representation to hide it.
- You can adjust the color scale in the usual way by navigating to VMD main → Graphics → Colors..., choosing the Color Scale tab and adjusting the scale midpoint.
Study this structure in some detail. If you wish, you could load and superimpose the DNA complexes to determine which conserved residues are in the vicinity of the double helix strands and potentially able to interact with backbone or bases. Note that the most highly conserved residues in the family alignment are all structurally conserved elements of the core. Solvent exposed residues that comprise the surface of the recognition helix are quite variable, especially at the binding site. You may also find - if you load the DNA molecules, that residues that contact the phosphate backbone in general tend to be more highly conserved than residues that contact bases.
Modelling the Ankyrin Domain Section
Creating an Ankyrin domain alignment
APSES domains are relatively easy to identify and annotate but we have had problems with the ankyrin domains in Mbp1 homologues. Both CDD as well as SMART have identified such domains, but while the domain model was based on the same Pfam profile for both, and both annotated approximately the same regions, the details of the alignments and the extent of the predicted region was different.
Mbp1 forms heterodimeric complexes with a homologue, Swi6. Swi6 does not have an APSES domain, thus it does not bind DNA. But it is similar to Mbp1 in the region spanning the ankyrin domains and in 1999 Foord et al. published its crystal structure (1SW6). This structure is a good model for Ankyrin repeats in Mbp1. For details, please refer to the consolidated Mbp1 annotation page I have prepared.
In what follows, we will use the program JALVIEW - a Java based multiple sequence alignment editor to load and align sequences and to consider structural similarity between yeast Mbp1 and its closest homologue in your organism.
In this part of the assignment,
- You will load sequences that are most similar to Mbp1 into an MSA editor;
- You will add sequences of ankyrin domain models;
- You will perform a multiple sequence alignment;
- You will try to improve the alignment manually;
Jalview, loading sequences
Geoff Barton's lab in Dundee has developed an integrated MSA editor and sequence annotation workbench with a number of very useful functions. It is written in Java and should run on Mac, Linux and Windows platforms without modifications.
| Waterhouse et al. (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189-91. (pmid: 19151095) |
|
[ PubMed ] [ DOI ] UNLABELLED: Jalview Version 2 is a system for interactive WYSIWYG editing, analysis and annotation of multiple sequence alignments. Core features include keyboard and mouse-based editing, multiple views and alignment overviews, and linked structure display with Jmol. Jalview 2 is available in two forms: a lightweight Java applet for use in web applications, and a powerful desktop application that employs web services for sequence alignment, secondary structure prediction and the retrieval of alignments, sequences, annotation and structures from public databases and any DAS 1.53 compliant sequence or annotation server. AVAILABILITY: The Jalview 2 Desktop application and JalviewLite applet are made freely available under the GPL, and can be downloaded from www.jalview.org. |
We will use this tool for this assignment and explore its features as we go along.
Task:
- Navigate to the Jalview homepage click on Download, install Jalview on your computer and start it. A number of windows that showcase the program's abilities will load, you can close these.
- Prepare homologous Mbp1 sequences for alignment:
- Open the Reference Mbp1 orthologues (all fungi) page. (This is the list of Mbp1 orthologs I mentioned above.)
- Copy the FASTA sequences of the reference proteins, paste them into a text file (TextEdit on the Mac, Notepad on Windows) and save the file; you could give it an extension of
.fa–but you don't have to. - Check whether the sequence for YFO is included in the list. If it is, fine. If it is not, retrieve it from NCBI, paste it into the file and edit the header like the other sequences. If the wrong sequence from YFO is included, replace it and let me know.
- Return to Jalview and select File → Input Alignment → from File and open your file. A window with sequences should appear.
- Copy the sequences for ankyrin domain models (below), click on the Jalview window, select File → Add sequences → from Textbox and paste them into the Jalview textbox. Paste two separate copies of the CD00204 consensus sequence and one copy of 1SW6.
- When all the sequences are present, click on Add.
Jalview now displays all the sequences, but of course this is not yet an alignment.
- Ankyrin domain models
>CD00204 ankyrin repeat consensus sequence from CDD NARDEDGRTPLHLAASNGHLEVVKLLLENGADVNAKDNDGRTPLHLAAKNGHLEIVKLLL EKGADVNARDKDGNTPLHLAARNGNLDVVKLLLKHGADVNARDKDGRTPLHLAAKNGHL
>1SW6 from PDB - unstructured loops replaced with xxxx GPIITFTHDLTSDFLSSPLKIMKALPSPVVNDNEQKMKLEAFLQRLLFxxxxSFDSLLQE VNDAFPNTQLNLNIPVDEHGNTPLHWLTSIANLELVKHLVKHGSNRLYGDNMGESCLVKA VKSVNNYDSGTFEALLDYLYPCLILEDSMNRTILHHIIITSGMTGCSAAAKYYLDILMGW IVKKQNRPIQSGxxxxDSILENLDLKWIIANMLNAQDSNGDTCLNIAARLGNISIVDALL DYGADPFIANKSGLRPVDFGAG
Computing alignments
try two MSA's algorithms and load them in Jalview. Locally: which one do you prefer? Modify the consensus. Annotate domains.
The EBI has a very convenient page to access a number of MSA algorithms. This is especially convenient when you want to compare, e.g. T-Coffee and Muscle and MAFFT results to see which regions of your alignment are robust. You could use any of these tools, just paste your sequences into a Webform, download the results and load into Jalview. Easy.
But even easier is to calculate the alignments directly from Jalview. available. (Not today. Bummer.)
No. Calculate an external alignment and import.
- Calculate a MAFFT alignment using the Jalview Web service option
Task:
- In Jalview, select Web Service → Alignment → MAFFT with defaults.... The alignment is calculated in a few minutes and displayed in a new window.
- Calculate a MAFFT alignment when the Jalview Web service is NOT available
Task:
- In Jalview, select File → Output to Textbox → FASTA
- Copy the sequences.
- Navigate to the MAFFT Input form at the EBI.
- Paste your sequences into the form.
- Click on Submit.
- Close the Jalview sequence window and either save your MAFFT alignment to file and load in Jalview, or simply 'File → Input Alignment → from Textbox, paste and click New Window.
In any case, you should now have an alignment.
Task:
- Choose Colour → Hydrophobicity and → by Conservation. Then adjust the slider left or right to see which columns are highly conserved. You will notice that the Swi6 sequence that was supposed to align only to the ankyrin domains was in fact aligned to other parts of the sequence as well. This is one part of the MSA that we will have to correct manually and a common problem when aligning sequences of different lengths.
Editing ankyrin domain alignments
A good MSA comprises only columns of residues that play similar roles in the proteins' mechanism and/or that evolve in a comparable structural context. Since the alignment reflects the result of biological selection and conservation, it has relatively few indels and the indels it has are usually not placed into elements of secondary structure or into functional motifs. The contiguous features annotated for Mbp1 are expected to be left intact by a good alignment.
A poor MSA has many errors in its columns; these contain residues that actually have different functions or structural roles, even though they may look similar according to a (pairwise!) scoring matrix. A poor MSA also may have introduced indels in biologically irrelevant positions, to maximize spurious sequence similarities. Some of the features annotated for Mbp1 will be disrupted in a poor alignment and residues that are conserved may be placed into different columns.
Often errors or inconsistencies are easy to spot, and manually editing an MSA is not generally frowned upon, even though this is not a strictly objective procedure. The main goal of manual editing is to make an alignment biologically more plausible. Most comonly this means to mimize the number of rare evolutionary events that the alignment suggests and/or to emphasize conservation of known functional motifs. Here are some examples for what one might aim for in manually editing an alignment:
- Reduce number of indels
From a Probcons alignment: 0447_DEBHA ILKTE-K-T---K--SVVK ILKTE----KTK---SVVK 9978_GIBZE MLGLN-PGLKEIT--HSIT MLGLNPGLKEIT---HSIT 1513_CANAL ILKTE-K-I---K--NVVK ILKTE----KIK---NVVK 6132_SCHPO ELDDI-I-ESGDY--ENVD ELDDI-IESGDY---ENVD 1244_ASPFU ----N-PGLREIC--HSIT -> ----NPGLREIC---HSIT 0925_USTMA LVKTC-PALDPHI--TKLK LVKTCPALDPHI---TKLK 2599_ASPTE VLDAN-PGLREIS--HSIT VLDANPGLREIS---HSIT 9773_DEBHA LLESTPKQYHQHI--KRIR LLESTPKQYHQHI--KRIR 0918_CANAL LLESTPKEYQQYI--KRIR LLESTPKEYQQYI--KRIR
Gaps marked in red were moved. The sequence similarity in the alignment does not change considerably, however the total number of indels in this excerpt is reduced to 13 from the original 22
- Move indels to more plausible position
From a CLUSTAL alignment: 4966_CANGL MKHEKVQ------GGYGRFQ---GTW MKHEKVQ------GGYGRFQ---GTW 1513_CANAL KIKNVVK------VGSMNLK---GVW KIKNVVK------VGSMNLK---GVW 6132_SCHPO VDSKHP-----------QID---GVW -> VDSKHPQ-----------ID---GVW 1244_ASPFU EICHSIT------GGALAAQ---GYW EICHSIT------GGALAAQ---GYW
The two characters marked in red were swapped. This does not change the number of indels but places the "Q" into a a column in which it is more highly conserved (green). Progressive alignments are especially prone to this type of error.
- Conserve motifs
From a CLUSTAL alignment: 6166_SCHPO --DKRVA---GLWVPP --DKRVA--G-LWVPP XBP1_SACCE GGYIKIQ---GTWLPM GGYIKIQ--G-TWLPM 6355_ASPTE --DEIAG---NVWISP -> ---DEIA--GNVWISP 5262_KLULA GGYIKIQ---GTWLPY GGYIKIQ--G-TWLPY
The first of the two residues marked in red is a conserved, solvent exposed hydrophobic residue that may mediate domain interactions. The second residue is the conserved glycine in a beta turn that cannot be mutated without structural disruption. Changing the position of a gap and insertion in one sequence improves the conservation of both motifs.
The Ankyrin domains are quite highly diverged, the boundaries not well defined and not even CDD, SMART and SAS agree on the precise annotations. We expect there to be alignment errors in this region. Nevertheless we would hope that a good alignment would recognize homology in that region and that ideally the required indels would be placed between the secondary structure elements, not in their middle. But judging from the sequence alignment alone, we cannot judge where the secondary structure elements ought to be. You should therefore add the following "sequence" to the alignment; it contains exactly as many characters as the Swi6 sequence above and annotates the secondary structure elements. I have derived it from the 1SW6 structure
>SecStruc 1SW6 E: strand t: turn H: helix _: irregular _EEE__tt___ttt______EE_____t___HHHHHHHHHHHHHHHH_xxxx_HHHHHHH HHHH_t_____t_____t____HHHHHHH__tHHHHHHHHH____t___tt____HHHHH HH__HHHH___HHHHHHHHHHHHHEE_t____HHHHHHHHH__t__HHHHHHHHHHHHHH HHHHHH__EEE_xxxx_HHHHHt_HHHHHHH______t____HHHHHHHH__HHHHHHHH H____t____t____HHHH___
To proceed:
- Manually align the Swi6 sequence with yeast Mbp1
- Bring the Secondary structure annotation into its correct alignment with Swi6
- Bring both CDD ankyrin profiles into the correct alignment with yeast Mbp1
Proceed along the following steps:
Task:
- Add the secondary structure annotation to the sequence alignment in Jalview. Copy the annotation, select File → Add sequences → from Textbox and paste the sequence.
- Select Help → Documentation and read about Editing Alignments, Cursor Mode and Key strokes.
- Click on the yeast Mbp1 sequence row to select the entire row. Then use the cursor key to move that sequence down, so it is directly above the 1SW6 sequence. Select the row of 1SW6 and use shift/mouse to move the sequence elements and edit the alignment to match yeast Mbp1. Refer to the alignment given in the Mbp1 annotation page for the correct alignment.
- Align the secondary structure elements with the 1SW6 sequence: Every character of 1SW6 should be matched with either E, t, H, or _. The result should be similar to the Mbp1 annotation page. If you need to insert gaps into all sequences in the alignment, simply drag your mouse over all row headers - movement of sequences is constrained to selected regions, the rest is locked into place to prevent inadvertent misalignments. Remember to save your project from time to time: File → save so you can reload a previous state if anything goes wrong and can't be fixed with Edit → Undo.
- Finally align the two CD00204 consensus sequences to their correct positions (again, refer to the Mbp1 annotation page).
- You can now consider the principles stated above and see if you can improve the alignment, for example by moving indels out of regions of secondary structure if that is possible without changing the character of the aligned columns significantly. Select blocks within which to work to leave the remaining alignment unchanged. So that this does not become tedious, you can restrict your editing to one Ankyrin repeat that is structurally defined in Swi6. You may want to open the 1SW6 structure in VMD to define the boundaries of one such repeat. You can copy and paste sections from Jalview into your assignment for documentation or export sections of the alignment to HTML (see the example below).
Editing ankyrin domain alignments - Sample
This sample was created by
- Editing the alignments as described above;
- Copying a block of aligned sequence;
- Pasting it To New Alignment;
- Colouring the residues by Hydrophobicity and setting the colour saturation according to Conservation;
- Choosing File → Export Image → HTML and pasting the resulting HTML source into this Wikipage.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Aligned sequences before editing. The algorithm has placed gaps into the Swi6 helix
LKWIIANand the four-residue gaps before the block of well aligned sequence on the right are poorly supported.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Aligned sequence after editing. A significant cleanup of the frayed region is possible. Now there is only one insertion event, and it is placed into the loop that connects two helices of the 1SW6 structure.
Final analysis of the ankyrin alignment
Task:
- Compare the distribution of indels in the ankyrin repeat regions of your alignments.
- Review whether the indels in this region are concentrated in segments that connect the helices, or if they are more or less evenly distributed along the entire region of similarity.
- Think about whether the assertion that indels should not be placed in elements of secondary structure has merit in your alignment.
- Recognize that an indel in an element of secondary structure could be interpreted in a number of different ways:
- The alignment is correct, the annotation is correct too: the indel is tolerated in that particular case, for example by extending the length of an α-helix or β-strand;
- The alignment algorithm has made an error, the structural annotation is correct: the indel should be moved a few residues;
- The alignment is correct, the structural annotation is wrong, this is not a secondary structure element after all;
- Both the algorithm and the annotation are probably wrong, but we have no data to improve the situation.
(NB: remember that the structural annotations have been made for the yeast protein and might have turned out differently for the other proteins...)
You should be able to analyse discrepancies between annotation and expectation in a structured and systematic way. In particular if you notice indels that have been placed into an annotated region of secondary structure, you should be able to comment on whether the location of the indel has strong support from aligned sequence motifs, or whether the indel could possibly be moved into a different location without much loss in alignment quality.
- Considering the whole alignment and your experience with editing, you should be able to state whether the position of indels relative to structural features of the ankyrin domains in your organism's Mbp1 protein is reliable. That would be the result of this task, in which you combine multiple sequence and structural information.
- You can also critically evaluate database information that you have encountered:
- Navigate to the CDD annotation for yeast Mbp1.
- You can check the precise alignment boundaries of the ankyrin domains by clicking on the (+) icon to the left of the matching domain definition.
- Confirm that CDD extends the ankyrin domain annotation beyond the 1SW6 domain boundaries. Given your assessment of conservation in the region beyond the structural annotation: do you think that extending the annotation is reasonable also in YFO's protein? Is there evidence for this in the alignment of the CD00204 consensus with well aligned blocks of sequence beyond the positions that match Swi6?
From Homology Modeling - Modeling alternative binding modes
Finding a similar protein-DNA complex
Remember that homologous sequences can have diverged to the point where their sequence similarity is no longer recognizable, however their structure may be quite well conserved. Thus if we could find similar structures in the PDB, these might provide us with some plausible hypotheses for how DNA is bound by APSES domains. We thus need a tool similar to BLAST, but not for the purpose of sequence alignment, but for structure alignment. A kind of BLAST for structures. Just like with sequence searches, we might not want to search with the entire protein, if we are interested in is a subdomain that binds to DNA. Attempting to match all structural elements in addition to the ones we are actually interested in is likely to make the search less specific - we would find false positives that are similar to some irrelevant part of our structure. However, defining too small of a subdomain would also lead to a loss of specificity: in the extreme it is easy to imagine that the search for e.g. a single helix would retrieve very many hits that would be quite meaningless.
At the NCBI, VAST is provided as a search tool for structural similarity search.
Task:
- Navigate to the VAST search interface page.
- Enter
1bm8as the PDB ID to search for and click Go. - Follow the link to Related Structures.
- Study the result.
You will see that VAST finds more than 3,000 partially similar structures, but it would be almost impossibly tedious to manually search through the list for structures of protein DNA complexes that are similar to the interacting core of the APSES domain. It turns out that our search is not specific enough in two ways: we have structural elements in our PDB file that are unnecessary for the question at hand, and thus cause the program to find irrelevant matches. But, if we constrain ourselves to just a single helix and strand (i.e. the 50-74 subdomain that has been implicated in DNA binding, the search will become too non-specific. Also we have no good way to retrieve functional information from these hits: which ones are DNA-binding proteins, that bind DNA through residues of this subdomain and for which the structure of a complex has been solved? It seems we need to define our question more precisely.
Task:
- Open VMD and load the 1BM8 structure or your YFO homology model.
- Display the backbone as a Trace (of CA atoms) and color by Index
- In the sequence viewer, highlight residues 50 to 74.
- In the representations window, find the yellow representation (with Color ID 4) that the sequence viewer has generated. Change the Drawing Method to NewCartoon.
- Now (using stereo), study the topology of the region. Focus on the helix at the N-terminus of the highlighted subdomain, it is preceded by a turn and another helix. This first helix makes interactions with the beta hairpin at the C-terminal end of the subdomain and is thus important for the orientation of these elements. (This is what is referred to as a helix-turn-helix motif, or HtH motif, it is very common in DNA-binding proteins.)
- Holding the shift key in the alignment viewer, extend your selection until you cover all of the first helix, and the residues that contact the beta hairpin. I think that the first residue of interest here is residue 33.
- Again holding the shift key, extend the selection at the C-terminus to include the residues of the beta hairpin to where they contact the helix at the N-terminus. I think that the last residue of interest here is residue 79.
- Study the topology and arrangement of this compact subdomain. It contains the DNA-binding elements and probably most of the interactions that establish its three-dimensional shape. This subdomain even has a name: it is a winged helix DNA binding motif, a member of a very large family of DNA-binding domains. I have linked a review by Gajiwala and Burley to the end of this page; note that their definition of a canonical winged helix motif is a bit larger than what we have here, with an additional helix at the N-terminus and a second "wing". )
Armed with this insight, we can attempt again to find meaningfully similar structures. At the EBI there are a number of very well designed structure analysis tools linked off the Structural Analysis page. As part of its MSD Services, PDBeFold provides a convenient interface for structure searches for our purpose
Task:
- Navigate to the PDBeFold search interface page.
- Enter
1bm8for the PDB code and choose Select range from the drop down menu. Select the residues you have defined above. - Note that you can enter the lowest acceptable match % separately for query and target. This means: what percentage of secondary structure elements would need to be matched in either query or target to produce a hit. Keep that value at 80 for our query, since we would want to find structures with almost all of the elements of the winged helix motif. Set the match to 10 % for the target, since we are interested in such domains even if they happen to be small subdomains of large proteins.
- Keep the Precision at normal. Precision and % query match could be relaxed if we wanted to find more structures.
- Finally click on: Submit your query.
- On the results page, click on the index number (in the left-hand column) of the top hit that is not one of our familiar Mbp1 structures to get a detailed view of the result. Most likely this is
1wq2:a, an enzyme. Click on View Superposed. This will open a window with the structure coordinates superimposed in the Jmol molecular viewer. Control-click anywhere in the window area to open a menu of viewing options. Select Style → Stereographic → Wall-eyed viewing. Select Trace as the rendering. Then study the superposition. You will note that the secondary structure elements match quite well, but does this mean we have a DNA-binding domain in this sulfite reductase?
All in all this appears to be well engineered software! It gives you many options to access result details for further processing. I think this can be put to very good use. But for our problem, we would have to search through too many structures because, once again, we can't tell which ones of the hits are DNA binding domains, especially domains for which the structure of a complex has been solved.
APSES domains represent one branch of the tree of helix-turn-helix (HTH) DNA binding modules. (A review on HTH proteins is linked from the resources section at the bottom of this page). Winged Helix domains typically bind their cognate DNA with a "recognition helix" which precedes the beta hairpin and binds into the major groove; additional stabilizing interactions are provided by the edge of a beta-strand binding into the minor groove. This is good news: once we have determined that the APSES domain is actually an example of a larger group of transcription factors, we can compare our model to a structure of a protein-DNA complex. Superfamilies of such structural domains are compiled in the CATH database. Unfortunately CATH itself does not provide information about whether the structures have been determined as complexes. But we can search the PDB with CATH codes and restrict the results to complexes. Essentially, this should give us a list of all winged helix domains for which the structure of complexes with DNA have been determined. This works as follows:
Task:
- For reference, access CATH domain superfamily 1.10.10.10; this is the CATH classification code we will use to find protein-DNA complexes. Click on Superfamily Superposition to get a sense of the structural core of the winged helix domain.
- Navigate to the PDB home page and follow the link to Advanced Search
- In the options menu for Choose a Query Type select Structure Features → CATH Classification Browser. A window will open that allows you to navigate down through the CATH tree. You can view the Class/Architecture/Topology names on the CATH page linked above. Click on the triangle icons (not the text) for Mainly Alpha → Orthogonal Bundle → ARC repressor mutant, subunit A then click on the link to winged helix repressor DNA binding domain. Or, just enter "winged helix" into the search field. This subquery should match more than 550 coordinate entries.
- Click on the (+) button behind Add search criteria to add an additional query. Select the option Structure Features → Macromolecule type. In the option menus that pop up, select Contains Protein→Yes, Contains DNA→Yes, Contains RNA→Ignore, Contains DNA/RNA hybrid→Ignore. This selects files that contain Protein-DNA complexes.
- Check the box below this subquery to Remove Similar Sequences at 90% identity and click on Submit Query. This query should retrieve more than 100 complexes.
- Scroll down to the beginning of the list of PDB codes and locate the Reports menu. Under the heading View select Gallery. This is a fast way to obtain an overview of the structures that have been returned. Adjust the number of Results to see all 100 images and choose Options→Resize medium.
- Finally we have a set of winged-helix domain/DNA complexes, for comparison. Scroll through the gallery and study how the protein binds DNA.
First of all you may notice that in fact not all of the structures are really different, despite having requested only to retrieve dissimilar sequences, and not all images show DNA. This appears to be a deficiency of the algorithm. But you can also easily recognize how in most of the the structures the recognition helix inserts into the major groove of B-DNA (eg. 1BC8, 1CF7) and the wing - if clearly visible at all in the image - appears to make accessory interactions with the DNA backbone.. There is one exception: the structure 1DP7 shows how the human RFX1 protein binds DNA in a non-canonical way, through the beta-strands of the "wing". This is interesting since it suggests there is more than one way for winged helix domains to bind to DNA. We can therefore use structural superposition of your homology model and two of the winged-helix proteins to decide whether the canonical or the non-canonical mode of DNA binding seems to be more plausible for Mbp1 orthologues.
Preparation and superposition of a canonical complex
The structure we shall use as a reference for the canonical binding mode is the Elk-1 transcription factor.
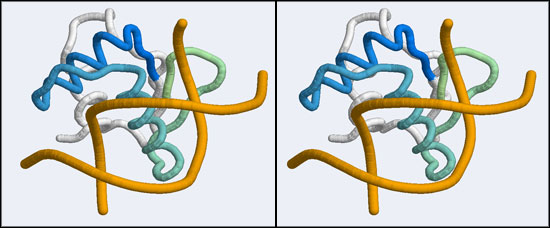
The 1DUX coordinate-file contains two protein domains and two B-DNA dimers in one asymmetric unit. For simplicity, you should delete the second copy of the complex from the PDB file. (Remember that PDB files are simply text files that can be edited.)
Task:
- Find the 1DUX structure in the image gallery and open the 1DUX structure explorer page in a separate window. Download the coordinates to your computer.
- Open the coordinate file in a text-editor (TextEdit or Notepad - NOT MS-Word!) and delete the coordinates for chains
D,EandF; you may also delete allHETATMrecords and theMASTERrecord. Save the file with a different name, e.g. 1DUX_monomer.pdb . - Open VMD and load your homology model. Turn off the axes, display the model as a Tube representation in stereo, and color it by Index. Then load your edited 1DUX file, display this coordinate set in a tube representation as well, and color it by ColorID in some color you like. It is important that you can distinguish easily which structure is which.
- You could use the Extensions→Analysis→RMSD calculator interface to superimpose the two strutcures IF you would know which residues correspond to each other. Sometimes it is useful to do exactly that: define exact correspondences between residue pairs and superimpose according to these selected pairs. For our purpose it is much simpler to use the Multiseq tool (and the structures are simple and small enough that the STAMP algorithm for structural alignment can define corresponding residue pairs automatically). Open the multiseq extension window, select the check-boxes next to both protein structures, and open the Tools→Stamp Structural Alignment interface.
- In the "'Stamp Alignment Options'" window, check the radio-button for Align the following ... Marked Structures and click on OK.
- In the Graphical Representations window, double-click on all "NewCartoon" representations for both molecules, to undisplay them.
- You should now see a superimposed tube model of your homology model and the 1DUX protein-DNA complex. You can explore it, display side-chains etc. and study some of the details of how a transcription factor recognizes and binds to its cognate DNA sequence. However, remember that your model's side-chain orientations have not been determined experimentally but inferred from the template, and that the template's structure was determined in the absence of bound DNA ligand.
- Orient and scale your superimposed structures so that their structural similarity is apparent, and the recognition helix can be clearly seen inserting into the DNA major groove. You may want to keep a copy of the image for future reference. Consider which parts of the structure appear to superimpose best. Note whether it is plausible that your model could bind a B-DNA double-helix in this orientation.
Preparation and superposition of a non-canonical complex
The structure displaying a non-canonical complex between a winged-helix domain and its cognate DNA binding site is the human Regulatory Factor X.
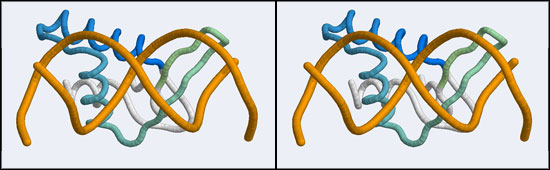
Before we can work with this however, we have to fix an annoying problem. If you download and view the 1DP7 structure in VMD, you will notice that there is only a single strand of DNA! Where is the second strand of the double helix? It is not in the coordinate file, because it happens to be exactly equivalent to the frist starnd, rotated around a two-fold axis of symmetry in the crystal lattice. We need to download and work with the so-called Biological Assembly instead. But there is a problem related to the way the PDB stores replicates in biological assemblies. The PDB generates the additional chains as copies of the original and delineates them with MODEL and ENDMDL records, just like in a multi-structure NMR file. The chain IDs and the atom numbers are the same as the original. The PDB file thus contains the same molecule in two different orientations, not two independent molecules. This is an important difference regarding how such molecules are displayed by VMD. If you try to use the biological unit file of the PDB, VMD does not recognize that there is a second molecule present and displays only one chain. And that looks exactly like the one we have seen before. We have to edit the file, extract the second DNA molecule, change its chain ID and then append it to the original 1DP7 structure[5]...
Task:
- On the structure explorer page for 1DP7, select the option Download Files → PDB File.
- Also select the option Download Files → Biological Assembly.
- Uncompress the biological assembly file.
- Open the file in a text editor.
- Delete everything except the second DNA molecule. This comes after the
MODEL 2line and has chain ID D. Keep theTERandENDlines. Save this with a new filename (e.g.1DP7_DNAonly.pdb). - Also delete all
HETATMrecords forHOH,PEGandEDO, as well as the entire second protein chain and theMASTERrecord. The resulting file should only contain the DNA chain and its copy and one protein chain. Save the file with a new name, eg.1DP7_BDNA.PDB. - Use a similar procedure as BIO_Assignment_Week_8#R code: renumbering the model in the last assignment to change the chain ID.
PDBin <- "1DP7_DNAonly.pdb"
PDBout <- "1DP7_DNAnewChain.pdb"
pdb <- read.pdb(PDBin)
pdb$atom[,"chain"] <- "E"
write.pdb(pdb=pdb,file=PDBout)- Use your text-editor to open both the
1DP7.pdbstructure file and the1DP7_DNAnewChain.pdb. Copy the DNA coordinates, paste them into the original file before theENDline and save. - Open the edited coordinate file with VMD. You should see one protein chain and a B-DNA double helix. (Actually, the BDNA helix has a gap, because the R-library did not read the BRDU nucleotide as DNA). Switch to stereo viewing and spend some time to see how amazingly beautiful the complementarity between the protein and the DNA helix is (you might want to display protein and nucleic in separate representations and color the DNA chain by Position → Radial for clarity) ... in particular, appreciate how not all positively charged side chains contact the phosphate backbone, but some pnetrate into the helix and make detailed interactions with the nucleobases!
- Then clear all molecules
- In VMD, open Extensions→Analysis→MultiSeq. When you run MultiSeq for the first time, you will be asked for a directory in which to store metadata. You can use the default, or a directory of your choice; you may subsequently skip all steps that ask you to install "required" databases locally since we will not need them for this task.
- Choose File→Import Data, browse to your directory and load one by one:
- -Your model;
- -The 1DUX complex;
- -The 1DP7 complex.
- Mark all three protein chains by selecting the checkbox next to their name and choose Tools→ STAMP structural alignment.
- Align the Marked Structures, choose a scanscore of 2 and scanslide of 5. Also choose Slow scan. You may have to play around with the setting to get the molecules to superimpose: but the can be superimposed quite well - at least the DNA-binding helices and the wings should line up.
- In the graphical representations window, double-click on the cartoon representations that multiseq has generated to undisplay them, also undisplay the Tube representation of 1DUX. Then create a Tube representation for 1DP7, and select a Color by ColorID (a different color that you like). The resulting scene should look similar to the one you have created above, only with 1DP7 in place of 1DUX and colored differently.
- Orient and scale your superimposed structures so that their structural similarity is apparent, and the differences in binding elements is clear. Perhaps visualizing a solvent accessible surface of the DNA will help understand the spatial requirements of the complex formation. You may want to keep a copy of the image for future reference. Note whether it is plausible that your model could bind a B-DNA double-helix in the "alternative" conformation.
Task:
- Spend some time studying the complex.
- Recapitulate in your mind how we have arrived at this comparison, in particular, how this was possible even though the sequence similarity between these proteins is low - none of these winged helix domains came up as a result of our previous BLAST search in the PDB.
- You should clearly think about the following question: considering the position of the two DNA helices relative to the YFO structural model, which binding mode appears to be more plausible for protein-DNA interactions in the YFO Mbp1 APSES domains? Is it the canonical, or the non-canonical binding mode? Is there evidence that allows you to distinguish between the two modes?
- Before you quit VMD, save the "state" of your session so you can reload it later. We will look at residue conservation once we have built phylogenetic trees. In the main VMD window, choose File→Save State....
Interpretation
Analysis of the ligand binding site:
- http://dnasite.limlab.ibms.sinica.edu.tw/
- http://proline.biochem.iisc.ernet.in/pocketannotate/
- http://www.biosolveit.de/PoseView/
- Comparison with seq2logo
| Chu et al. (2009) ProteDNA: a sequence-based predictor of sequence-specific DNA-binding residues in transcription factors. Nucleic Acids Res 37:W396-401. (pmid: 19483101) |
|
[ PubMed ] [ DOI ] This article presents the design of a sequence-based predictor named ProteDNA for identifying the sequence-specific binding residues in a transcription factor (TF). Concerning protein-DNA interactions, there are two types of binding mechanisms involved, namely sequence-specific binding and nonspecific binding. Sequence-specific bindings occur between protein sidechains and nucleotide bases and correspond to sequence-specific recognition of genes. Therefore, sequence-specific bindings are essential for correct gene regulation. In this respect, ProteDNA is distinctive since it has been designed to identify sequence-specific binding residues. In order to accommodate users with different application needs, ProteDNA has been designed to operate under two modes, namely, the high-precision mode and the balanced mode. According to the experiments reported in this article, under the high-precision mode, ProteDNA has been able to deliver precision of 82.3%, specificity of 99.3%, sensitivity of 49.8% and accuracy of 96.5%. Meanwhile, under the balanced mode, ProteDNA has been able to deliver precision of 60.8%, specificity of 97.6%, sensitivity of 60.7% and accuracy of 95.4%. ProteDNA is available at the following websites: http://protedna.csbb.ntu.edu.tw/, http://protedna.csie.ntu.edu.tw/, http://bio222.esoe.ntu.edu.tw/ProteDNA/. |
- protedna server PMID: 19483101
- http://serv.csbb.ntu.edu.tw/ProteDNA/
- http://protedna.csie.ntu.edu.tw/
- Multi Harmony
| Brandt et al. (2010) Multi-Harmony: detecting functional specificity from sequence alignment. Nucleic Acids Res 38:W35-40. (pmid: 20525785) |
|
[ PubMed ] [ DOI ] Many protein families contain sub-families with functional specialization, such as binding different ligands or being involved in different protein-protein interactions. A small number of amino acids generally determine functional specificity. The identification of these residues can aid the understanding of protein function and help finding targets for experimental analysis. Here, we present multi-Harmony, an interactive web sever for detecting sub-type-specific sites in proteins starting from a multiple sequence alignment. Combining our Sequence Harmony (SH) and multi-Relief (mR) methods in one web server allows simultaneous analysis and comparison of specificity residues; furthermore, both methods have been significantly improved and extended. SH has been extended to cope with more than two sub-groups. mR has been changed from a sampling implementation to a deterministic one, making it more consistent and user friendly. For both methods Z-scores are reported. The multi-Harmony web server produces a dynamic output page, which includes interactive connections to the Jalview and Jmol applets, thereby allowing interactive analysis of the results. Multi-Harmony is available at http://www.ibi.vu.nl/ programs/shmrwww. |
EC
Enzyme Commission Codes ...
GO
Introduction
| Harris (2008) Developing an ontology. Methods Mol Biol 452:111-24. (pmid: 18563371) |
|
[ PubMed ] [ DOI ] In recent years, biological ontologies have emerged as a means of representing and organizing biological concepts, enabling biologists, bioinformaticians, and others to derive meaning from large datasets.This chapter provides an overview of formal principles and practical considerations of ontology construction and application. Ontology development concepts are illustrated using examples drawn from the Gene Ontology (GO) and other OBO ontologies. |
| Hackenberg & Matthiesen (2010) Algorithms and methods for correlating experimental results with annotation databases. Methods Mol Biol 593:315-40. (pmid: 19957156) |
|
[ PubMed ] [ DOI ] An important procedure in biomedical research is the detection of genes that are differentially expressed under pathologic conditions. These genes, or at least a subset of them, are key biomarkers and are thought to be important to describe and understand the analyzed biological system (the pathology) at a molecular level. To obtain this understanding, it is indispensable to link those genes to biological knowledge stored in databases. Ontological analysis is nowadays a standard procedure to analyze large gene lists. By detecting enriched and depleted gene properties and functions, important insights on the biological system can be obtained. In this chapter, we will give a brief survey of the general layout of the methods used in an ontological analysis and of the most important tools that have been developed. |
GO
The Gene Ontology project is the most influential contributor to the definition of function in computational biology and the use of GO terms and GO annotations is ubiquitous.
|
GO: the Gene Ontology project [ link ] [ page ] Ontologies are important tools to organize and compute with non-standardized information, such as gene annotations. The Gene Ontology project (GO) constructs ontologies for gene and gene product attributes across numerous species. Three major ontologies are being developed: molecular process, biological function and cellular location. Each includes terms, their definition, and their relationships. In addition, genes and gene products are being been annotated with their GO terms and the type of evidence that underlies the annotation. A number of tools such as the AmiGO browser are available to analyse relationships, construct ontologies and curate annotations. Data can be freely downloaded in formats that are convenient for computation. |  |
| du Plessis et al. (2011) The what, where, how and why of gene ontology--a primer for bioinformaticians. Brief Bioinformatics 12:723-35. (pmid: 21330331) |
|
[ PubMed ] [ DOI ] With high-throughput technologies providing vast amounts of data, it has become more important to provide systematic, quality annotations. The Gene Ontology (GO) project is the largest resource for cataloguing gene function. Nonetheless, its use is not yet ubiquitous and is still fraught with pitfalls. In this review, we provide a short primer to the GO for bioinformaticians. We summarize important aspects of the structure of the ontology, describe sources and types of functional annotations, survey measures of GO annotation similarity, review typical uses of GO and discuss other important considerations pertaining to the use of GO in bioinformatics applications. |
The GO actually comprises three separate ontologies:
- Molecular function
- ...
- Biological Process
- ...
- Cellular component
- ...
GO terms
GO terms comprise the core of the information in the ontology: a carefully crafted definition of a term in any of GO's separate ontologies.
GO relationships
The nature of the relationships is as much a part of the ontology as the terms themselves. GO uses three categories of relationships:
- is a
- part of
- regulates
GO annotations
The GO terms are conceptual in nature, and while they represent our interpretation of biological phenomena, they do not intrinsically represent biological objects, such a specific genes or proteins. In order to link molecules with these concepts, the ontology is used to annotate genes. The annotation project is referred to as GOA.
| Dimmer et al. (2007) Methods for gene ontology annotation. Methods Mol Biol 406:495-520. (pmid: 18287709) |
|
[ PubMed ] [ DOI ] The Gene Ontology (GO) is an established dynamic and structured vocabulary that has been successfully used in gene and protein annotation. Designed by biologists to improve data integration, GO attempts to replace the multiple nomenclatures used by specialised and large biological knowledgebases. This chapter describes the methods used by groups to create new GO annotations and how users can apply publicly available GO annotations to enhance their datasets. |
GO evidence codes
Annotations can be made according to literature data or computational inference and it is important to note how an annotation has been justified by the curator to evaluate the level of trust we should have in the annotation. GO uses evidence codes to make this process transparent. When computing with the ontology, we may want to filter (exclude) particular terms in order to avoid tautologies: for example if we were to infer functional relationships between homologous genes, we should exclude annotations that have been based on the same inference or similar, and compute only with the actual experimental data.
The following evidence codes are in current use; if you want to exclude inferred anotations you would restrict the codes you use to the ones shown in bold: EXP, IDA, IPI, IMP, IEP, and perhaps IGI, although the interpretation of genetic interactions can require assumptions.
- Automatically-assigned Evidence Codes
- IEA: Inferred from Electronic Annotation
- Curator-assigned Evidence Codes
- Experimental Evidence Codes
- EXP: Inferred from Experiment
- IDA: Inferred from Direct Assay
- IPI: Inferred from Physical Interaction
- IMP: Inferred from Mutant Phenotype
- IGI: Inferred from Genetic Interaction
- IEP: Inferred from Expression Pattern
- Computational Analysis Evidence Codes
- ISS: Inferred from Sequence or Structural Similarity
- ISO: Inferred from Sequence Orthology
- ISA: Inferred from Sequence Alignment
- ISM: Inferred from Sequence Model
- IGC: Inferred from Genomic Context
- IBA: Inferred from Biological aspect of Ancestor
- IBD: Inferred from Biological aspect of Descendant
- IKR: Inferred from Key Residues
- IRD: Inferred from Rapid Divergence
- RCA: inferred from Reviewed Computational Analysis
- Author Statement Evidence Codes
- TAS: Traceable Author Statement
- NAS: Non-traceable Author Statement
- Curator Statement Evidence Codes
- IC: Inferred by Curator
- ND: No biological Data available
For further details, see the Guide to GO Evidence Codes and the GO Evidence Code Decision Tree.
GO tools
For many projects, the simplest approach will be to download the GO ontology itself. It is a well constructed, easily parseable file that is well suited for computation. For details, see Computing with GO on this wiki.
AmiGO
practical work with GO: at first via the AmiGO browser AmiGO is a GO browser developed by the Gene Ontology consortium and hosted on their website.
AmiGO - Gene products
Task:
- Navigate to the GO homepage.
- Enter
SOX2into the search box to initiate a search for the human SOX2 transcription factor (WP, HUGO) (as gene or protein name). - There are a number of hits in various organisms: sulfhydryl oxidases and (sex determining region Y)-box genes. Check to see the various ways by which you could filter and restrict the results.
- Select Homo sapiens as the species filter and set the filter. Note that this still does not give you a unique hit, but ...
- ... you can identify the Transcription factor SOX-2 and follow its gene product information link. Study the information on that page.
- Later, we will need Entrez Gene IDs. The GOA information page provides these as GeneID in the External references section. Note it down. With the same approach, find and record the Gene IDs (a) of the functionally related Oct4 (POU5F1) protein, (b) the human cell-cycle transcription factor E2F1, (c) the human bone morphogenetic protein-4 transforming growth factor BMP4, (d) the human UDP glucuronosyltransferase 1 family protein 1, an enzyme that is differentially expressed in some cancers, UGT1A1, and (d) as a positive control, SOX2's interaction partner NANOG, which we would expect to be annotated as functionally similar to both Oct4 and SOX2.
AmiGO - Associations
GO annotations for a protein are called associations.
Task:
- Open the associations information page for the human SOX2 protein via the link in the right column in a separate tab. Study the information on that page.
- Note that you can filter the associations by ontology and evidence code. You have read about the three GO ontologies in your previous assignment, but you should also be familiar with the evidence codes. Click on any of the evidence links to access the Evidence code definition page and study the definitions of the codes. Make sure you understand which codes point to experimental observation, and which codes denote computational inference, or say that the evidence is someone's opinion (TAS, IC etc.). Note: it is good practice - but regrettably not universally implemented standard - to clearly document database semantics and keep definitions associated with database entries easily accessible, as GO is doing here. You won't find this everywhere, but as a user please feel encouraged to complain to the database providers if you come across a database where the semantics are not clear. Seriously: opaque semantics make database annotations useless.
- There are many associations (around 60) and a good way to select which ones to pursue is to follow the most specific ones. Set
IDAas a filter and among the returned terms selectGO:0035019– somatic stem cell maintenance in the Biological Process ontology. Follow that link. - Study the information available on that page and through the tabs on the page, especially the graph view.
- In the Inferred Tree View tab, find the genes annotated to this go term for homo sapiens. There should be about 55. Click on the number behind the term. The resulting page will give you all human proteins that have been annotated with this particular term. Note that the great majority of these is via the
IEAevidence code.
Semantic similarity
A good, recent overview of ontology based functional annotation is found in the following article. This is not a formal reading assignment, but do familiarize yourself with section 3: Derivation of Semantic Similarity between Terms in an Ontology as an introduction to the code-based annotations below.
| Gan et al. (2013) From ontology to semantic similarity: calculation of ontology-based semantic similarity. ScientificWorldJournal 2013:793091. (pmid: 23533360) |
|
[ PubMed ] [ DOI ] Advances in high-throughput experimental techniques in the past decade have enabled the explosive increase of omics data, while effective organization, interpretation, and exchange of these data require standard and controlled vocabularies in the domain of biological and biomedical studies. Ontologies, as abstract description systems for domain-specific knowledge composition, hence receive more and more attention in computational biology and bioinformatics. Particularly, many applications relying on domain ontologies require quantitative measures of relationships between terms in the ontologies, making it indispensable to develop computational methods for the derivation of ontology-based semantic similarity between terms. Nevertheless, with a variety of methods available, how to choose a suitable method for a specific application becomes a problem. With this understanding, we review a majority of existing methods that rely on ontologies to calculate semantic similarity between terms. We classify existing methods into five categories: methods based on semantic distance, methods based on information content, methods based on properties of terms, methods based on ontology hierarchy, and hybrid methods. We summarize characteristics of each category, with emphasis on basic notions, advantages and disadvantages of these methods. Further, we extend our review to software tools implementing these methods and applications using these methods. |
Practical work with GO: bioconductor.
The bioconductor project hosts the GOSemSim package for semantic similarity.
Task:
- Work through the following R-code. If you have problems, discuss them on the mailing list. Don't go through the code mechanically but make sure you are clear about what it does.
# GOsemanticSimilarity.R
# GO semantic similarity example
# B. Steipe for BCB420, January 2014
setwd("~/your-R-project-directory")
# GOSemSim is an R-package in the bioconductor project. It is not installed via
# the usual install.packages() comand (via CRAN) but via an installation script
# that is run from the bioconductor Website.
source("http://bioconductor.org/biocLite.R")
biocLite("GOSemSim")
library(GOSemSim)
# This loads the library and starts the Bioconductor environment.
# You can get an overview of functions by executing ...
browseVignettes()
# ... which will open a listing in your Web browser. Open the
# introduction to GOSemSim PDF. As the introduction suggests,
# now is a good time to execute ...
help(GOSemSim)
# The simplest function is to measure the semantic similarity of two GO
# terms. For example, SOX2 was annotated with GO:0035019 (somatic stem cell
# maintenance), QSOX2 was annotated with GO:0045454 (cell redox homeostasis),
# and Oct4 (POU5F1) with GO:0009786 (regulation of asymmetric cell division),
# among other associations. Lets calculate these similarities.
goSim("GO:0035019", "GO:0009786", ont="BP", measure="Wang")
goSim("GO:0035019", "GO:0045454", ont="BP", measure="Wang")
# Fair enough. Two numbers. Clearly we would appreciate an idea of the values
# that high similarity and low similarity can take. But in any case -
# we are really less interested in the similarity of GO terms - these
# are a function of how the Ontology was constructed. We are more
# interested in the functional similarity of our genes, and these
# have a number of GO terms associated with them.
# GOSemSim provides the functions ...
?geneSim()
?mgeneSim()
# ... to compute these values. Refer to the vignette for details, in
# particular, consider how multiple GO terms are combined, and how to
# keep/drop evidence codes.
# Here is a pairwise similarity example: the gene IDs are the ones you
# have recorded previously. Note that this will download a package
# of GO annotations - you might not want to do this on a low-bandwidth
# connection.
geneSim("6657", "5460", ont = "BP", measure="Wang", combine = "BMA")
# Another number. And the list of GO terms that were considered.
# Your task: use the mgeneSim() function to calculate the similarities
# between all six proteins for which you have recorded the GeneIDs
# previously (SOX2, POU5F1, E2F1, BMP4, UGT1A1 and NANOG) in the
# biological process ontology.
# This will run for some time. On my machine, half an hour or so.
# Do the results correspond to your expectations?GO reading and resources
- General
| Sauro & Bergmann (2008) Standards and ontologies in computational systems biology. Essays Biochem 45:211-22. (pmid: 18793134) |
|
[ PubMed ] [ DOI ] With the growing importance of computational models in systems biology there has been much interest in recent years to develop standard model interchange languages that permit biologists to easily exchange models between different software tools. In the present chapter two chief model exchange standards, SBML (Systems Biology Markup Language) and CellML are described. In addition, other related features including visual layout initiatives, ontologies and best practices for model annotation are discussed. Software tools such as developer libraries and basic editing tools are also introduced, together with a discussion on the future of modelling languages and visualization tools in systems biology. |
- Phenotype etc. Ontologies
See also:
| Köhler et al. (2014) The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 42:D966-74. (pmid: 24217912) |
|
[ PubMed ] [ DOI ] The Human Phenotype Ontology (HPO) project, available at http://www.human-phenotype-ontology.org, provides a structured, comprehensive and well-defined set of 10,088 classes (terms) describing human phenotypic abnormalities and 13,326 subclass relations between the HPO classes. In addition we have developed logical definitions for 46% of all HPO classes using terms from ontologies for anatomy, cell types, function, embryology, pathology and other domains. This allows interoperability with several resources, especially those containing phenotype information on model organisms such as mouse and zebrafish. Here we describe the updated HPO database, which provides annotations of 7,278 human hereditary syndromes listed in OMIM, Orphanet and DECIPHER to classes of the HPO. Various meta-attributes such as frequency, references and negations are associated with each annotation. Several large-scale projects worldwide utilize the HPO for describing phenotype information in their datasets. We have therefore generated equivalence mappings to other phenotype vocabularies such as LDDB, Orphanet, MedDRA, UMLS and phenoDB, allowing integration of existing datasets and interoperability with multiple biomedical resources. We have created various ways to access the HPO database content using flat files, a MySQL database, and Web-based tools. All data and documentation on the HPO project can be found online. |
| Schriml et al. (2012) Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res 40:D940-6. (pmid: 22080554) |
|
[ PubMed ] [ DOI ] The Disease Ontology (DO) database (http://disease-ontology.org) represents a comprehensive knowledge base of 8043 inherited, developmental and acquired human diseases (DO version 3, revision 2510). The DO web browser has been designed for speed, efficiency and robustness through the use of a graph database. Full-text contextual searching functionality using Lucene allows the querying of name, synonym, definition, DOID and cross-reference (xrefs) with complex Boolean search strings. The DO semantically integrates disease and medical vocabularies through extensive cross mapping and integration of MeSH, ICD, NCI's thesaurus, SNOMED CT and OMIM disease-specific terms and identifiers. The DO is utilized for disease annotation by major biomedical databases (e.g. Array Express, NIF, IEDB), as a standard representation of human disease in biomedical ontologies (e.g. IDO, Cell line ontology, NIFSTD ontology, Experimental Factor Ontology, Influenza Ontology), and as an ontological cross mappings resource between DO, MeSH and OMIM (e.g. GeneWiki). The DO project (http://diseaseontology.sf.net) has been incorporated into open source tools (e.g. Gene Answers, FunDO) to connect gene and disease biomedical data through the lens of human disease. The next iteration of the DO web browser will integrate DO's extended relations and logical definition representation along with these biomedical resource cross-mappings. |
| Evelo et al. (2011) Answering biological questions: querying a systems biology database for nutrigenomics. Genes Nutr 6:81-7. (pmid: 21437033) |
|
[ PubMed ] [ DOI ] The requirement of systems biology for connecting different levels of biological research leads directly to a need for integrating vast amounts of diverse information in general and of omics data in particular. The nutritional phenotype database addresses this challenge for nutrigenomics. A particularly urgent objective in coping with the data avalanche is making biologically meaningful information accessible to the researcher. This contribution describes how we intend to meet this objective with the nutritional phenotype database. We outline relevant parts of the system architecture, describe the kinds of data managed by it, and show how the system can support retrieval of biologically meaningful information by means of ontologies, full-text queries, and structured queries. Our contribution points out critical points, describes several technical hurdles. It demonstrates how pathway analysis can improve queries and comparisons for nutrition studies. Finally, three directions for future research are given. |
| Oti et al. (2009) The biological coherence of human phenome databases. Am J Hum Genet 85:801-8. (pmid: 20004759) |
|
[ PubMed ] [ DOI ] Disease networks are increasingly explored as a complement to networks centered around interactions between genes and proteins. The quality of disease networks is heavily dependent on the amount and quality of phenotype information in phenotype databases of human genetic diseases. We explored which aspects of phenotype database architecture and content best reflect the underlying biology of disease. We used the OMIM-based HPO, Orphanet, and POSSUM phenotype databases for this purpose and devised a biological coherence score based on the sharing of gene ontology annotation to investigate the degree to which phenotype similarity in these databases reflects related pathobiology. Our analyses support the notion that a fine-grained phenotype ontology enhances the accuracy of phenome representation. In addition, we find that the OMIM database that is most used by the human genetics community is heavily underannotated. We show that this problem can easily be overcome by simply adding data available in the POSSUM database to improve OMIM phenotype representations in the HPO. Also, we find that the use of feature frequency estimates--currently implemented only in the Orphanet database--significantly improves the quality of the phenome representation. Our data suggest that there is much to be gained by improving human phenome databases and that some of the measures needed to achieve this are relatively easy to implement. More generally, we propose that curation and more systematic annotation of human phenome databases can greatly improve the power of the phenotype for genetic disease analysis. |
| Groth et al. (2007) PhenomicDB: a new cross-species genotype/phenotype resource. Nucleic Acids Res 35:D696-9. (pmid: 16982638) |
|
[ PubMed ] [ DOI ] Phenotypes are an important subject of biomedical research for which many repositories have already been created. Most of these databases are either dedicated to a single species or to a single disease of interest. With the advent of technologies to generate phenotypes in a high-throughput manner, not only is the volume of phenotype data growing fast but also the need to organize these data in more useful ways. We have created PhenomicDB (freely available at http://www.phenomicdb.de), a multi-species genotype/phenotype database, which shows phenotypes associated with their corresponding genes and grouped by gene orthologies across a variety of species. We have enhanced PhenomicDB recently by additionally incorporating quantitative and descriptive RNA interference (RNAi) screening data, by enabling the usage of phenotype ontology terms and by providing information on assays and cell lines. We envision that integration of classical phenotypes with high-throughput data will bring new momentum and insights to our understanding. Modern analysis tools under development may help exploiting this wealth of information to transform it into knowledge and, eventually, into novel therapeutic approaches. |
- Semantic similarity
| Wu et al. (2013) Improving the measurement of semantic similarity between gene ontology terms and gene products: insights from an edge- and IC-based hybrid method. PLoS ONE 8:e66745. (pmid: 23741529) |
|
[ PubMed ] [ DOI ] BACKGROUND: Explicit comparisons based on the semantic similarity of Gene Ontology terms provide a quantitative way to measure the functional similarity between gene products and are widely applied in large-scale genomic research via integration with other models. Previously, we presented an edge-based method, Relative Specificity Similarity (RSS), which takes the global position of relevant terms into account. However, edge-based semantic similarity metrics are sensitive to the intrinsic structure of GO and simply consider terms at the same level in the ontology to be equally specific nodes, revealing the weaknesses that could be complemented using information content (IC). RESULTS AND CONCLUSIONS: Here, we used the IC-based nodes to improve RSS and proposed a new method, Hybrid Relative Specificity Similarity (HRSS). HRSS outperformed other methods in distinguishing true protein-protein interactions from false. HRSS values were divided into four different levels of confidence for protein interactions. In addition, HRSS was statistically the best at obtaining the highest average functional similarity among human-mouse orthologs. Both HRSS and the groupwise measure, simGIC, are superior in correlation with sequence and Pfam similarities. Because different measures are best suited for different circumstances, we compared two pairwise strategies, the maximum and the best-match average, in the evaluation. The former was more effective at inferring physical protein-protein interactions, and the latter at estimating the functional conservation of orthologs and analyzing the CESSM datasets. In conclusion, HRSS can be applied to different biological problems by quantifying the functional similarity between gene products. The algorithm HRSS was implemented in the C programming language, which is freely available from http://cmb.bnu.edu.cn/hrss. |
| Gan et al. (2013) From ontology to semantic similarity: calculation of ontology-based semantic similarity. ScientificWorldJournal 2013:793091. (pmid: 23533360) |
|
[ PubMed ] [ DOI ] Advances in high-throughput experimental techniques in the past decade have enabled the explosive increase of omics data, while effective organization, interpretation, and exchange of these data require standard and controlled vocabularies in the domain of biological and biomedical studies. Ontologies, as abstract description systems for domain-specific knowledge composition, hence receive more and more attention in computational biology and bioinformatics. Particularly, many applications relying on domain ontologies require quantitative measures of relationships between terms in the ontologies, making it indispensable to develop computational methods for the derivation of ontology-based semantic similarity between terms. Nevertheless, with a variety of methods available, how to choose a suitable method for a specific application becomes a problem. With this understanding, we review a majority of existing methods that rely on ontologies to calculate semantic similarity between terms. We classify existing methods into five categories: methods based on semantic distance, methods based on information content, methods based on properties of terms, methods based on ontology hierarchy, and hybrid methods. We summarize characteristics of each category, with emphasis on basic notions, advantages and disadvantages of these methods. Further, we extend our review to software tools implementing these methods and applications using these methods. |
| Alvarez & Yan (2011) A graph-based semantic similarity measure for the gene ontology. J Bioinform Comput Biol 9:681-95. (pmid: 22084008) |
|
[ PubMed ] [ DOI ] Existing methods for calculating semantic similarities between pairs of Gene Ontology (GO) terms and gene products often rely on external databases like Gene Ontology Annotation (GOA) that annotate gene products using the GO terms. This dependency leads to some limitations in real applications. Here, we present a semantic similarity algorithm (SSA), that relies exclusively on the GO. When calculating the semantic similarity between a pair of input GO terms, SSA takes into account the shortest path between them, the depth of their nearest common ancestor, and a novel similarity score calculated between the definitions of the involved GO terms. In our work, we use SSA to calculate semantic similarities between pairs of proteins by combining pairwise semantic similarities between the GO terms that annotate the involved proteins. The reliability of SSA was evaluated by comparing the resulting semantic similarities between proteins with the functional similarities between proteins derived from expert annotations or sequence similarity. Comparisons with existing state-of-the-art methods showed that SSA is highly competitive with the other methods. SSA provides a reliable measure for semantics similarity independent of external databases of functional-annotation observations. |
| Jain & Bader (2010) An improved method for scoring protein-protein interactions using semantic similarity within the gene ontology. BMC Bioinformatics 11:562. (pmid: 21078182) |
|
[ PubMed ] [ DOI ] BACKGROUND: Semantic similarity measures are useful to assess the physiological relevance of protein-protein interactions (PPIs). They quantify similarity between proteins based on their function using annotation systems like the Gene Ontology (GO). Proteins that interact in the cell are likely to be in similar locations or involved in similar biological processes compared to proteins that do not interact. Thus the more semantically similar the gene function annotations are among the interacting proteins, more likely the interaction is physiologically relevant. However, most semantic similarity measures used for PPI confidence assessment do not consider the unequal depth of term hierarchies in different classes of cellular location, molecular function, and biological process ontologies of GO and thus may over-or under-estimate similarity. RESULTS: We describe an improved algorithm, Topological Clustering Semantic Similarity (TCSS), to compute semantic similarity between GO terms annotated to proteins in interaction datasets. Our algorithm, considers unequal depth of biological knowledge representation in different branches of the GO graph. The central idea is to divide the GO graph into sub-graphs and score PPIs higher if participating proteins belong to the same sub-graph as compared to if they belong to different sub-graphs. CONCLUSIONS: The TCSS algorithm performs better than other semantic similarity measurement techniques that we evaluated in terms of their performance on distinguishing true from false protein interactions, and correlation with gene expression and protein families. We show an average improvement of 4.6 times the F1 score over Resnik, the next best method, on our Saccharomyces cerevisiae PPI dataset and 2 times on our Homo sapiens PPI dataset using cellular component, biological process and molecular function GO annotations. |
| Yu et al. (2010) GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26:976-8. (pmid: 20179076) |
|
[ PubMed ] [ DOI ] SUMMARY: The semantic comparisons of Gene Ontology (GO) annotations provide quantitative ways to compute similarities between genes and gene groups, and have became important basis for many bioinformatics analysis approaches. GOSemSim is an R package for semantic similarity computation among GO terms, sets of GO terms, gene products and gene clusters. Four information content (IC)- and a graph-based methods are implemented in the GOSemSim package, multiple species including human, rat, mouse, fly and yeast are also supported. The functions provided by the GOSemSim offer flexibility for applications, and can be easily integrated into high-throughput analysis pipelines. AVAILABILITY: GOSemSim is released under the GNU General Public License within Bioconductor project, and freely available at http://bioconductor.org/packages/2.6/bioc/html/GOSemSim.html. |
- GO
| Gene Ontology Consortium (2012) The Gene Ontology: enhancements for 2011. Nucleic Acids Res 40:D559-64. (pmid: 22102568) |
|
[ PubMed ] [ DOI ] The Gene Ontology (GO) (http://www.geneontology.org) is a community bioinformatics resource that represents gene product function through the use of structured, controlled vocabularies. The number of GO annotations of gene products has increased due to curation efforts among GO Consortium (GOC) groups, including focused literature-based annotation and ortholog-based functional inference. The GO ontologies continue to expand and improve as a result of targeted ontology development, including the introduction of computable logical definitions and development of new tools for the streamlined addition of terms to the ontology. The GOC continues to support its user community through the use of e-mail lists, social media and web-based resources. |
| Bastos et al. (2011) Application of gene ontology to gene identification. Methods Mol Biol 760:141-57. (pmid: 21779995) |
|
[ PubMed ] [ DOI ] Candidate gene identification deals with associating genes to underlying biological phenomena, such as diseases and specific disorders. It has been shown that classes of diseases with similar phenotypes are caused by functionally related genes. Currently, a fair amount of knowledge about the functional characterization can be found across several public databases; however, functional descriptors can be ambiguous, domain specific, and context dependent. In order to cope with these issues, the Gene Ontology (GO) project developed a bio-ontology of broad scope and wide applicability. Thus, the structured and controlled vocabulary of terms provided by the GO project describing the biological roles of gene products can be very helpful in candidate gene identification approaches. The method presented here uses GO annotation data in order to identify the most meaningful functional aspects occurring in a given set of related gene products. The method measures this meaningfulness by calculating an e-value based on the frequency of annotation of each GO term in the set of gene products versus the total frequency of annotation. Then after selecting a GO term related to the underlying biological phenomena being studied, the method uses semantic similarity to rank the given gene products that are annotated to the term. This enables the user to further narrow down the list of gene products and identify those that are more likely of interest. |
| Gene Ontology Consortium (2010) The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res 38:D331-5. (pmid: 19920128) |
|
[ PubMed ] [ DOI ] The Gene Ontology (GO) Consortium (http://www.geneontology.org) (GOC) continues to develop, maintain and use a set of structured, controlled vocabularies for the annotation of genes, gene products and sequences. The GO ontologies are expanding both in content and in structure. Several new relationship types have been introduced and used, along with existing relationships, to create links between and within the GO domains. These improve the representation of biology, facilitate querying, and allow GO developers to systematically check for and correct inconsistencies within the GO. Gene product annotation using GO continues to increase both in the number of total annotations and in species coverage. GO tools, such as OBO-Edit, an ontology-editing tool, and AmiGO, the GOC ontology browser, have seen major improvements in functionality, speed and ease of use. |
Carol Goble on the tension between purists and pragmatists in life-science ontology construction. Plenary talk at SOFG2...
| Goble & Wroe (2004) The Montagues and the Capulets. Comp Funct Genomics 5:623-32. (pmid: 18629186) |
|
[ PubMed ] [ DOI ] Two households, both alike in dignity, In fair Genomics, where we lay our scene, (One, comforted by its logic's rigour, Claims ontology for the realm of pure, The other, with blessed scientist's vigour, Acts hastily on models that endure), From ancient grudge break to new mutiny, When 'being' drives a fly-man to blaspheme. From forth the fatal loins of these two foes, Researchers to unlock the book of life; Whole misadventured piteous overthrows, Can with their work bury their clans' strife. The fruitful passage of their GO-mark'd love, And the continuance of their studies sage, Which, united, yield ontologies undreamed-of, Is now the hour's traffic of our stage; The which if you with patient ears attend, What here shall miss, our toil shall strive to mend. |
| Gajiwala & Burley (2000) Winged helix proteins. Curr Opin Struct Biol 10:110-6. (pmid: 10679470) |
|
[ PubMed ] [ DOI ] The winged helix proteins constitute a subfamily within the large ensemble of helix-turn-helix proteins. Since the discovery of the winged helix/fork head motif in 1993, a large number of topologically related proteins with diverse biological functions have been characterized by X-ray crystallography and solution NMR spectroscopy. Recently, a winged helix transcription factor (RFX1) was shown to bind DNA using unprecedented interactions between one of its eponymous wings and the major groove. This surprising observation suggests that the winged helix proteins can be subdivided into at least two classes with radically different modes of DNA recognition. |
| Aravind et al. (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231-62. (pmid: 15808743) |
|
[ PubMed ] [ DOI ] The helix-turn-helix (HTH) domain is a common denominator in basal and specific transcription factors from the three super-kingdoms of life. At its core, the domain comprises of an open tri-helical bundle, which typically binds DNA with the 3rd helix. Drawing on the wealth of data that has accumulated over two decades since the discovery of the domain, we present an overview of the natural history of the HTH domain from the viewpoint of structural analysis and comparative genomics. In structural terms, the HTH domains have developed several elaborations on the basic 3-helical core, such as the tetra-helical bundle, the winged-helix and the ribbon-helix-helix type configurations. In functional terms, the HTH domains are present in the most prevalent transcription factors of all prokaryotic genomes and some eukaryotic genomes. They have been recruited to a wide range of functions beyond transcription regulation, which include DNA repair and replication, RNA metabolism and protein-protein interactions in diverse signaling contexts. Beyond their basic role in mediating macromolecular interactions, the HTH domains have also been incorporated into the catalytic domains of diverse enzymes. We discuss the general domain architectural themes that have arisen amongst the HTH domains as a result of their recruitment to these diverse functions. We present a natural classification, higher-order relationships and phyletic pattern analysis of all the major families of HTH domains. This reconstruction suggests that there were at least 6-11 different HTH domains in the last universal common ancestor of all life forms, which covered much of the structural diversity and part of the functional versatility of the extant representatives of this domain. In prokaryotes the total number of HTH domains per genome shows a strong power-equation type scaling with the gene number per genome. However, the HTH domains in two-component signaling pathways show a linear scaling with gene number, in contrast to the non-linear scaling of HTH domains in single-component systems and sigma factors. These observations point to distinct evolutionary forces in the emergence of different signaling systems with HTH transcription factors. The archaea and bacteria share a number of ancient families of specific HTH transcription factors. However, they do not share any orthologous HTH proteins in the basal transcription apparatus. This differential relationship of their basal and specific transcriptional machinery poses an apparent conundrum regarding the origins of their transcription apparatus. |
Links and resources
Footnotes and references
- ↑ That's assuming the worst case in that the attacker needs to know the pattern with which the password is formed, i.e. the number of characters and the alphabet that we chose from. But note that there is an even worse case: if the attacker had access to our code and the seed to our random number generator. When the random number generator starts up, a new seed is generated from system time, thus the possible space of seeds can be devastatingly small. But even if a seed is set explicitly with the
set.seed()function, that seed is a 32-bit integer and thus can take only a bit more than 4*109 values, six orders of magnitude less than the 1015 password complexity we thought we had. It turns out that the code may be a much greater vulnerability than the password itself. Keep that in mind. Keep it secret. Keep it safe. - ↑ (Taylor et al. (2000) Biochemistry 39: 3943-3954 and Deleeuw et al. (2008) Biochemistry. 47:6378-6385)
- ↑ Think of the ribosome or DNA-polymerase as extreme examples.
- ↑ Otherwise, you need to study the PDB Web page for the structure, or the text in the PDB file itself, to identify which part of the complex is labeled with which chain ID. For example, immunoglobulin structures some time label the light- and heavy chain fragments as "L" and "H", and sometimes as "A" and "B"–there are no fixed rules. You can also load the structure in VMD, color "by chain" and use the mouse to click on residues in each chain to identify it.
- ↑ My apologies if this is tedious. But in the real world, we encounter such problems a lot and I would be remiss not to use this opportunity to let you practice how to fix the issue that could otherwise be a roadblock in a project of yours.
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.