BIO Assignment Week 4
Assignment for Week 4
Sequence alignment
| < Assignment 3 | Assignment 5 > |
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz.
Contents
- 1 Introduction
- 2 The Mutation Data Matrix
- 3 The DNA binding site
- 4 R code: coloring the alignment by quality
- 5 Retrieve
- 6 MSA (formerly A5)
- 7 Defining the sequence to search with
- 8 Selecting species for a PSI-BLAST search
- 9 Executing the PSI-BLAST search
- 10 Introduction
- 11 Orthologs and Paralogs revisited
- 12 Defining orthologs
- 13 Align and Annotate
- 14 R code: load alignment and compute information scores
- 15 Links and resources
- 16 Footnotes and references
- 17 Ask, if things don't work for you!
Introduction
In this assignment we will perform an optimal global and local sequence alignment, and use R to plot the alignment quality as a colored bar-graph.
Optimal sequence alignments
Online programs for optimal sequence alignment are part of the EMBOSS tools. The programs take FASTA files as input.
- Local optimal SEQUENCE alignment "water"
Task:
- Retrieve the FASTA file for the YFO Mbp1 protein and for Saccharomyces cerevisiae.
- Save the files as text files to your computer, (if you haven't done so already). You could give them an extension of
.fa. - Access the EMBOSS Explorer site (if you haven't done so yet, you might want to bookmark it.)
- Look for ALIGNMENT LOCAL, click on water, paste your FASTA sequences and run the program with default parameters.
- Study the results. You will probably find that the alignment extends over most of the protein, but does not include the termini.
- Considering the sequence identy cutoff we discussed in class (25% over the length of a domain), do you believe that the APSES domains are homologous?
- Change the Gap opening and Gap extension parameters to high values (e.g. 30 and 5). Then run the alignment again.
- Note what is different.
- You could try getting only an alignment for the ankyrin domains that you have found in the last assignment, by deleting the approximate region of the APSES domains from your input.
- Global optimal SEQUENCE alignment "needle"
Task:
- Look for ALIGNMENT GLOBAL, click on needle, paste your FASTA sequences and run the program with default parameters.
- Study the results. You will find that the alignment extends over the entire protein, likely with long indels at the termini.
- Change the Output alignment format to FASTA pairwise simple, to retrieve the aligned FASTA files with indels.
- Copy the aligned sequences (with indels) and save them to your computer. You could give them an extension of
.falto remind you that they are aligned FASTA sequences.
The Mutation Data Matrix
The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the BLOSUM62 matrix[1]. Access that site and download the BLOSUM62 matrix to your computer. You could give it a filename of BLOSUM62.mdm.
It should look like this.
# Matrix made by matblas from blosum62.iij
# * column uses minimum score
# BLOSUM Clustered Scoring Matrix in 1/2 Bit Units
# Blocks Database = /data/blocks_5.0/blocks.dat
# Cluster Percentage: >= 62
# Entropy = 0.6979, Expected = -0.5209
A R N D C Q E G H I L K M F P S T W Y V B Z X *
A 4 -1 -2 -2 0 -1 -1 0 -2 -1 -1 -1 -1 -2 -1 1 0 -3 -2 0 -2 -1 0 -4
R -1 5 0 -2 -3 1 0 -2 0 -3 -2 2 -1 -3 -2 -1 -1 -3 -2 -3 -1 0 -1 -4
N -2 0 6 1 -3 0 0 0 1 -3 -3 0 -2 -3 -2 1 0 -4 -2 -3 3 0 -1 -4
D -2 -2 1 6 -3 0 2 -1 -1 -3 -4 -1 -3 -3 -1 0 -1 -4 -3 -3 4 1 -1 -4
C 0 -3 -3 -3 9 -3 -4 -3 -3 -1 -1 -3 -1 -2 -3 -1 -1 -2 -2 -1 -3 -3 -2 -4
Q -1 1 0 0 -3 5 2 -2 0 -3 -2 1 0 -3 -1 0 -1 -2 -1 -2 0 3 -1 -4
E -1 0 0 2 -4 2 5 -2 0 -3 -3 1 -2 -3 -1 0 -1 -3 -2 -2 1 4 -1 -4
G 0 -2 0 -1 -3 -2 -2 6 -2 -4 -4 -2 -3 -3 -2 0 -2 -2 -3 -3 -1 -2 -1 -4
H -2 0 1 -1 -3 0 0 -2 8 -3 -3 -1 -2 -1 -2 -1 -2 -2 2 -3 0 0 -1 -4
I -1 -3 -3 -3 -1 -3 -3 -4 -3 4 2 -3 1 0 -3 -2 -1 -3 -1 3 -3 -3 -1 -4
L -1 -2 -3 -4 -1 -2 -3 -4 -3 2 4 -2 2 0 -3 -2 -1 -2 -1 1 -4 -3 -1 -4
K -1 2 0 -1 -3 1 1 -2 -1 -3 -2 5 -1 -3 -1 0 -1 -3 -2 -2 0 1 -1 -4
M -1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 0 -2 -1 -1 -1 -1 1 -3 -1 -1 -4
F -2 -3 -3 -3 -2 -3 -3 -3 -1 0 0 -3 0 6 -4 -2 -2 1 3 -1 -3 -3 -1 -4
P -1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7 -1 -1 -4 -3 -2 -2 -1 -2 -4
S 1 -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4 1 -3 -2 -2 0 0 0 -4
T 0 -1 0 -1 -1 -1 -1 -2 -2 -1 -1 -1 -1 -2 -1 1 5 -2 -2 0 -1 -1 0 -4
W -3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 -4 -3 -2 11 2 -3 -4 -3 -2 -4
Y -2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7 -1 -3 -2 -1 -4
V 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4 -3 -2 -1 -4
B -2 -1 3 4 -3 0 1 -1 0 -3 -4 0 -3 -3 -2 0 -1 -4 -3 -3 4 1 -1 -4
Z -1 0 0 1 -3 3 4 -2 0 -3 -3 1 -1 -3 -1 0 -1 -3 -2 -2 1 4 -1 -4
X 0 -1 -1 -1 -2 -1 -1 -1 -1 -1 -1 -1 -1 -1 -2 0 0 -2 -1 -1 -1 -1 -1 -4
* -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 1
Task:
- Study this and make sure you understand what this table is, how it can be used, and what a reasonable range of values for identities and pairscores for non-identical, similar and dissimilar residues is. Ask on the mailing list in case you have questions.
The DNA binding site
Now, that you know how YFO Mbp1 aligns with yeast Mbp1, you can evaluate functional conservation in these homologous proteins. You probably already downloaded the two Biochemistry papers by Taylor et al. (2000) and by Deleeuw et al. (2008) that we encountered in Assignment 2. These discuss the residues involved in DNA binding[2]. In particular the residues between 50-74 have been proposed to comprise the DNA recognition domain.
Task:
- Using the APSES domain alignment you have just constructed, find the YFO Mbp1 residues that correspond to the range 50-74 in yeast.
- Note whether the sequences are especially highly conserved in this region.
- Using Chimera, look at the region. Use the sequence window to make sure that the sequence numbering between the paper and the PDB file are the same (they are often not identical!). Then select the residues - the proposed recognition domain - and color them differently for emphasis. Study this in stereo to get a sense of the spatial relationships. Check where the conserved residues are.
- A good representation is stick - but other representations that include sidechains will also serve well.
- Calculate a solvent accessible surface of the protein in a separate representation and make it transparent.
- You could combine three representations: (1) the backbone (in ribbon view), (2) the sidechains of residues that presumably contact DNA, distinctly colored, and (3) a transparent surface of the entire protein. This image should show whether residues annotated as DNA binding form a contiguous binding interface.
DNA binding interfaces are expected to comprise a number of positively charged amino acids, that might form salt-bridges with the phosphate backbone.
Task:
- Study and consider whether this is the case here and which residues might be included.
R code: coloring the alignment by quality
Task:
- Study this code carefully, execute it, section by section and make sure you understand all of it. Ask on the list if anything is not clear.
# BiostringsExample.R
# Short tutorial on sequence alignment with the Biostrings package.
# Boris Steipe for BCH441, 2013 - 2014
#
setwd("~/path/to/your/R_files/")
setwd("~/Documents/07.TEACHING/37-BCH441 Bioinformatics 2014/05-Materials/Assignment_5 data")
# Biostrings is a package within the bioconductor project.
# bioconducter packages have their own installation system,
# they are normally not installed via CRAN.
# First, you load the BioConductor installer...
source("http://bioconductor.org/biocLite.R")
# Then you can install the Biostrings package and all of its dependencies.
biocLite("Biostrings")
# ... and load the library.
library(Biostrings)
# Some basic (technical) information is available ...
library(help=Biostrings)
# ... but for more in depth documentation, use the
# so called "vignettes" that are provided with every R package.
browseVignettes("Biostrings")
# In this code, we mostly use functions that are discussed in the
# pairwise alignement vignette.
# Read in two fasta files - you will need to edit this for YFO
sacce <- readAAStringSet("mbp1-sacce.fa", format="fasta")
# "USTMA" is used only as an example here - modify for YFO :-)
ustma <- readAAStringSet("mbp1-ustma.fa", format="fasta")
sacce
names(sacce)
names(sacce) <- "Mbp1 SACCE"
names(ustma) <- "Mbp1 USTMA" # Example only ... modify for YFO
width(sacce)
as.character(sacce)
# Biostrings takes a sophisticated approach to sequence alignment ...
?pairwiseAlignment
# ... but the use in practice is quite simple:
ali <- pairwiseAlignment(sacce, ustma, substitutionMatrix = "BLOSUM50")
ali
pattern(ali)
subject(ali)
writePairwiseAlignments(ali)
p <- aligned(pattern(ali))
names(p) <- "Mbp1 SACCE aligned"
s <- aligned(subject(ali))
names(s) <- "Mbp1 USTMA aligned"
# don't overwrite your EMBOSS .fal files
writeXStringSet(p, "mbp1-sacce.R.fal", append=FALSE, format="fasta")
writeXStringSet(s, "mbp1-ustma.R.fal", append=FALSE, format="fasta")
# Done.- Compare the alignments you received from the EMBOSS server, and that you co puted using R. Are they aproximately the same? Exactly? You did use different matrices and gap aameters, so minor differences are to be expected. But by and large you should get the same alignments.
We will now use the aligned sequences to compute a graphical display of alignment quality.
Task:
- Study this code carefully, execute it, section by section and make sure you understand all of it. Ask on the list if anything is not clear.
# aliScore.R
# Evaluating an alignment with a sliding window score
# Boris Steipe, October 2012. Update October 2013
setwd("~/path/to/your/R_files/")
# Scoring matrices can be found at the NCBI.
# ftp://ftp.ncbi.nih.gov/blast/matrices/BLOSUM62
# It is good practice to set variables you might want to change
# in a header block so you don't need to hunt all over the code
# for strings you need to update.
#
fa1 <- "mbp1-sacce.R.fal"
fa2 <- "mbp1-ustma.R.fal"
code1 <- "SACCE"
code2 <- "USTMA"
mdmFile <- "BLOSUM62.mdm"
window <- 9 # window-size (should be an odd integer)
# ================================================
# Read data files
# ================================================
# read fasta datafiles using seqinr function read.fasta()
install.packages("seqinr")
library(seqinr)
tmp <- unlist(read.fasta(fa1, seqtype="AA", as.string=FALSE, seqonly=TRUE))
seq1 <- unlist(strsplit(as.character(tmp), split=""))
tmp <- unlist(read.fasta(fa2, seqtype="AA", as.string=FALSE, seqonly=TRUE))
seq2 <- unlist(strsplit(as.character(tmp), split=""))
if (length(seq1) != length(seq2)) {
print("Error: Sequences have unequal length!")
}
lSeq <- length(seq1)
# ================================================
# Read scoring matrix
# ================================================
MDM <- read.table(mdmFile, skip=6)
# This is a dataframe. Study how it can be accessed:
MDM
MDM[1,]
MDM[,1]
MDM[5,5] # Cys-Cys
MDM[20,20] # Val-Val
MDM[,"W"] # the tryptophan column
MDM["R","W"] # Arg-Trp pairscore
MDM["W","R"] # Trp-Arg pairscore: pairscores are symmetric
colnames(MDM) # names of columns
rownames(MDM) # names of rows
colnames(MDM)[3] # third column
rownames(MDM)[12] # twelfth row
# change the two "*" names to "-" so we can use them to score
# indels of the alignment. This is a bit of a hack, since this
# does not reflect the actual indel penalties (which is, as you)
# remember from your lectures, calculated as a gap opening
# + gap extension penalty; it can't be calculated in a pairwise
# manner) EMBOSS defaults for BLODSUM62 are opening -10 and
# extension -0.5 i.e. a gap of size 3 (-11.5) has approximately
# the same penalty as a 3-character score of "-" matches (-12)
# so a pairscore of -4 is not entirely unreasonable.
colnames(MDM)[24]
rownames(MDM)[24]
colnames(MDM)[24] <- "-"
rownames(MDM)[24] <- "-"
colnames(MDM)[24]
rownames(MDM)[24]
MDM["Q", "-"]
MDM["-", "D"]
# so far so good.
# ================================================
# Tabulate pairscores for alignment
# ================================================
# It is trivial to create a pairscore vector along the
# length of the aligned sequences.
PS <- vector()
for (i in 1:lSeq) {
aa1 <- seq1[i]
aa2 <- seq2[i]
PS[i] = MDM[aa1, aa2]
}
PS
# The same vector could be created - albeit perhaps not so
# easy to understand - with the expression ...
MDM[cbind(seq1,seq2)]
# ================================================
# Calculate moving averages
# ================================================
# In order to evaluate the alignment, we will calculate a
# sliding window average over the pairscores. Somewhat surprisingly
# R doesn't (yet) have a native function for moving averages: options
# that are quoted are:
# - rollmean() in the "zoo" package http://rss.acs.unt.edu/Rdoc/library/zoo/html/rollmean.html
# - MovingAverages() in "TTR" http://rss.acs.unt.edu/Rdoc/library/TTR/html/MovingAverages.html
# - ma() in "forecast" http://robjhyndman.com/software/forecast/
# But since this is easy to code, we shall implement it ourselves.
PSma <- vector() # will hold the averages
winS <- floor(window/2) # span of elements above/below the centre
winC <- winS+1 # centre of the window
# extend the vector PS with zeros (virtual observations) above and below
PS <- c(rep(0, winS), PS , rep(0, winS))
# initialize the window score for the first position
winScore <- sum(PS[1:window])
# write the first score to PSma
PSma[1] <- winScore
# Slide the window along the sequence, and recalculate sum()
# Loop from the next position, to the last position that does not exceed the vector...
for (i in (winC + 1):(lSeq + winS)) {
# subtract the value that has just dropped out of the window
winScore <- winScore - PS[(i-winS-1)]
# add the value that has just entered the window
winScore <- winScore + PS[(i+winS)]
# put score into PSma
PSma[i-winS] <- winScore
}
# convert the sums to averages
PSma <- PSma / window
# have a quick look at the score distributions
boxplot(PSma)
hist(PSma)
# ================================================
# Plot the alignment scores
# ================================================
# normalize the scores
PSma <- (PSma-min(PSma))/(max(PSma) - min(PSma) + 0.0001)
# spread the normalized values to a desired range, n
nCol <- 10
PSma <- floor(PSma * nCol) + 1
# Assign a colorspectrum to a vector (with a bit of colormagic,
# don't worry about that for now). Dark colors are poor scores,
# "hot" colors are high scores
spect <- colorRampPalette(c("black", "red", "yellow", "white"), bias=0.4)(nCol)
# Color is an often abused aspect of plotting. One can use color to label
# *quantities* or *qualities*. For the most part, our pairscores measure amino
# acid similarity. That is a quantity and with the spectrum that we just defined
# we associte the measured quantities with the color of a glowing piece
# of metal: we start with black #000000, then first we ramp up the red
# (i.e. low-energy) part of the visible spectrum to red #FF0000, then we
# add and ramp up the green spectrum giving us yellow #FFFF00 and finally we
# add blue, giving us white #FFFFFF. Let's have a look at the spectrum:
s <- rep(1, nCol)
barplot(s, col=spect, axes=F, main="Color spectrum")
# But one aspect of our data is not quantitatively different: indels.
# We valued indels with pairscores of -4. But indels are not simply poor alignment,
# rather they are non-alignment. This means stretches of -4 values are really
# *qualitatively* different. Let's color them differently by changing the lowest
# level of the spectrum to grey.
spect[1] <- "#CCCCCC"
barplot(s, col=spect, axes=F, main="Color spectrum")
# Now we can display our alignment score vector with colored rectangles.
# Convert the integers in PSma to color values from spect
PScol <- vector()
for (i in 1:length(PSma)) {
PScol[i] <- spect[ PSma[i] ] # this is how a value from PSma is used as an index of spect
}
# Plot the scores. The code is similar to the last assignment.
# Create an empty plot window of appropriate size
plot(1,1, xlim=c(-100, lSeq), ylim=c(0, 2) , type="n", yaxt="n", bty="n", xlab="position in alignment", ylab="")
# Add a label to the left
text (-30, 1, adj=1, labels=c(paste("Mbp1:\n", code1, "\nvs.\n", code2)), cex=0.9 )
# Loop over the vector and draw boxes without border, filled with color.
for (i in 1:lSeq) {
rect(i, 0.9, i+1, 1.1, border=NA, col=PScol[i])
}
# Note that the numbers along the X-axis are not sequence numbers, but numbers
# of the alignment, i.e. sequence number + indel length. That is important to
# realize: if you would like to add the annotations from the last assignment
# which I will leave as an exercise, you need to map your sequence numbering
# into alignment numbering. Let me know in case you try that but need some help.
- That is all.
==Choosing the Sequence (formerly A3)
One of the foundations of bioinformatics is the empirical observation that related sequences conserve structure, and often function. This is the basis on which we can make inferences from well-studied model organisms in species that have not been studied as deeply. The model case for our assignments is to take annotations from baker's yeast, Saccharomyces cerevisiae and apply them to YFO.
Therefore, in this assignment we will
- use the sequence search program BLAST to retrieve a sequence similar to yeast Mbp1 in YFO;
- use a number of tools to annotate the sequence.
Keeping with our theme of sequence analysis, we will
- explore EMBOSS tools;
- compute and plot relative amino acid frequencies in R;
- and (optionally) use Chimera to explore H-bond patterns in the Mbp1 APSES domain structure.
Retrieve
In Assignment 2 you looked at sequences in YFO that are related to yeast Mbp1, by following a link from the RefSeq record. I mentioned that there are more principled ways to find related proteins: that principle is to search for similar sequences. Exactly how this works will be the subject of later lectures, but the tool that is most commonly used for this task is called BLAST (Basic Local Alignment And Search Tool). The task of this assignment is to perform a number of sequence annotations to the sequence from YFO that is most similar to Mbp1, or, more precisely, that contains an APSES domain that is most similar[3].
Search input
First, we need to define the sequence we will search with, as the search input.
Defining the sequence to search with
I have highlighted the extent of the APSES domain sequence in the previous assignment, but when you explored the corresponding structure in Chimera, you saw that the structured protein domain is larger and the additional secondary structure elements are in fact well integrated into the overall domain. This is not surprising: canonical domain definitions are compiled from many species and examples, and they generally comprise only the common core. Looking up the source of the domain annotations for Mbp1 is very easy:
Task:
- Access the RefSeq record for yeast Mbp1.
- While you are here, download a FASTA formatted version of the sequence to your R working directory and give it a filename of
mbp1-sacce.fa. We will need it later. It should be straightforward from the NCBI page how to achieve that. As a hint, you need to use the Send to... link to actually download the file. - On the RefSeq page, look for the link Related Information → CDD Search Results and follow it.
This is a domain annotation: CDD is the NCBI's Conserved Domain Database and the annotation was done by a tool that scanned the sequence of Mbp1 for segments that are similar to any of the domain definitions stored in the CDD. We will return to CDD in the next assignment.
- Click on the blue box labeled Kila-N in the graph to access the CDD entry for this domain.
- Read the abstract. You should understand the relationship between Kila-N and APSES domains. One is a subfamily of the other.
- Confirm that the domain definition – as applied to the Mbp1 sequence (which is labeled as "query") – corresponds to the region we highlighted in the last assignment.
What precisely constitutes an APSES domain however is a matter of definition, as you can explore in the following (optional) task.
There is a rather important lesson in this: domain definitions may be fluid, and their boundaries may be computationally derived from sequence comparisons across many families, and do not necessarily correspond to individual structures. Make sure you understand this well.
Given this, it seems appropriate to search the sequence database with the sequence of an Mbp1 structure–this being a structured, stable, subdomain of the whole that presumably contains the protein's most unique and specific function. Let us retrieve this sequence. All PDB structures have their sequences stored in the NCBI protein database. They can be accessed simply via the PDB-ID, which serves as an identifier both for the NCBI and the PDB databases. However there is a small catch (isn't there always?). PDB files can contain more than one protein, e.g. if the crystal structure contains a complex[4]. Each of the individual proteins gets a so-called chain ID–a one letter identifier– to identify them uniquely. To find their unique sequence in the database, you need to know the PDB ID as well as the chain ID. If the file contains only a single protein (as in our case), the chain ID is always A[5]. make sure you understand the concept of protein chains, and chain IDs.
Task:
- Back at the RefSeq record for yeast Mbp1, enter the PDB-ID, an underscore, and the chain ID for one of the crystal structures into the search field. You can use
1MB1_Aor1BM8_A, but don't use1L3G: this NMR structure includes a large stretch of unstructured residues. - Click on Display settings and choose FASTA (text). You should get something like:
>gi|157830387|pdb|1BM8|A Chain A, Dna-Binding Domain Of Mbp1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKETHEKVQGGFGKY QGTWVPLNIAKQLAEKFSVYDQLKPLFDF
- Save this sequence in your notebook, in case we need it later.
Next, we use this sequence to find its most similar relative in YFO using BLAST.
BLAST search
Task:
- Navigate to the BLAST entry page at the NCBI.
- Click on protein blast as the BLAST program to run.
- Paste the sequence of the yeast Mbp1 DNA-binding domain into the search field.
- Set the following parameters:
- As Database option choose Reference proteins (refseq_protein)
- As Organism enter the binomial name of YFO. Make sure you spell it right, the page will try to autocomplete your entry. Species level is detailed enough, you don't have to specify the strain (e.g. I would specify "Ustilago maydis" not "Ustilago maydis 521").
- Then click on the BLAST button and wait for the result to appear. You will first see a graph of any conserved domains in your query sequence, this is not yet what you are waiting for...
- Patience.
- Patience. The database is large.
- Patience. Execution times vary greatly by time of day.
- The top "hit" on the results page is what you are looking for. Its alignment and alignment score are shown in the Alignments section a bit further down the page. Your hit should have on the order of more than 40% identities to the query and match at least 80 residues or so. If your match seems less and worse than that, please eMail me to troubleshoot.
- The first item for each hit is a link to its database entry, right next to the checkbox. It says something like
ref|NP_123456789orref|XP_123456789... follow that link. - Note the RefSeq ID, and save the sequence in FASTA format into your R working directory, as you did for Mbp1 at the beginning of the assignment. Give this a filename of
mbp1-xxxxx.fa, but replacexxxxxwith its short species label for YFO. For simplicity I will refer to this sequence as "YFO Mbp1" in the future.
MSA (formerly A5)
- Take care of things, and they will take care of you.
- Shunryu Suzuki
Anyone can click buttons on a Web page, but to use the powerful sequence database search tools right often takes considerable more care, caution and consideration.
Much of what we know about a protein's physiological function is based on the conservation of that function as the species evolves. We assess conservation by comparing sequences between related proteins. Conservation - or its opposite: variation - is a consequence of selection under constraints: protein sequences change as a consequence of DNA mutations, this changes the protein's structure, this in turn changes functions and that has the multiple effects on a species' fitness function. Detrimental variants may be removed. Variation that is tolerated is largely neutral and therefore found only in positions that are neither structurally nor functionally critical. Conservation patterns can thus provide evidence for many different questions: structural conservation among proteins with similar 3D-structures, functional conservation among homologues with comparable roles, or amino acid propensities as predictors for protein engineering and design tasks.
Measuring conservation requires alignment. Therefore a carefully done multiple sequence alignment (MSA) is a cornerstone for the annotation of the essential properties a gene or protein. MSAs are also useful to resolve ambiguities in the precise placement of indels and to ensure that columns in alignments actually contain amino acids that evolve in a similar context. MSAs serve as input for
- functional annotation;
- protein homology modeling;
- phylogenetic analyses, and
- sensitive homology searches in databases.
In order to perform a multiple sequence alignment, we obviously need a set of homologous sequences. This is where the trouble begins. All interpretation of MSA results depends absolutely on how the input sequences were chosen. Should we include only orthologs, or paralogs as well? Should we include only species with fully sequenced genomes, or can we tolerate that some orthologous genes are possibly missing for a species? Should we include all sequences we can lay our hands on, or should we restrict the selection to a manageable number of representative sequences? All of these choices influence our interpretation:
- orthologs are expected to be functionally and structurally conserved;
- paralogs may have divergent function but have similar structure;
- missing genes may make paralogs look like orthologs; and
- selection bias may weight our results toward sequences that are over-represented and do not provide a fair representation of evolutionary divergence.
In this assignment, we will set ourselves the task to use PSI-BLAST and find all orthologs and paralogs of the APSES domain containing transcription factors in YFO. We will use these sequences later for multiple alignments, calculation of conservation etc. The methodical problem we will address is: how do we perform a sensitive PSI-BLAST search in one organism. There is an issue to consider:
- If we restrict the PSI-BLAST search to YFO, PSI-BLAST has little chance of building a meaningful profile - the number of homologues that actually are in YFO is too small. Thus the search will not become very sensitive.
- If we don't restrict our search, but search in all species, the number of hits may become too large. It becomes increasingly difficult to closely check all hits as to whether they have good coverage, and how will we evaluate the fringe cases of marginal E-value, where we need to decide whether to include a new sequence in the profile, or whether to hold off on it for one or two iterations, to see whether the E-value drops significantly. Profile corruption would make the search useless. This is maybe still manageable if we restrict our search to fungi, but imagine you are working with a bacterial protein, or a protein that is conserved across the entire tree of life: your search will find thousands of sequences. And by next year, thousands more will have been added.
Therefore we have to find a middle ground: add enough species (sequences) to compile a sensitive profile, but not so many that we can no longer individually assess the sequences that contribute to the profile.
Thus in practice, a sensitive PSI-BLAST search needs to address two issues before we begin:
- We need to define the sequence we are searching with; and
- We need to define the dataset we are searching in.
Defining the sequence to search with
Consider again the task we set out from: find all orthologs and paralogs of the APSES domain containing transcription factors in YFO.
Task:
What query sequence should you use? Should you ...
- Search with the full-length Mbp1 sequence from Saccharomyces cerevisiae?
- Search with the full-length Mbp1 homolog that you found in YFO?
- Search with the structurally defined S. cerevisiae APSES domain sequence?
- Search with the APSES domain sequence from the YFO homolog, that you have defined by sequence alignment with the yeast protein?
- Search with the KilA-N domain sequence?
Selecting species for a PSI-BLAST search
As discussed in the introduction, in order to use our sequence set for studying structural and functional features and conservation patterns of our APSES domain proteins, we should start with a well selected dataset of APSES domain containing homologs in YFO. Since these may be quite divergent, we can't rely on BLAST to find all of them, we need to use the much more sensitive search of PSI-BLAST instead. But even though you are interested only in YFO's genes, it would be a mistake to restrict the PSI-BLAST search to YFO. PSI-BLAST becomes more sensitive if the profile represents more diverged homologs. Therefore we should always search with a broadly representative set of species, even if we are interested only in the results for one of the species. This is important. Please reflect on this for a bit and make sure you understand the rationale why we include sequences in the search that we are not actually interested in.
But you can also search with too many species: if the number of species is large and PSI-BLAST finds a large number of results:
- it becomes unwieldy to check the newly included sequences at each iteration, inclusion of false-positive hits may result, profile corruption and loss of specificity. The search will fail.
- since genomes from some parts of the Tree Of Life are over represented, the inclusion of all sequences leads to selection bias and loss of sensitivity.
We should therefore try to find a subset of species
- that represent as large a range as possible on the evolutionary tree;
- that are as well distributed as possible on the tree; and
- whose genomes are fully sequenced.
These criteria are important. Again, reflect on them and understand their justification. Choosing your species well for a PSI-BLAST search can be crucial to obtain results that are robust and meaningful.
How can we define a list of such species, and how can we use the list?
The definition is a rather typical bioinformatics task for integrating datasources: "retrieve a list of representative fungi with fully sequenced genomes". Unfortunately, to do this in a principled way requires tools that you can't (yet) program: we would need to use a list of genome sequenced fungi, estimate their evolutionary distance and select a well-distributed sample. Regrettably you can't combine such information easily with the resources available from the NCBI.
We will use an approach that is conceptually similar: selecting a set of species according to their shared taxonomic rank in the tree of life. Biological classification provides a hierarchical system that describes evolutionary relatedness for all living entities. The levels of this hierarchy are so called taxonomic ranks. These ranks are defined in Codes of Nomenclature that are curated by the self-governed international associations of scientists working in the field. The number of ranks is not specified: there is a general consensus on seven principal ranks (see below, in bold) but many subcategories exist and may be newly introduced. It is desired–but not mandated–that ranks represent clades (a group of related species, or a "branch" of a phylogeny), and it is desired–but not madated–that the rank is sharply defined. The system is based on subjective dissimilarity. Needless to say that it is in flux.
If we follow a link to an entry in the NCBI's Taxonomy database, eg. Saccharomyces cerevisiae S228c, the strain from which the original "yeast genome" was sequenced in the late 1990s, we see the following specification of its taxonomic lineage:
cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya;
Ascomycota; Saccharomyceta; Saccharomycotina; Saccharomycetes;
Saccharomycetales; Saccharomycetaceae; Saccharomyces; Saccharomyces cerevisiae
These names can be mapped into taxonomic ranks ranks, since the suffixes of these names e.g. -mycotina, -mycetaceae are specific to defined ranks. (NCBI does not provide this mapping, but Wikipedia is helpful here.)
| Rank | Suffix | Example |
| Domain | Eukaryota (Eukarya) | |
| Subdomain | Opisthokonta | |
| Kingdom | Fungi | |
| Subkingdom | Dikarya | |
| Phylum | Ascomycota | |
| rankless taxon[6] | -myceta | Saccharomyceta |
| Subphylum | -mycotina | Saccharomycotina |
| Class | -mycetes | Saccharomycetes |
| Subclass | -mycetidae | |
| Order | -ales | Saccharomycetales |
| Family | -aceae | Saccharomycetaceae |
| Subfamily | -oideae | |
| Tribe | -eae | |
| Subtribe | -ineae | |
| Genus | Saccharomyces | |
| Species | Saccharomyces cerevisiae |
- Capnodiales Zymoseptoria tritici
- Erysiphales Blumeria graminis
- Eurotiales Aspergillus nidulans
- Glomerellales Glomerella graminicola
- Hypocreales Trichoderma reesei
- Magnaporthales Magnaporthe oryzae
- Microbotryales Microbotryum violaceum
- Pezizales Tuber melanosporum
- Pleosporales Phaeosphaeria nodorum
- Pucciniales Puccinia graminis
- Saccharomycetales Saccharomyces cerevisiae
- Schizosaccharomycetales Schizosaccharomyces pombe
- Sclerotiniaceae Sclerotinia sclerotiorum
- Sordariales Neurospora crassa
- Tremellales Cryptococcus neoformans
- Ustilaginales Ustilago maydis
Aspergillus nidulans[orgn]
OR Blumeria graminis[orgn]
OR Cryptococcus neoformans[orgn]
OR Glomerella graminicola[orgn]
OR Magnaporthe oryzae[orgn]
OR Microbotryum violaceum[orgn]
OR Neurospora crassa[orgn]
OR Phaeosphaeria nodorum[orgn]
OR Puccinia graminis[orgn]
OR Sclerotinia sclerotiorum[orgn]
OR Trichoderma reesei[orgn]
OR Tuber melanosporum[orgn]
OR Saccharomyces cerevisiae[orgn]
OR Schizosaccharomyces pombe[orgn]
OR Ustilago maydis[orgn]
OR Zymoseptoria tritici[orgn]
Executing the PSI-BLAST search
We have a list of species. Good. Next up: how do we use it.
Task:
- Navigate to the BLAST homepage.
- Select protein BLAST.
- Paste the APSES domain sequence into the search field.
- Select refseq as the database.
- Copy the organism restriction list from above and enter the correct name for YFO into the list if it is not there already. Obviously, you can't find sequences in YFO if YFO is not included in your search space. Paste the list into the Entrez Query field.
- In the Algorithm section, select PSI-BLAST.
- Click on BLAST.
Evaluate the results carefully. Since we used default parameters, the threshold for inclusion was set at an E-value of 0.005 by default, and that may be a bit too lenient. If you look at the table of your hits– in the Sequences producing significant alignments... section– there may also be a few sequences that have a low query coverage of less than 80%. Let's exclude these from the profile initially: not to worry, if they are true positives, the will come back with improved E-values and greater coverage in subsequent iterations. But if they were false positives, their E-values will rise and they should drop out of the profile and not contaminate it.
Task:
- In the header section, click on Formatting options and in the line "Format for..." set the with inclusion threshold to
0.001(This means E-values can't be above 10-03 for the sequence to be included.) - Click on the Reformat button (top right).
- In the table of sequence descriptions (not alignments!), click on the Query coverage to sort the table by coverage, not by score.
- Copy the rows with a coverage of less than 80% and paste them into some text editor so you can compare what happens with these sequences in the next iteration.
- Deselect the check mark next to these sequences in the right-hand column Select for PSI blast. (For me these are six sequences, but with YFO included that may be a bit different.)
- Then scroll to Run PSI-BLAST iteration 2 ... and click on
Go.
This is now the "real" PSI-BLAST at work: it constructs a profile from all the full-length sequences and searches with the profile, not with any individual sequence. Note that we are controlling what goes into the profile in two ways:
- we are explicitly removing sequences with poor coverage; and
- we are requiring a more stringent minimum E-value for each sequence.
Task:
- Again, study the table of hits. Sequences highlighted in yellow have met the search criteria in the second iteration. Note that the coverage of (some) of the previously excluded sequences is now above 80%.
- Let's exclude partial matches one more time. Again, deselect all sequences with less than 80% coverage. Then run the third iteration.
- Iterate the search in this way until no more "New" sequences are added to the profile. Then scan the list of excluded hits ... are there any from YFO that seem like they could potentially make the list? Marginal E-value perhaps, or reasonable E-value but less coverage? If that's the case, try returning the E-value threshold to the default 0.005 and see what happens...
Once no "new" sequences have been added, if we were to repeat the process again and again, we would always get the same result because the profile stays the same. We say that the search has converged. Good. Time to harvest.
Task:
- At the header, click on Taxonomy reports and find YFO in the Organism Report section. These are your APSES domain homologs. All of them. Actually, perhaps more than all: the report may also include sequences with E-values above the inclusion threshold.
- From the report copy the sequence identifiers
- from YFO,
- with E-values above your defined threshold.
For example, the list of Saccharomyces genes is the following:
Saccharomyces cerevisiae S288c [ascomycetes] taxid 559292
ref|NP_010227.1| Mbp1p [Saccharomyces cerevisiae S288c] [ 131] 1e-38
ref|NP_011036.1| Swi4p [Saccharomyces cerevisiae S288c] [ 123] 1e-35
ref|NP_012881.1| Phd1p [Saccharomyces cerevisiae S288c] [ 91] 1e-25
ref|NP_013729.1| Sok2p [Saccharomyces cerevisiae S288c] [ 93] 3e-25
ref|NP_012165.1| Xbp1p [Saccharomyces cerevisiae S288c] [ 40] 5e-07
Xbp1 is a special case. It has only very low coverage, but that is because it has a long domain insertion and the N-terminal match often is not recognized by alignment because the gap scores for long indels are unrealistically large. For now, I keep that sequence with the others.
Next we need to retrieve the sequences. Tedious to retrieve them one by one, but we can get them all at the same time:
Task:
- Return to the BLAST results page and again open the Formatting options.
- Find the Limit results section and enter YFO's name into the field. For example
Saccharomyces cerevisiae [ORGN] - Click on Reformat
- Scroll to the Descriptions section, check the box at the left-hand margin, next to each sequence you want to keep. Then click on Download → FASTA complete sequence → Continue.
Introduction
In the last assignment we discovered homologs to S. cerevisiae Mbp1 in YFO. Some of these will be orthologs to Mbp1, some will be paralogs. Some will have similar function, some will not. We discussed previously that genes that evolve under continuously similar evolutionary pressure should be most similar in sequence, and should have the most similar "function".
In this assignment we will define the YFO gene that is the most similar ortholog to S. cerevisiae Mbp1, and perform a multiple sequence alignment with it.
Let us briefly review the basic concepts.
Orthologs and Paralogs revisited
- All related genes are homologs.
Two central definitions about the mutual relationships between related genes go back to Walter Fitch who stated them in the 1970s:
- Orthologs have diverged after speciation.
- Paralogs have diverged after duplication.
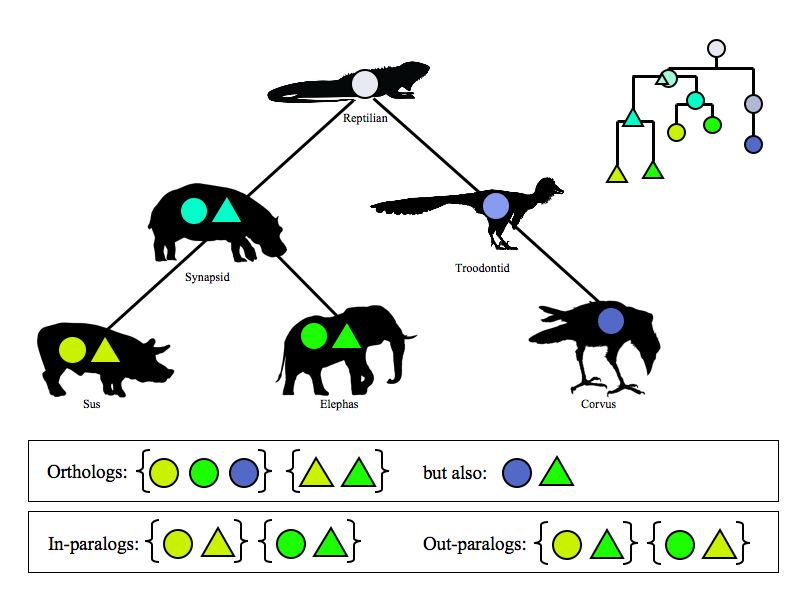
Defining orthologs
To be reasonably certain about orthology relationships, we would need to construct and analyze detailed evolutionary trees. This is computationally expensive and the results are not always unambiguous either, as we will see in a later assignment. But a number of different strategies are available that use precomputed results to define orthologs. These are especially useful for large, cross genome surveys. They are less useful for detailed analysis of individual genes. Pay the sites a visit and try a search.
- Orthologs by eggNOG
- The eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) database contains orthologous groups of genes at the EMBL. It seems to be continuously updtaed, the search functionality is reasonable and the results for yeast Mbp1 show many genes from several fungi. Importantly, there is only one gene annotated for each species. Alignments and trees are also available, as are database downloads for algorithmic analysis.
| Powell et al. (2014) eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231-9. (pmid: 24297252) |
- Orthologs at OrthoDB
- OrthoDB includes a large number of species, among them all of our protein-sequenced fungi. However the search function (by keyword) retrieves many paralogs together with the orthologs, for example, the yeast Soc2 and Phd1 proteins are found in the same orthologous group these two are clearly paralogs.
- Orthologs at OMA
OMA (the Orthologous Matrix) maintained at the Swiss Federal Institute of Technology contains a large number of orthologs from sequenced genomes. Searching with MBP1_YEAST (this is the Swissprot ID) as a "Group" search finds the correct gene in EREGO, KLULA, CANGL and SACCE. But searching with the sequence of the Ustilago maydis ortholog does not find the yeast protein, but the orthologs in YARLI, SCHPO, LACCBI, CRYNE and USTMA. Apparently the orthologous group has been split into several subgroups across the fungi. However as a whole the database is carefully constructed and available for download and API access; a large and useful resource.
- Orthologs by syntenic gene order conservation
- We will revisit this when we explore the UCSC genome browser.
- Orthologs by RBM
- Defining it yourself. RBM (or: Reciprocal Best Match) is easy to compute and half of the work you have already done in Assignment 3. Get the ID for the gene which you have identified and annotated as the best BLAST match for Mbp1 in YFO and confirm that this gene has Mbp1 as the most significant hit in the yeast proteome. The results are unambiguous, but there may be residual doubt whether these two best-matching sequences are actually the most similar orthologs.
Task:
- Navigate to the BLAST homepage.
- Paste the YFO RefSeq sequence identifier into the search field. (You don't have to search with sequences–you can search directly with an NCBI identifier IF you want to search with the full-length sequence.)
- Set the database to refseq, and restrict the species to Saccharomyces cerevisiae.
- Run BLAST.
- Keep the window open for the next task.
The top hit should be yeast Mbp1 (NP_010227). E mail me your sequence identifiers if it is not. If it is, you have confirmed the RBM or BBM criterion (Reciprocal Best Match or Bidirectional Best Hit, respectively).
Technically, this is not perfectly true since you have searched with the APSES domain in one direction, with the full-length sequence in the other. For this task I wanted you to try the search-with-accession-number. Therefore the procedural laxness, I hope it is permissible. In fact, performing the reverse search with the YFO APSES domain should actually be more stringent, i.e. if you find the right gene with the longer sequence, you are even more likely to find the right gene with the shorter one.
- Orthology by annotation
- The NCBI precomputes BLAST results and makes them available at the RefSeq database entry for your protein.
Task:
- In your BLAST result page, click on the RefSeq link for your query to navigate to the RefSeq database entry for your protein.
- Follow the Blink link in the right-hand column under Related information.
- Restrict the view RefSeq under the "Display options" and to Fungi.
You should see a number of genes with low E-values and high coverage in other fungi - however this search is problematic since the full length gene across the database finds mostly Ankyrin domains.
You will find that all of these approaches yield some of the orthologs. But none finds them all. The take home message is: precomputed results are good for large-scale survey-type investigations, where you can't humanly process the information by hand. But for more detailed questions, careful manual searches are still indsipensable.
- Orthology by crowdsourcing
- Luckily a crowd of willing hands has prepared the necessary sequences for you: in the section below you will find a link to the annotated and verified Mbp1 orthologs from last year's course :-)
Align and Annotate
Review of domain annotations
APSES domains are relatively easy to identify and annotate but we have had problems with the ankyrin domains in Mbp1 homologues. Both CDD as well as SMART have identified such domains, but while the domain model was based on the same Pfam profile for both, and both annotated approximately the same regions, the details of the alignments and the extent of the predicted region was different.
Mbp1 forms heterodimeric complexes with a homologue, Swi6. Swi6 does not have an APSES domain, thus it does not bind DNA. But it is similar to Mbp1 in the region spanning the ankyrin domains and in 1999 Foord et al. published its crystal structure (1SW6). This structure is a good model for Ankyrin repeats in Mbp1. For details, please refer to the consolidated Mbp1 annotation page I have prepared.
In what follows, we will use the program JALVIEW - a Java based multiple sequence alignment editor to load and align sequences and to consider structural similarity between yeast Mbp1 and its closest homologue in your organism.
In this part of the assignment,
- You will load sequences that are most similar to Mbp1 into an MSA editor;
- You will add sequences of ankyrin domain models;
- You will perform a multiple sequence alignment;
- You will try to improve the alignment manually;
Jalview, loading sequences
Geoff Barton's lab in Dundee has developed an integrated MSA editor and sequence annotation workbench with a number of very useful functions. It is written in Java and should run on Mac, Linux and Windows platforms without modifications.
| Waterhouse et al. (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189-91. (pmid: 19151095) |
We will use this tool for this assignment and explore its features as we go along.
Task:
- Navigate to the Jalview homepage click on Download, install Jalview on your computer and start it. A number of windows that showcase the program's abilities will load, you can close these.
- Prepare homologous Mbp1 sequences for alignment:
- Open the Reference Mbp1 orthologues (all fungi) page. (This is the list of Mbp1 orthologs I mentioned above.)
- Copy the FASTA sequences of the reference proteins, paste them into a text file (TextEdit on the Mac, Notepad on Windows) and save the file; you could give it an extension of
.fa–but you don't have to. - Check whether the sequence for YFO is included in the list. If it is, fine. If it is not, retrieve it from NCBI, paste it into the file and edit the header like the other sequences. If the wrong sequence from YFO is included, replace it and let me know.
- Return to Jalview and select File → Input Alignment → from File and open your file. A window with sequences should appear.
- Copy the sequences for ankyrin domain models (below), click on the Jalview window, select File → Add sequences → from Textbox and paste them into the Jalview textbox. Paste two separate copies of the CD00204 consensus sequence and one copy of 1SW6.
- When all the sequences are present, click on Add.
Jalview now displays all the sequences, but of course this is not yet an alignment.
- Ankyrin domain models
>CD00204 ankyrin repeat consensus sequence from CDD NARDEDGRTPLHLAASNGHLEVVKLLLENGADVNAKDNDGRTPLHLAAKNGHLEIVKLLL EKGADVNARDKDGNTPLHLAARNGNLDVVKLLLKHGADVNARDKDGRTPLHLAAKNGHL
>1SW6 from PDB - unstructured loops replaced with xxxx GPIITFTHDLTSDFLSSPLKIMKALPSPVVNDNEQKMKLEAFLQRLLFxxxxSFDSLLQE VNDAFPNTQLNLNIPVDEHGNTPLHWLTSIANLELVKHLVKHGSNRLYGDNMGESCLVKA VKSVNNYDSGTFEALLDYLYPCLILEDSMNRTILHHIIITSGMTGCSAAAKYYLDILMGW IVKKQNRPIQSGxxxxDSILENLDLKWIIANMLNAQDSNGDTCLNIAARLGNISIVDALL DYGADPFIANKSGLRPVDFGAG
Computing alignments
The EBI has a very convenient page to access a number of MSA algorithms. This is especially convenient when you want to compare, e.g. T-Coffee and Muscle and MAFFT results to see which regions of your alignment are robust. You could use any of these tools, just paste your sequences into a Webform, download the results and load into Jalview. Easy.
But even easier is to calculate the alignments directly from Jalview. available. (Not today. Bummer.)
- Calculate a MAFFT alignment using the Jalview Web service option
Task:
- In Jalview, select Web Service → Alignment → MAFFT with defaults.... The alignment is calculated in a few minutes and displayed in a new window.
- Calculate a MAFFT alignment when the Jalview Web service is NOT available
Task:
- In Jalview, select File → Output to Textbox → FASTA
- Copy the sequences.
- Navigate to the MAFFT Input form at the EBI.
- Paste your sequences into the form.
- Click on Submit.
- Close the Jalview sequence window and either save your MAFFT alignment to file and load in Jalview, or simply 'File → Input Alignment → from Textbox, paste and click New Window.
In any case, you should now have an alignment.
Task:
- Choose Colour → Hydrophobicity and → by Conservation. Then adjust the slider left or right to see which columns are highly conserved. You will notice that the Swi6 sequence that was supposed to align only to the ankyrin domains was in fact aligned to other parts of the sequence as well. This is one part of the MSA that we will have to correct manually and a common problem when aligning sequences of different lengths.
Editing ankyrin domain alignments
A good MSA comprises only columns of residues that play similar roles in the proteins' mechanism and/or that evolve in a comparable structural context. Since the alignment reflects the result of biological selection and conservation, it has relatively few indels and the indels it has are usually not placed into elements of secondary structure or into functional motifs. The contiguous features annotated for Mbp1 are expected to be left intact by a good alignment.
A poor MSA has many errors in its columns; these contain residues that actually have different functions or structural roles, even though they may look similar according to a (pairwise!) scoring matrix. A poor MSA also may have introduced indels in biologically irrelevant positions, to maximize spurious sequence similarities. Some of the features annotated for Mbp1 will be disrupted in a poor alignment and residues that are conserved may be placed into different columns.
Often errors or inconsistencies are easy to spot, and manually editing an MSA is not generally frowned upon, even though this is not a strictly objective procedure. The main goal of manual editing is to make an alignment biologically more plausible. Most comonly this means to mimize the number of rare evolutionary events that the alignment suggests and/or to emphasize conservation of known functional motifs. Here are some examples for what one might aim for in manually editing an alignment:
- Reduce number of indels
From a Probcons alignment: 0447_DEBHA ILKTE-K-T---K--SVVK ILKTE----KTK---SVVK 9978_GIBZE MLGLN-PGLKEIT--HSIT MLGLNPGLKEIT---HSIT 1513_CANAL ILKTE-K-I---K--NVVK ILKTE----KIK---NVVK 6132_SCHPO ELDDI-I-ESGDY--ENVD ELDDI-IESGDY---ENVD 1244_ASPFU ----N-PGLREIC--HSIT -> ----NPGLREIC---HSIT 0925_USTMA LVKTC-PALDPHI--TKLK LVKTCPALDPHI---TKLK 2599_ASPTE VLDAN-PGLREIS--HSIT VLDANPGLREIS---HSIT 9773_DEBHA LLESTPKQYHQHI--KRIR LLESTPKQYHQHI--KRIR 0918_CANAL LLESTPKEYQQYI--KRIR LLESTPKEYQQYI--KRIR
Gaps marked in red were moved. The sequence similarity in the alignment does not change considerably, however the total number of indels in this excerpt is reduced to 13 from the original 22
- Move indels to more plausible position
From a CLUSTAL alignment: 4966_CANGL MKHEKVQ------GGYGRFQ---GTW MKHEKVQ------GGYGRFQ---GTW 1513_CANAL KIKNVVK------VGSMNLK---GVW KIKNVVK------VGSMNLK---GVW 6132_SCHPO VDSKHP-----------QID---GVW -> VDSKHPQ-----------ID---GVW 1244_ASPFU EICHSIT------GGALAAQ---GYW EICHSIT------GGALAAQ---GYW
The two characters marked in red were swapped. This does not change the number of indels but places the "Q" into a a column in which it is more highly conserved (green). Progressive alignments are especially prone to this type of error.
- Conserve motifs
From a CLUSTAL alignment: 6166_SCHPO --DKRVA---GLWVPP --DKRVA--G-LWVPP XBP1_SACCE GGYIKIQ---GTWLPM GGYIKIQ--G-TWLPM 6355_ASPTE --DEIAG---NVWISP -> ---DEIA--GNVWISP 5262_KLULA GGYIKIQ---GTWLPY GGYIKIQ--G-TWLPY
The first of the two residues marked in red is a conserved, solvent exposed hydrophobic residue that may mediate domain interactions. The second residue is the conserved glycine in a beta turn that cannot be mutated without structural disruption. Changing the position of a gap and insertion in one sequence improves the conservation of both motifs.
The Ankyrin domains are quite highly diverged, the boundaries not well defined and not even CDD, SMART and SAS agree on the precise annotations. We expect there to be alignment errors in this region. Nevertheless we would hope that a good alignment would recognize homology in that region and that ideally the required indels would be placed between the secondary structure elements, not in their middle. But judging from the sequence alignment alone, we cannot judge where the secondary structure elements ought to be. You should therefore add the following "sequence" to the alignment; it contains exactly as many characters as the Swi6 sequence above and annotates the secondary structure elements. I have derived it from the 1SW6 structure
>SecStruc 1SW6 E: strand t: turn H: helix _: irregular _EEE__tt___ttt______EE_____t___HHHHHHHHHHHHHHHH_xxxx_HHHHHHH HHHH_t_____t_____t____HHHHHHH__tHHHHHHHHH____t___tt____HHHHH HH__HHHH___HHHHHHHHHHHHHEE_t____HHHHHHHHH__t__HHHHHHHHHHHHHH HHHHHH__EEE_xxxx_HHHHHt_HHHHHHH______t____HHHHHHHH__HHHHHHHH H____t____t____HHHH___
To proceed:
- Manually align the Swi6 sequence with yeast Mbp1
- Bring the Secondary structure annotation into its correct alignment with Swi6
- Bring both CDD ankyrin profiles into the correct alignment with yeast Mbp1
Proceed along the following steps:
Task:
- Add the secondary structure annotation to the sequence alignment in Jalview. Copy the annotation, select File → Add sequences → from Textbox and paste the sequence.
- Select Help → Documentation and read about Editing Alignments, Cursor Mode and Key strokes.
- Click on the yeast Mbp1 sequence row to select the entire row. Then use the cursor key to move that sequence down, so it is directly above the 1SW6 sequence. Select the row of 1SW6 and use shift/mouse to move the sequence elements and edit the alignment to match yeast Mbp1. Refer to the alignment given in the Mbp1 annotation page for the correct alignment.
- Align the secondary structure elements with the 1SW6 sequence: Every character of 1SW6 should be matched with either E, t, H, or _. The result should be similar to the Mbp1 annotation page. If you need to insert gaps into all sequences in the alignment, simply drag your mouse over all row headers - movement of sequences is constrained to selected regions, the rest is locked into place to prevent inadvertent misalignments. Remember to save your project from time to time: File → save so you can reload a previous state if anything goes wrong and can't be fixed with Edit → Undo.
- Finally align the two CD00204 consensus sequences to their correct positions (again, refer to the Mbp1 annotation page).
- You can now consider the principles stated above and see if you can improve the alignment, for example by moving indels out of regions of secondary structure if that is possible without changing the character of the aligned columns significantly. Select blocks within which to work to leave the remaining alignment unchanged. So that this does not become tedious, you can restrict your editing to one Ankyrin repeat that is structurally defined in Swi6. You may want to open the 1SW6 structure in VMD to define the boundaries of one such repeat. You can copy and paste sections from Jalview into your assignment for documentation or export sections of the alignment to HTML (see the example below).
Editing ankyrin domain alignments - Sample
This sample was created by
- Editing the alignments as described above;
- Copying a block of aligned sequence;
- Pasting it To New Alignment;
- Colouring the residues by Hydrophobicity and setting the colour saturation according to Conservation;
- Choosing File → Export Image → HTML and pasting the resulting HTML source into this Wikipage.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Aligned sequences before editing. The algorithm has placed gaps into the Swi6 helix
LKWIIANand the four-residue gaps before the block of well aligned sequence on the right are poorly supported.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Aligned sequence after editing. A significant cleanup of the frayed region is possible. Now there is only one insertion event, and it is placed into the loop that connects two helices of the 1SW6 structure.
Final analysis
Task:
- Compare the distribution of indels in the ankyrin repeat regions of your alignments.
- Review whether the indels in this region are concentrated in segments that connect the helices, or if they are more or less evenly distributed along the entire region of similarity.
- Think about whether the assertion that indels should not be placed in elements of secondary structure has merit in your alignment.
- Recognize that an indel in an element of secondary structure could be interpreted in a number of different ways:
- The alignment is correct, the annotation is correct too: the indel is tolerated in that particular case, for example by extending the length of an α-helix or β-strand;
- The alignment algorithm has made an error, the structural annotation is correct: the indel should be moved a few residues;
- The alignment is correct, the structural annotation is wrong, this is not a secondary structure element after all;
- Both the algorithm and the annotation are probably wrong, but we have no data to improve the situation.
(NB: remember that the structural annotations have been made for the yeast protein and might have turned out differently for the other proteins...)
You should be able to analyse discrepancies between annotation and expectation in a structured and systematic way. In particular if you notice indels that have been placed into an annotated region of secondary structure, you should be able to comment on whether the location of the indel has strong support from aligned sequence motifs, or whether the indel could possibly be moved into a different location without much loss in alignment quality.
- Considering the whole alignment and your experience with editing, you should be able to state whether the position of indels relative to structural features of the ankyrin domains in your organism's Mbp1 protein is reliable. That would be the result of this task, in which you combine multiple sequence and structural information.
- You can also critically evaluate database information that you have encountered:
- Navigate to the CDD annotation for yeast Mbp1.
- You can check the precise alignment boundaries of the ankyrin domains by clicking on the (+) icon to the left of the matching domain definition.
- Confirm that CDD extends the ankyrin domain annotation beyond the 1SW6 domain boundaries. Given your assessment of conservation in the region beyond the structural annotation: do you think that extending the annotation is reasonable also in YFO's protein? Is there evidence for this in the alignment of the CD00204 consensus with well aligned blocks of sequence beyond the positions that match Swi6?
R code: load alignment and compute information scores
As discussed in the lecture, Shannon information is calculated as the difference between expected and observed entropy, where entropy is the negative sum over probabilities times the log of those probabilities:
Here we compute Shannon information scores for aligned positions of the APSES domain, and plot the values in R. You can try this with any part of your alignment, but I have used only the aligned residues for the APSES domain for my example. This is a good choice for a first try, since there are (almost) no gaps.
Task:
- Export only the sequences of the aligned APSES domains to a file on your computer, in FASTA format as explained below. You could call this:
Mbp1_All_APSES.fa.- Use your mouse and clik and drag to select the aligned APSES domains in the alignment window.
- Copy your selection to the clipboard.
- Use the main menu (not the menu of your alignment window) and select File → Input alignment → from Textbox; paste the selection into the textbox and click New Window.
- Use File → save as to save the aligned siequences in multi-FASTA format under the filename you want in your R project directory.
- Explore the R-code below. Be sure that you understand it correctly. Note that this code does not implement any sampling bias correction, so positions with large numbers of gaps will receive artificially high scores (the alignment looks like the gap charecter were a conserved character).
# CalculateInformation.R
# Calculate Shannon information for positions in a multiple sequence alignment.
# Requires: an MSA in multi FASTA format
# It is good practice to set variables you might want to change
# in a header block so you don't need to hunt all over the code
# for strings you need to update.
#
setwd("/your/R/working/directory")
mfa <- "MBP1_All_APSES.fa"
# ================================================
# Read sequence alignment fasta file
# ================================================
# read MFA datafile using seqinr function read.fasta()
library(seqinr)
tmp <- read.alignment(mfa, format="fasta")
MSA <- as.matrix(tmp) # convert the list into a characterwise matrix
# with appropriate row and column names using
# the seqinr function as.matrix.alignment()
# You could have a look under the hood of this
# function to understand beter how to convert a
# list into something else ... simply type
# "as.matrix.alignment" - without the parentheses
# to retrieve the function source code (as for any
# function btw).
### Explore contents of and access to the matrix of sequences
MSA
MSA[1,]
MSA[,1]
length(MSA[,1])
# ================================================
# define function to calculate entropy
# ================================================
entropy <- function(v) { # calculate shannon entropy for the aa vector v
# Note: we are not correcting for small sample sizes
# here. Thus if there are a large number of gaps in
# the alignment, this will look like small entropy
# since only a few amino acids are present. In the
# extreme case: if a position is only present in
# one sequence, that one amino acid will be treated
# as 100% conserved - zero entropy. Sampling error
# corrections are discussed eg. in Schneider et al.
# (1986) JMB 188:414
l <- length(v)
a <- rep(0, 21) # initialize a vector with 21 elements (20 aa plus gap)
# the set the name of each row to the one letter
# code. Through this, we can access a row by its
# one letter code.
names(a) <- unlist(strsplit("acdefghiklmnpqrstvwy-", ""))
for (i in 1:l) { # for the whole vector of amino acids
c <- v[i] # retrieve the character
a[c] <- a[c] + 1 # increment its count by one
} # note: we could also have used the table() function for this
tot <- sum(a) - a["-"] # calculate number of observed amino acids
# i.e. subtract gaps
a <- a/tot # frequency is observations of one amino acid
# divided by all observations. We assume that
# frequency equals probability.
a["-"] <- 0
for (i in 1:length(a)) {
if (a[i] != 0) { # if a[i] is not zero, otherwise leave as is.
# By definition, 0*log(0) = 0 but R calculates
# this in parts and returns NaN for log(0).
a[i] <- a[i] * (log(a[i])/log(2)) # replace a[i] with
# p(i) log_2(p(i))
}
}
return(-sum(a)) # return Shannon entropy
}
# ================================================
# calculate entropy for reference distribution
# (from UniProt, c.f. Assignment 2)
# ================================================
refData <- c(
"A"=8.26,
"Q"=3.93,
"L"=9.66,
"S"=6.56,
"R"=5.53,
"E"=6.75,
"K"=5.84,
"T"=5.34,
"N"=4.06,
"G"=7.08,
"M"=2.42,
"W"=1.08,
"D"=5.45,
"H"=2.27,
"F"=3.86,
"Y"=2.92,
"C"=1.37,
"I"=5.96,
"P"=4.70,
"V"=6.87
)
### Calculate the entropy of this distribution
H.ref <- 0
for (i in 1:length(refData)) {
p <- refData[i]/sum(refData) # convert % to probabilities
H.ref <- H.ref - (p * (log(p)/log(2)))
}
# ================================================
# calculate information for each position of
# multiple sequence alignment
# ================================================
lAli <- dim(MSA)[2] # length of row in matrix is second element of dim(<matrix>).
I <- rep(0, lAli) # initialize result vector
for (i in 1:lAli) {
I[i] = H.ref - entropy(MSA[,i]) # I = H_ref - H_obs
}
### evaluate I
I
quantile(I)
hist(I)
plot(I)
# you can see that we have quite a large number of columns with the same,
# high value ... what are these?
which(I > 4)
MSA[,which(I > 4)]
# And what is in the columns with low values?
MSA[,which(I < 1.5)]
# ===================================================
# plot the information
# (c.f. Assignment 5, see there for explanations)
# ===================================================
IP <- (I-min(I))/(max(I) - min(I) + 0.0001)
nCol <- 15
IP <- floor(IP * nCol) + 1
spect <- colorRampPalette(c("#DD0033", "#00BB66", "#3300DD"), bias=0.6)(nCol)
# lets set the information scores from single informations to grey. We
# change the highest level of the spectrum to grey.
#spect[nCol] <- "#CCCCCC"
Icol <- vector()
for (i in 1:length(I)) {
Icol[i] <- spect[ IP[i] ]
}
plot(1,1, xlim=c(0, lAli), ylim=c(-0.5, 5) ,
type="n", bty="n", xlab="position in alignment", ylab="Information (bits)")
# plot as rectangles: height is information and color is coded to information
for (i in 1:lAli) {
rect(i, 0, i+1, I[i], border=NA, col=Icol[i])
}
# As you can see, some of the columns reach very high values, but they are not
# contiguous in sequence. Are they contiguous in structure? We will find out in
# a later assignment, when we map computed values to structure.
- That is all.
Links and resources
Footnotes and references
- ↑ The directory also contains sourcecode to generte the PAM matrices. This may be of interest for you if you ever want to produce scoring matrices from your own datasets.
- ↑ (Taylor et al. (2000) Biochemistry 39: 3943-3954 and Deleeuw et al. (2008) Biochemistry. 47:6378-6385)
- ↑ As you will see later on in the assignment, Mbp1-related proteins contain "Ankyrin" domains, a very widely distributed protein-protein interaction motif that may give rise to false-positive similarities for full-length sequence searches. Therefore, we search only with the DNA binding domain sequence, since this is the functionality that best characterizes the "function" of the protein we are interested in.
- ↑ Think of the ribosome or DNA-polymerase as extreme examples.
- ↑ Otherwise, you need to study the PDB Web page for the structure, or the text in the PDB file itself, to identify which part of the complex is labeled with which chain ID. For example, immunoglobulin structures some time label the light- and heavy chain fragments as "L" and "H", and sometimes as "A" and "B"–there are no fixed rules. You can also load the structure in VMD, color "by chain" and use the mouse to click on residues in each chain to identify it.
- ↑ The -myceta are well supported groups above the Class rank. See Leotiomyceta for details and references.
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.
| < Assignment 3 | Assignment 5 > |
