User:Boris/Temp/APB
Note! This assignment is currently active. All significant changes will be announced on the mailing list.
Contents
Assignment 3 - Multiple Sequence Alignment
Preparation, submission and due date
- Read carefully.
- Be sure you have understood all parts of the assignment and cover all questions in your answers! Sadly, we always get assignments back in which people have simply overlooked crucial questions. Sadly, we always get assignments back in which people have not described procedural details. If you did not notice that the above were two different sentences, you are still not reading carefully enough.
Review the guidelines for preparation and submission of BCH441 assignments.
The due date for the assignment is Monday, October 27. at 10:00 in the morning.
- Your documentation for the procedures you follow in this assignment will be worth 1 mark.
Introduction
- Take care of things, and they will take care of you.
- Shunryu Suzuki
Much of what we know about a protein's physiological function is based on the conservation of that function as the species evolves. We assess conservation by comparison to related proteins. Conservation - or variability - is a consequence of selection under constraints: the multiple effects on a species' fitness function that are induced through changes to the structural or functional features of a protein. Conservation patterns can thus provide evidence for many different questions: structural conservation among proteins with similar 3D-structures, functional conservation among homologues with comparable roles, peaks of sequence variability that indicate domain boundaries in multi-domain proteins, or amino acid propensities as predictors for protein engineering and design tasks.
Measuring conservation requires alignment. Therefore a carefully done multiple sequence alignment (MSA) is a cornerstone for the annotation of the essential properties a gene or protein. MSAs are also useful to resolve ambiguities in the precise placement of indels and to ensure that columns in alignments actually contain amino acids that evolve in a similar context. MSAs serve as input for
- functional annotation;
- protein homology modeling;
- phylogenetic analyses, and
- sensitive homology searches in databases.
As a first step, we will explore the search and retrieval of fungal proteins that are orthologous to yeast Mbp1, and of the APSES domains they contain. Each student is being assigned one genome-sequenced fungus. Briefly, you will
- Collect sequence identifiers for all APSES domain transcription factors in your assigned species;
- Retrieve the sequences;
- Perform a multiple sequence alignment with these, and a number of reference domains;
- Edit the alignment and annotate.
Multiple Sequence Alignment is not a solved, computational problem and a significant number of alignment tools exist, each with different strengths and objectives. It is remarkable that by far the most frequently used MSA algorithm is CLUSTAL, a procedure that was first published for the microprocessors of the late 1980s, surpassed in performance many times, and shown to be significantly inferior to more modern approaches when aligning sequences with 30% identity or less. In this assignment we will encounter various approaches to multiple alignment:
- A model-based approach (based on the PSSM that PSI-BLAST generates)
- Progressive alignments - CLUSTAL and MAFFT
- Consistency based alignment - T-Coffee and MUSCLE
(1) Mbp1 homologues
(1.1) Retrieving sequences
In Assignment 2 you retrieved the protein sequences of saccharomyces cerevisiae Mbp1 and defined its APSES (KilA-N) domain. Let us now search for an orthologue of this sequence in Your Species More precisely, you should identify prtoteins that fulfill the Reciprocal Best Match criterion.
First, we need to define the sequence you will use to find Mbp1 homologues. Since Mbp1 contains the very widely distributed Ankyrin motifs, a BLAST search with full length sequences will pick up a large number of Ankyrin-repeat containing proteins that are otherwise unrelated to our query. We will instead search for homologues using only the APSES domain as a query. However, the Pfam definition of the APSES domain (or KilA-N family, as it is now called) does not cover the entire length of the domain that has been crystallized. Therefore, we will use the sequence of the crystallized protein instead of the Pfam alignment. One of the results of our analysis will be whether APSES domains in fungi all have the same length as the Mbp1 domain, or whether some are indeed much shorter, as sugested by the Pfam alignment. To remind you, here is the full sequence of the 1MB1 structure (Note that the C-terminal His6 tag that has been added for purification is not part of the Mbp1 protein sequence.) ...
>PDB:1MB1 MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKETHEKVQGGFGKYQGTWVPL NIAKQLAEKFSVYDQLKPLFDFTQTDGSASPPPAPKHHHASKVDHHHHHH
... and, for comparison, this is the corresponding alignment with the Pfam KilA-N model obtained from a RPS-BLAST search of the above sequence against the CDD database:
10 20 30 40 50 60 70 80 ....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....| 1MB1 19 IHSTGSIMKRKKDDWVNATHILKAANFAKaKRTRILEKEVLKETHEKVQ----------------GGFGKYQGTWVPLNI 82 Cdd:pfam04383 3 YNDFEIIIRRDKDGYINATKLCKAAGATK-RFRNWLRLESTKELIEELSkennidvliievenkkGKNGRLQGTYVHPDL 81 90 ....*....|....* 1MB1 83 AKQLA----EKFSVY 93 Cdd:pfam04383 82 ALAIAswisPEFALK 96
As you can see, the Pfam alignment is 18 amino acids shorter at the N-terminus and 31 amino acids shorter at the C-terminus.
- Find APSES domain proteins in your species
- Access the species list and identify the species that has been assigned to you.
- Navigate to the NCBI's main page.
- In the left-hand menu of links, follow the link to Genomes & Maps.
- Under the Databases tab, follow the link to Genome.
- In the Genome tools section of that page, follow the link to Genomic groups BLAST.
- Click on link to the eukaryotic genomes tree, then on the link for the text table. This produces a BLAST interface to a list of species for which whole-genome sequences have been sequenced, annotated and entered into the various databases.
- Paste the FASTA sequence of the structurally defined Mbp1 APSES domain (e.g. from 1MB1) into the search field (excluding the His-tag, of course), set the parameters correctly for a Protein search against Protein sequences using blastp. Then find your assigned species in the table and check the box next to its name. Remember to record the parameters for your search. I expect you to understand which parameters would be needed in order to make this search reproducible. Run the search.
- On the next screen, check the box next to Format for: PSI-BLAST. Then click on View report to show the results of the first PSI-BLAST iteration.
- Run subsequent iterations of PSI-BLAST simply by clicking on Go after checking the sequences that have been included.
- Iterate the PSI-BLAST search until convergence (i.e. until no more new sequences are added); make sure to include only sequences for which the E-value is small (smaller than about 10e-03 should be safe). Sequences with borderline E-values that improve significantly in an iteration are probably homologues. Sequences with borderline E-values that do not improve much, or for which the E-value increases are probably not homologues. If this step does not work for you or the results are not what you expect, please contact your TA right away.
- Note: Please spend a little time on each page to understand its contents. Ask, if the page contains resources or features you don't understand. Think about what you are doing. If you simply click on the links I provide, you will miss the opportunity to understand how the resources fit into the workflow you are working on, and to be able to execute similar processes yourself. Questions on page contents can potentially appear on quizzes and exam.
Familiarize yourself with the output form you obtain, this is by far the most frequently used bioinformatics result page. You may want to refer to the NCBI explanation.
Here is a list of things to look for, all of which I expect you to know and understand. (However you do not need to comment on these points in your submission.)
- On the alignment image
- What do the different colored bars mean?
- What is the information you get when you "mouse-over" a colored bar on the alignment image.
- What happens when you click on one of the bars?
- In the description list
- Where does the link next to an identifier take you?
- Where does the link in the "score" column take you?
- What does the icon at the end of each row mean? What other icons could appear there?
- In the alignment section
- What do the alignment metrics mean:
- Score?
- Expect (E-value)?
- Identities?
- Positives?
- Gaps?
- What is the alignment length?
- Which sequence is labeled Query and which one is labelled Sbjct?
- Next
- retrieve the sequences that have E-values low enough to make you conclude they contain APSES domain homologues.
- Review the sequences you have found: they should all be significantly similar to the query profile. In some of the assigned species you will find one hit for each distinct sequence in the genome, in others, you will find several versions of essentially the same gene (e.g. refseq and other accession numbers).
- Explore the relationship between the hits by clicking on select all sequences, then choosing Distance tree of results at the top or bottom of your search results to visualize a tree representation of similarity. Highly similar sequences will be collapsed into the same node in the distance tree; you can expand those nodes to list all the node's members.
- Identify one representative for each distinct protein you have found. If possible, use proteins with refseq identifiers. Avoid duplicates or nearly identical variants. If there are length differences, use the longer version (shorter versions may contain only partial sequences). Click on the checkbox next to each protein you have identified.
- Click on get selected sequences at the top or bottom of the page. Note and record the GIs for your sequences that are listed in the Search details box, you can use them to easily reproduce your results by pasting them into any Entrez search. Also note the URL that this has produced (in your browser's URL bar). As you see, you can retrieve a list of sequences from NCBI simply by adding a list of comma-separated GI numbers to the URL of the protein database.
- Click on Display settings and choose FASTA (text).
If you want, for comparison, you can run a multiple alignment with an NCBI-developed MSA tool: COBALT. On the sequence list page, in the right-hand column, in the section Analyze these sequences, click on Align sequences with COBALT. It is a convenient way to get a quick first look at an alignment of NCBI retrieved sequences.
You now have a collection of APSES domain-containing homologues in your organism. There are two more tasks we need to address before we can compute alignments and analyze them. (A) we need to rename our sequences, and (B) we need to define the boundaries of their APSES domains.
(1.2) Renaming Sequences
A phylogenetic tree or multiple alignment is not really informative if it that displays GI numbers or other abstract identifiers as labels of rows or nodes. The relationship between species is fundamental to the variation we observe and we need to make this relationship explicit.
Imagine that the rows in an MSA were completely unlabeled, or the nodes in the tree would be just circles: we would have a very hard time relating the computed relationships back to the biology they represent. Abstract identifiers like NP_010227 are not much better.
Typically, the information that programs use to label sequences is taken from the FASTA header. This provides us with an easy way to make sure they display the information we need and that we can interpret. Typically such programs will use the first few (often ten) characters they find. We will therefore design short strings strings that identify potential gene family relationships as well as species.
- Species codes
The scientific name of a species is formed according to Linnaean binomial nomenclature and Swissprot has for a long time condensed species names into mnemonic five-character codes, taking the first three from the genus name and the last two from the specific name. For example Saccharomyces cerevisiae is abbreviated as SACCE and Lachancea thermotolerans is LACTH. For the most part, this creates unique strings that are good mnemonic labels for the species. I have added these "codes" to the Species list.
- Gene families
Most yeast genes have traditional names, like mbp1 or sok2. These names are convenient family labels since saccharomyces cerevisiae is one of the best studied model organisms. Therefore, once we identify a protein family that includes a yeast gene, we can easily access expert knowledge in textbooks or manuscripts. Of course, such labels are arbitrary - whether we call a gene Mbp1 or WXYZ makes no difference - as long as all genes that we presume to be family members carry the same label. For higher eukaryotes, I would probably choose human gene names as a reference point, for bacteria I would choose E. coli.
To define which gene belongs into which family, we can align all newly found genes with all yeast APSES domain homologues, to find out which ones they are most similar to. This creates common family labels. We can use these as provisional family names for the encoded proteins, even though we may want to revise them once we have mapped out explicit phylogenetic trees.
- Identifying APSES domains (general procedure).
In order to identify the APSES domain boundaries, you can simply run a multiple sequence alignment of the structurally defined APSES domain sequence (e.g. taken from PDB-ID 1MB1) against all sequences you have found. The boundaries of the aligned APSES domain then define the domain boundaries in the aligned proteins.
- Identifiying family relationships (in the same run)
However, for efficiency, we can also determine family relationships in the same alignment that we use to define domain boundaries, if we simply include all yeast APSES domains in the MSA. Then we can judge similarity simply from examining the guide tree of the alignment and label the families accordingly. This has the added advantage that the domain boundaries are more securely defined, since we include more sequence information into the alignment.
- Proceed as follows.
- Open the Muscle MSA input page at the EBI.
- Access the Yeast APSES domain collection I have prepared and copy the FASTA sequences. Paste them into the sequence field of the MUSCLE program input form.
- Copy the FASTA sequenced of the full length APSES domain protein sequence collection from your PSI-BLAST search (above) and paste them into the MUSCLE input form as well.
- Set the following parameters:
OUTPUT FORMAT: CLUSTALW2 OUTPUT TREE: from second iteration OUTPUT ORDER: aligned
- Click on Submit.
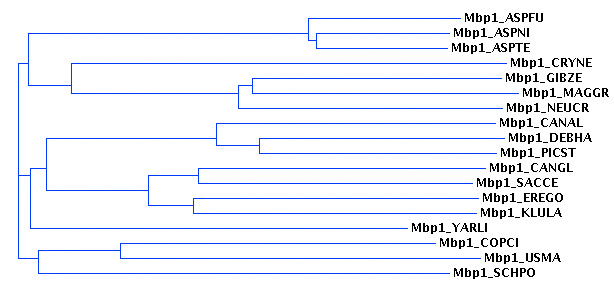
The output should show the MSA. The overlap of the yeast APSES domains with your sequences defines the domain boundaries. Moreover, a tree has been calculated and you can view the tree to identify family relationships.
- Visualize the alignment tree and decide on names
Click on the link to the Guide tree. This is the so-called Newick tree format and there are a large number of online tree viewers to visualize such trees. The MUSCLE form will display one tree for you,
You could also navigate (for example) to the proWeb Tree viewer and paste the tree data into the User-supplied Newick Tree input field. Choose any graphics format your browser can handle (JPEG is a pretty safe bet) and click on View tree.
- Interpret the tree to decide on the protein family names for your sequences:
- If a yeast protein is grouped with exactly one of your proteins, your protein gets the same name.
- If a yeast protein is grouped with more than one of your proteins, replace the number in the yeast protein with a, b, c ..., from most similar to least similar for your protein. For example: if one Aspergillus fumigatus protein is most similar to yeast Mbp1, you will give it the name MBP1_ASPFU. If two proteins are both most similar to yeast Sok2, you will name them SOKA_ASPFU and SOKB_ASPFU. Try to get it approximately right but remember that this is a process of estimation - we are not accurately measuring distances (yet).
That done, edit your FASTA headers and save your APSES domain sequence set. We will need them for the next assignment.
(2) Mbp1 orthologue
(2.1) Input data for multiple alignments
You have identified homologues to yeast Mbp1 in your species, but which one of these (if any) is an orthologue?
- Perform a reciprocal BLAST search with your highest scoring hit and note whether the reciprocal best match criterion has been fulfilled.
- Repeat this procedure (yeast → your species → yeast) but restict the query sequence to the Mbp1 APSES domain that you have defined in Assignment 2.
This should retrieve a number of APSES domain proteins in your species, apparently related to Mbp1.
- Analysis (1 mark)
- Based on your results, comment briefly on whether your species appears to have an orthologue of the entire Mbp1 gene and/or only an APSES domain orthologous to Mbp1.
Next, compare the (empirical, local) BLAST alignment with a (optimal, global) Needleman-Wunsch sequence alignment. Use the correct algorithm from the set of EMBOSS tools:
- Retrieve the full-length sequence of the orthologue to yeast Mbp1 in your species, and generate an optimal global alignment between this and S. cerevisiae Mbp1. You have to figure out where to find a Web service that does such alignments, what the name of the algorithm is that you should use and how to define reasonable parameters for the alignment.
- Review if and how the alignments are different, or whether the two alignment algorithms have given essentially the same results.
- Note: When I instruct you to review..., I do not require you to include your conclusions in the submitted assignment. However I expect you to be familiar with the analysis and to be able to answer questions on the process and the conclusions.
(2.1) Input data for multiple alignments
Preparing a set of sequences as input for a multiple sequence alignment usually follows from a BLAST search like the one you have performed above, or perhaps a PSI-BLAST search for added sensitivity. This includes
- searching a query sequence across a database subset of interest,
- retrieving orthologue- and or paralogue- sequences,
- validating BLAST alignments (if needed to distinguish orthologues and paralogues),
- trimming the sequences to a particular region of interest (if needed to remove non-homologous domains that would otherwise corrupt the alignment), and
- saving the result as a multi-FASTA formatted file.
I have generated a reference list of Mbp1 orthologue sequences, using the canonical procedure defined below: (departures from the procedure are noted below the table). Please check whether your orthologue search has identified the same sequence as the one listed in the table. In case the sequence identifiers differ, check whether the actual sequences are the same and in case the actual sequences differ, let me know.
- Procedure
- Retrieved the Mbp1 protein sequence by searching Entrez for
Mbp1 AND "saccharomyces cerevisiae"[species] - Clicked on the RefSeq tab to find the RefSeq ID "
NP_010227" - Accessed the BLAST form, followed the link to the list of all genomic BLAST databases and clicked on the (B) icon, next to Fungi to navigate to the Fungi Genomic BLAST page.
- Pasted "
NP_010227" into the query field. Chose Protein for both Query and Database, kept default parameters but set the Filter option to none. Clicked on the check-box of each of the fungal species we have considered in the previous assignment. Run BLAST. - On the results page, checked the checkbox next to the alignment to select the most significant hit from each species we are studying.
- Clicked on the "Get selected sequences" button.
- Separately searched for sequences from species that were either not included in the list or for which no hits were reported. Verified all ambiguous cases, as explained in the notes below.
- Verified that each of these sequences finds Mbp1 as the best match in the saccharomyces cerevisiae genome by clicking on each "Blink" (click for example) in the retrieved list. Scrolled down the list to confirm that the top hit of a saccharomyces cerevisiae protein is indeed Mbp1 (
NP_010227). - Obtained UniProt accessions for all sequences, with a single query using the UniProt ID mapping service. This service accepts a comma delimited list of RefSeq IDs, GI numbers or GenPept accession numbers and returns a list of Uniprot accession numbers.
Since it was thus confirmed that each of these sequences is the protein that is most similar to yeast Mbp1 in its respective species' genome, and that yeast Mbp1 is the most similar yeast protein to each of them, they all fulfil the criterion of a reciprocal best match with yeast Mbp1. Accordingly we can postulate that this list contains the fungal orthologues to Mbp1.
| Mbp1 and its orthologues | |||||
| Species | CODE |
GI | NCBI | Uniprot | Most similar yeast gene |
| Aspergillus fumigatus | ASPFU |
70999021 | XP_754232 | Q4WYQ9_ASPFU | Mbp1 |
| Aspergillus nidulans | ASPNI |
67525393 | XP_660758 | Q5B8H6_EMENI | Mbp1 |
| Aspergillus terreus | ASPTE |
115391425 | XP_001213217 | Q0CQJ5_ASPTN | Mbp1 |
| Candida albicans | CANAL |
68465714 | XP_722925 | Q5ANP5_CANAL | Mbp1 |
| Candida glabrata | CANGL |
50286059 | XP_445458 | Q6FWD6_CANGA | Mbp1 |
| Coprinopsis cinerea | COPCI |
169861520 | XP_001837394 | A8NYC6 | Mbp1 |
| Cryptococcus neoformans | CRYNE |
134110416 | XP_776035 | Q5KHS0_CRYNE | Mbp1 |
| Debaryomyces hansenii | DEBHA |
50420495 | XP_458784 | Q6BSN6_DEBHA | Mbp1 |
| Eremothecium gossypii | EREGO |
45199118 | NP_986147 | Q752H3_ASHGO | Mbp1 |
| Gibberella zeae | GIBZE |
46116756 | XP_384396 | UPI000023DBF3 | Mbp1 |
| Kluyveromyces lactis | KLULA |
50308375 | XP_454189 | MBP1_KLULA | Mbp1 |
| Magnaporthe grisea | MAGGR |
74274844 | ABA02072 | Q3S405_MAGGR | Mbp1* |
| Neurospora crassa | NEUCR |
164424100 | XP_962967 | Q7SBG9 | Mbp1 |
| Pichia stipitis | PICST |
126275256 | XP_001386821 | A3GHD6_PICST | Mbp1 |
| Saccharomyces cerevisiae | SACCE |
6320147 | NP_010227 | MBP1_YEAST | Mbp1 |
| Schizosaccharomyces pombe | SCHPO |
19113944 | NP_593032 | RES2_SCHPO | Mbp1 |
| Ustilago maydis | USTMA |
71024227 | XP_762343 | Q4P117_USTMA | Mbp1 |
| Yarrowia lipolytica | YARLI |
50545439 | XP_500257 | Q6CGF5_YARLI | Mbp1 |
Table of yeast Mbp1 orthologues in genome-sequenced fungi. Columns from left to right: Systematic name, species code (simply a string that lets us identify the species in alignments), GI number, RefSeq ID (if existing) or GenPept accession, Uniprot accession, most similar yeast protein.
The procedure described above needed to be adapted for some cases and that is to be expected i practice. You should be familiar with exceptions such as the ones described below and know how to deal with them. A good way to do that is to repeat some of the procedures and see if you arrive at the same conclusions.
Note: for Aspergillus fumigatus and Aspergillus nidulans, the top BLAST hit is not the best match. The reason is that the best matching protein has a deletion just C-terminal to the APSES domain. This causes BLAST to split the HSP into two parts, and even though the APSES domain alone has a higher % identity, its E-value turns out to be lower because it is a shorter sequence. Global alignment of each sequence with yeast Mbp1, as well as alignment of only the APSES domains were consistent in showing that for both Aspergillus species the second highest BLAST score is indeed the most similar protein. The take-home message is that the comparison of BLAST scores can be misleading if we apply them to sequences of different length. For the record: Aspergillus fumigatus highest BLAST score is with XP_748947, second highest BLAST score is with XP_754232; the latter has higher global identity (25.7% vs. 22.6%) and higher identity in the APSES domain (55% vs. 45%). Aspergillus nidulans highest BLAST score is with XP_664319, second highest BLAST score is with XP_660758; the latter has higher global identity (26.7% vs. 22.8%) and higher identity in the APSES domain (59.5% vs. 50.6%). Interestingly, the Aspergillus terreus orthologue has the same deletion, but it provided the highest BLAST score to begin with.
Note: For Giberella zeae XP_384396 no UniProt ID was returned as cross-reference. EBI-BLAST retrieved FG04220 which is largely identical, except for short stretches that are absent in GenPept: apparently UniProt has a different gene-model for this protein.
Note: The Magnaporthe grisea protein ABA02072 has greater local C-terminal similarity to the yeast protein Swi6 than to Mbp1, whereas the N-terminal APSES domain is most similar to yeast Mbp1. However a global Needleman-Wunsch alignment (BLOSUM 30, gaps: 8.0/1.0) shows greater overall similarity to yeast Mbp1 than to Swi6. Accordingly I consider this an orthologue to Mbp1 even though its database annotation calls ABA02072 the M. grisea Swi6 homologue.
Note: For Pichia stipitis, BLAST finds two very similar sequences in GenPept as candidate Mbp1 orthologues; the RefSeq sequence XP_001386821.1 is translated according to the standard code, the entry EAZ62798.2 is translated according to the alternative nuclear code 12. The question had to be considered which translation appears to be correct. This required looking at the conservation of the residues in question in the BLAST alignment; better conservation indeed supports the alternative code translation.
Note: The Ustilago maydis protein XP_762343 (the protein with the sytematic name UM06196) is only the second-best hit in the original BLAST list as performed on the genomic BLAST page for the species, however local optimal alignment (EMBOSS water) shows a much higher percentage of identity to yeast Mbp1 in the APSES domain than the top BLAST hit (XP_761485, systematic name UM05338) and global alignment (after trimming the N- and C- terminal extensions, respectively) also shows a slightly higher degree of similarity for the latter. Accordingly, XP_762343 is considered the Mbp1 orthologue, even though it is the second highest hit according to BLAST. The situation is similar as with the Aspergillus species, one protein was reported as a single HSP and one protein was broken into two HSPs. This emphasizes the fact that optimal sequence alignments are not entirely equivalent to BLAST alignments. Further, performing the same search against the "nr" database and applying an Species filter for Ustilago maydis resulted in both proteins being split and the correct orthologue having the highest BLAST score in the list. This emphasizes the fact that searches in species databases are not entirely equivalent to searches in the global database, even if the results are filtered.
To obtain all FASTA sequences based on a list of identifiers and to save them in a format in which we can use them as input for other programs or services is easy. We can simply paste all GI numbers as a comma separated list into the Entrez search form and on the results page, select Display FASTA and send to Text; then save the contents as a text file. This is a multi-Fasta file, suitable for input into MSA programs.
- Review the resulting multi-FASTA file for the Mbp1 proteins (linked here) and make sure you understand the procedure that led to it. Depending on your personal learning style you may either carefully review the described procedure, reproduce key steps of the procedure, reproduce the entire procedure paying special attention to the problem cases discussed in the notes, or develop your own procedure. Whatever you do, you must be confident in the end that you could have produced the same input file. (You do not need to submit documentation for this part of the assignment, but you do need to understand the process.)
As you have seen from the results of your BLAST searches, Mbp1 orthologues are not the only proteins that contain APSES domains. In order to find all the rest, a PSI-BLAST search was performed using the yeast Mbp1 APSES domain as query. From the list of hits, the APSES domains were extracted and summarized in a file.
- Review the resulting file for the APSES domains (linked here) and make sure you understand the procedure that was used in its construction, as above.
(3) Align and annotate
(3.1) Review of domain annotations
Let us first review some of the features of the yeast Mbp1 protein that we have defined in the second assignment (and some structural features I have compiled from various sources). Below is an annotated yeast Mbp1, compiled according to the following procedure.
- Performed CDD search with yeast Mbp1 protein sequence. This retrieves alignments of Mbp1 with the APSES and the ANKYRIN domains. These are profile based alignments and thus they are more reliable than pairwise alignments.
- Performed SMART search with yeast Mbp1 protein sequence. This retrieved the APSES domain, annotated a number of low-complexity regions and a stretch of coiled coil.
- Performed a SAS search with yeast Mbp1 protein sequence. This retrieved pairwise alignments with the structures 1MB1 (APSES) and chain D of 1IKN (ankyrin domains of Ikappab), together with their respective secondary structure annotations.
- Copied GenPept sequence into Word-processor.
- Transferred annotations of low complexity and coiled-coil regions from SMART.
- Transferred annotations of APSES secondary structure from SAS (this is a direct annotation, since the experimentally determined structure 1MB1 is a fagment of of the Mbp1 protein). The central helix that was proposed to be part of the DNA binding region is slightly distorted and SAS annotates a break in the helix, this break was bridged with lowercase "h" in the annotation.
- Ankyrin domain annotation was not as straightforward. While CDD, SMART and SAS all annotate the same general regions, they disagree in details of the domain boundaries and on the precise alignment. Used the profile-based CDD alignment of 1IKN. Transferred annotations of secondary structure from SAS output for 1IKN to sequence (this is a transferred annotation, the original annotation was for 1IKN and we assume that it applies to Mbp1 as well).
MBP1_SACCE
Annotations based on
- CDD domain analysis,
- SAS structure annotation and
- literature data on binding region
Keys:
C Coiled coil regions predicted by Coils2 program
x Low complexity region
* Proposed binding region
+ positively charged residues, oriented for possible DNA binding interactions
- negatively charged residues, oriented for possible DNA binding interactions
E beta strand
H alpha helix
t beta turn
10 20 30 40 50 60
MSNQIYSARY SGVDVYEFIH STGSIMKRKK DDWVNATHIL KAANFAKAKR TRILEKEVLK
1MB1 ----EEEEEt t-EEEEEEEE t-EEEEEEtt ---EEHHHHH HH----HHHH HHHHhhhHHH
* *+**-+****
70 80 90 100 110 120
ETHEKVQGGF GKYQGTWVPL NIAKQLAEKF SVYDQLKPLF DFTQTDGSAS PPPAPKHHHA
1MB1 ---EEE---- tt--EEEE-H HHHHHHHHH- --HHHHtt- xxx xxxxxxxxxx
**+*+***** ****
130 140 150 160 170 180
SKVDRKKAIR SASTSAIMET KRNNKKAEEN QFQSSKILGN PTAAPRKRGR PVGSTRGSRR
x
190 200 210 220 230 240
KLGVNLQRSQ SDMGFPRPAI PNSSISTTQL PSIRSTMGPQ SPTLGILEEE RHDSRQQQPQ
xxxxx
250 260 270 280 290 300
QNNSAQFKEI DLEDGLSSDV EPSQQLQQVF NQNTGFVPQQ QSSLIQTQQT ESMATSVSSS
x xx xxxxxxxxxx xxxxxxxxxx
310 320 330 340 350 360
PSLPTSPGDF ADSNPFEERF PGGGTSPIIS MIPRYPVTSR PQTSDINDKV NKYLSKLVDY
xxxxxxx
370 380 390 400 410 420
FISNEMKSNK SLPQVLLHPP PHSAPYIDAP IDPELHTAFH WACSMGNLPI AEALYEAGTS
ANKYRIN -- t----HHHHH HH---HHHHH t-t--t-t--
430 440 450 460 470 480
IRSTNSQGQT PLMRSSLFHN SYTRRTFPRI FQLLHETVFD IDSQSQTVIH HIVKRKSTTP
ANKYRIN t----t---- HHHHHHHH-- -------HHH HHHHHH-ttH HH-----HHH HHHH--tH--
490 500 510 520 530 540
SAVYYLDVVL SKIKDFSPQY RIELLLNTQD KNGDTALHIA SKNGDVVFFN TLVKMGALTT
ANKYRIN HHHHHHHHH- ---------- -----t---- tt---HHHHH HH---HHHHH HHH--t-tt-
550 560 570 580 590 600
ISNKEGLTAN EIMNQQYEQM MIQNGTNQHV NSSNTDLNIH VNTNNIETKN DVNSMVIMSP
ANKYRIN ---t----HH HHHHHH--HH HHH-t--HHH -t----HHHH HHH--tHHHH HHHHHH---t
610 620 630 640 650 660
VSPSDYITYP SQIATNISRN IPNVVNSMKQ MASIYNDLHE QHDNEIKSLQ KTLKSISKTK
ANKYRIN ---tt----H HHHHHH---H HHHHHHH CCCCCCCC CCCCCCCCCC CCCCC
670 680 690 700 710 720
IQVSLKTLEV LKESSKDENG EAQTNDDFEI LSRLQEQNTK KLRKRLIRYK RLIKQKLEYR
x xxxxxxxxxx xxxxxxx
730 740 750 760 770 780
QTVLLNKLIE DETQATTNNT VEKDNNTLER LELAQELTML QLQRKNKLSS LVKKFEDNAK
790 800 810 820 830
IHKYRRIIRE GTEMNIEEVD SSLDVILQTL IANNNKNKGA EQIITISNAN SHA
A good MSA comprises only columns of residues that play similar roles in the proteins' mechanism and/or that evolve in a comparable structural context. Since it is a result of biological selection and conservation, it has relatively few indels and the indels it has are usually not placed into elements of secondary structure or into functional motifs. The contiguous features annotated for Mbp1 are left intact.
A poor MSA has many errors in its columns, they contain residues that actuallly have diffferent functions or structural roles, even though they may look similar to a scoring matrix. It also may have introduced indels in biologically irrelevant positions, to maximize spurious sequence similarities. Some of the features annotated for Mbp1 will be disrupted.
- Produce a similar set of annotations for your Mbp1 orthologue protein.
(3.2) Computing alignments
Multiple sequence alignments are compute-intensive and this used to require downloading and installing software on your own computer. While most tools were available on the Web in principle, many groups have restricted the total number of sequences or the total number of characters to be aligned. The EBI however offers three of the most commonly used tools with few limitations which I have used to produce reference alignments for Mbp1 orthologues and APSES domains.
- CLUSTAL-W is a progressive alignment program, it is the most popular, most widely referenced MSA algorithm, it is reasonably fast and easy to use. But alignment errors that are made early in the process can't get corrected and thus CLUSTAL is prone to misalign sets of sequences that have poor (<30% ID) local similarity. CLUSTAL is no longer considered state-of-the-art for carefully done alignments.
- MUSCLE essentially starts out from a CLUSTAL like alignment as a draft, then identifies similar groups of sequences from which it calculates profiles, it then re-aligns the group to the profile. This procedure is iterated.
- T-Coffee is one of my favourites - the tradeoffs appear to be especially well balanced. It too starts from a set of pairwise global alignments, like CLUSTAL, then additionally calculates sets of best local alignments. Global and local alignments are then combined to a similarity matrix and based on this matrix a guide-tree is constructed. This determines the order of steps in which sequences are added to the multiple alignment. A nice feature of T-Coffee is color coded output that allows you to quickly judge the local reliability of the alignment.
Multiple sequence alignments were performed for all 18 Mbp1 orthologues. I have posted the reference alignments here. (Of course you are welcome to run an alignment on your own for your own learning experience, or to find an alternative program, but I do not require this for the assignment.)
The first alignment was run with CLUSTAL.
Subseqently, sequence alignments were performed with T-Coffee and MUSCLE. For these two, the input files were re-ordered to correspond to the order of the CLUSTAL output, and the option to order the alignments according to the input sequences was chosen on the form. This makes it much easier to compare alignments, since all MSAs are displayed in the same relative order.
Finally I have merged the domain annotations for the yeast Mbp1 protein into the output files.
The result files are linked here:
- Mbp1 proteins CLUSTAL aligned
- Mbp1 proteins MUSCLE aligned
- Mbp1 proteins T-Coffee aligned (text version) or Mbp1 proteins T-Coffee aligned (coloured according to scores)
Globally speaking, the alignments are quite similar. Let's first look at the common themes, before we discuss details of the results. The (score-colored T-COFFEE alignment) is well suited to look at general relationships between the sequences, since outliers can be easily identified. For example, if one of the sequences would have a low-scoring domain that aligns poorly to the others of the group, it may be possible that that domain has been acquired in a separate evolutionary event and is not homologous to all others. We would notice an isolated stretch of poorly alignable sequence, i.e. it should be a segment coloured with a low score in a set of otherwise high-scoring segments. Also a gene may have acquired significant lengths of N- or C-terminal extensions which may not be homologous (unless they are the result of an internal duplication).
- Review the (score-colored T-COFFEE alignment). Based on this alignment, how do you feel about our initial assertion that these 18 proteins should be considered orthologous over their entire length? You do not need to discuss this in the assignment but you should study the evidence in the alignment. Note that this question does not ask about the general level of conservation, but about whether significant segments (of about the length of a domain) do not appear related/alignable at all in regions where the rest of the group are reasonably well conserved.
(4) Mbp1 orthologues: analysis of full length MSAs
(4.1) APSES domains
The APSES domains in all of our Mbp1 orthologues are highly conserved and pretty much any alignment program must be able to align such obviously similar regions.
- Consider the CLUSTAL, Muscle and T-Coffee alignments of the Mbp1 orthologues. Orient yourselves as to where the APSES domains are located. For one alignment, refer to the specific residues annoted with (+) or (-) and review whether the charged residues in the proposed binding region are wholly conserved (marked *) or partially conserved (marked : or .) across all 18 proteins. Remember that these are surface, solvent exposed residues that would be expected to be highly variable if not constrained for functional reasons.
(4.2) Ankyrin domains
The Ankyrin domains are more highly diverged, the boundaries are less well defined and not even CDD, SMART and SAS agree on the precise annotations. Nevertheless we would hope that a good alignment would recognize homology in that region and that ideally the required indels would be placed between the secondary structure elements, not in their middle.
- Compare the distribution of indels in the ankyrin repeat regions of all three alignments. Review whether the indels in this region are concentrated in segments that connect the helices, or if they are more or less evenly distributed along the entire region of similarity. Think about whether the assertion that indels should not be placed in elements of secondary structure has merit in each alignment. Recognize that an indel in an element of secondary structure could be interpreted in a number of different ways:
- The alignment is correct, the annotation is correct too: the indel is tolerated in that particular case;
- the alignment algorithm has made an error, the structural anotation is correct: the indel should be moved a few residues;
- the alignment is correct, the structural annotation is wrong, this is not a secondary structure element after all;
- both the algorithm and the annotation are probably wrong, but we have no data to improve the situation
(NB: remember that the structural annotations have been made for the yeast protein and might have turned out differently for the other proteins...)
You should be able to analyse discrepancies in such a structured and systematic way. In particular if you notice indels that have been placed into structurally annotated regions of secondary structure, to consider whether the location of the indel has strong support from aligned sequence motifs, or whether it could apparently be placed into a different location whithout much loss in alignment quality.
Considering the different alignments, please note in your assignment which alignment you consider more reliable regarding the position of indels relative to structural features.
(4.3) Other features
Aligning functional features like coiled coil domains, intrinsically disorderd regions, or low complexity regions is difficult, since these features are to a large degree a property of the amino acid composition, not of the precise sequence. Thus there may be no recognizable similarity between aligned pairs of amino acids and the correspondence between sequences in such regions may be lost. In such cases, we may be able to detect conserved features in the absence of conserved sequence.
- Analysis (1 mark)
I have annotated four low complexity regions of the yeast Mbp1 sequence.
- Refer to your annotation of your species' Mbp1 orthologue. Comment on one of the multiple sequence alignments: does your protein have a similar distribution of low complexity regions as
Mbp1_SACCEdoes, and have these regions been aligned with the yeast protein by the MSA algorithm? Briefly describe the situation: state whether these segments are found in the same general region, in the same detailed location, or perhaps even conserved in sequence, when you compare them to the saccharomyces cerevisiae protein. Backup your conclusions with specific reference to particular elements of the alignment.
- Briefly discuss whether this situation implies that disorder in these proteins appears to be a conserved functional feature, i.e. that disorder is selected for in evolution. If this is the case, consider whether the disordered segments appear to be homologous or analogous or whether the data does not allow a conlusion.
(5) APSES domain homologues: analysis of domain MSAs
You have read how to generate a source sequence file based on the results of a PSI-BLAST search for all APSES domains in fungi. Of course, since PSI-BLAST has detected these sequences due to their high-similarity to a sequence profile, this similarity implies an alignment: a model based MSA because the sequences are aligned to a model (the sequence profile) and not to each other.
To align such highly diverged sequences the MUSCLE server is the tool of choice. For comparison, a CLUSTAL alignment has been computed as well.
- The resulting alignment derived from the PSI-BLAST profile as an example of a model-based alignment. Note that PSI-BLAST has not been optimized to work as an alignment program, thus the conclusion that model-based alignments are inferior because this example is a poor alignment is not justified.
- The CLUSTAL-W alignment as an example of a progressive alignment.
- The MUSCLE alignment as an example of a consistency-based alignment.
If we compare the alignments, we notice immediately that they disagree over siginficant portions of the sequences.
(5.1) Patterns of residue conservation
Whenever we use a program that optimizes something, we have to be aware of whether the program's objective function actually matches our needs for analysis. For example, if an MSA displays variability in a particular column of residues, this may mean that a residue has changed in evolution - but sometimes the column should have been conserved and the alignment has matched residues with a higher score at the expense of positions that we believe to be biologically important. MSAs can only take sequence information into account, while we may have complementary information available on structural and functional conservation patterns. This may include secondary structure (gaps should be moved out of regions of secondary structure, where possible), structurally required residues (these are expected to be conserved accross all structurally similar sequences), and functionally conserved residues (these are expected to have a high likelyhood of being conserved within groups of orthologues, but to vary between paralogues).
In terms of structural conservation, we expect motif or consistency based alignments to be more accurate since they align to the "big picture". In terms of functional variation we expect progressive alignments to be more accurate, since they align to local similarities.
Let us consider the alignments in terms of their biological relevance. I have annotated the ligand-binding residues for the yeast Mbp1 APSES domain in the multiple sequence alignments by color coding the charged residues that putatively could bind DNA red (-) and blue (+). Thus these residues label columns of the alignment in which we expect functional conservation. I have also highlighted two residues that are associated with important structural features of the APSES domain in green. These two residues are G75, a glycine required in the third position of a particular type of beta-turn, and W77W77, a key component of the domain's hydrophobic core. Thus these two residues label columns in which we expect structural conservation. Let's assume (i) that all the APSES domains fold into similar structures and (ii) that they all bind DNA, but (iii) they do not necessarily bind the same cognate sequence, as a consequence of the functional diversification of paralogues. This should allow you to discuss the following questions:
Consider any one of the three APSES domain alignments.
- Review whether the patterns of sequence variation for functionally conserved residues are compatible with the notion that orthologues have conserved binding specificities and paralogues have acquired new functions by binding to different sequences.
- Review whether the patterns of sequence variation for structurally conserved residues are compatible with the notion that all APSES domains have a common fold.
To approach these questions systematically, define (with reference to specific sequences and residues) what you would expect (hypothesis) and whether the alignment supports or contradicts your expectations (observation). We have determined that the sequences labelled as Mbp1 are orthologues, and the other labels were constructed to identify the yeast gene that each sequence is most similar to. This means you may group Mbp1 sequences as orthologues, Swi4, Sok2, and Phd1 sequences are presumably orthologous, and all sequences originating from the same species are of course groups of paralogues. Labels such as MbpA, MbpB etc. are paralogous to e.g. Mbp1 but not necessarily orthologous to each other.
(5.2) Visualization and analysis of alignment with VMD
VMD offers a very well constructed set of tools for the analyis of sequence and structural conservation: the MultiSeq extension. In this part of the assignment you will use VMD to analyse and visualize conservation patterns and comment on the alignments the servers have produced. I highly recommend to familiarize yourself with MultiSeq and the developers have produced an excellent tutorial on the evolution of tRNA synthetases to showcase the program's capabilities. However I am not requiring that you go through the tutorial and we will be using only a subset of the available Multiseq functions. The tool is intuitive enough, beginning to use it should require no more than following the steps below.
Proceed through the following steps:
- (1) Save an alignment of the APSES domains on your computer.
- (A) Access the MUSCLE alignment of all APSES domains, copy it from the Wiki page and save it on your computer, as a text file with some convenient filename and the extension .aln . This is a CLUSTAL formatted input file.
- (B) Edit the file to remove any header lines and lines containing the conservation symbols
.:*. Leave the gene-names and aligned sequences as they are. Make sure you are not saving the file in MS-Word binary format (.doc) and that the extension is not changed (depending on how your computer is configured, it may silently append a.txtextension that will cause trouble later on).
- (2) Open the Multiseq extension in VMD.
- (A) start VMD and load the 1MB1 APSES domain structure.
- (B) choose a stereo representation that will show you the fold of the domain and the sidechains of key residues. For example you could use a Tube representation for the protein backbone and a Licorice representation for the selection
((sidechain or type CA) and not element H) and resid 30 to 90. (And switch the axes display off! The axes carry no information you need). - (C) On the VMD Main form navigate to Extensions → Analysis → MultiSeq
- (D) When you run MultiSeq for the first time, you will be asked for a directory in which to store metadata. You can use the default or a directory of your choice; you may subsequently skip all steps that ask you to install "required" databases locally since we will not need them for this task.
- (E) A window will appear - the MultiSeq window - it contains the sequence of the APSES domain you are visualizing. MultiSeq will also generate an additional cartoon representation of the structure.
- (3) Load the APSES alignment.
- (A) In the MultiSeq Window, navigate to File → Import Data...; Choose "From Files" and Browse to the location of the alignment you have saved. The File navigation window gives you options which files to enable: choose to Enable ALN files (these are CLUSTAL formatted multiple sequence alignments).
- (B) Open the alignment file, click on Ok to Import Data, it will take a short while to load. If the data can't be loaded, the file may have the wrong extension:
.alnis required. - (C) find the Mbp1_SACCE sequence in the list, click on it and move it to the top of the Sequences list with your mouse (the list is not static, you can re-order the sequences in any way you like).
You will see that the 1MB1 sequence and the APSES domain sequence do not match; at the beginning the structure has extra sequence extending its N-terminus, and in the middle the APSES sequences have gaps inserted.
- (4) Bring the 1MB1 sequence in register with the APSES alignment.
- (A) MultiSeq supports typical text-editor selection mechanisms. Clicking on a residue selects it, clicking on a row selects the whole sequence. Dragging with the mouse selects several residues, shift-clicking selects ranges, and option-clicking toggles the selection on or off for individual residues. Using the mouse and/or the shift key as required, select the entire first column of the sequences you have imported.
- (B) Select Edit → Enable Editing... → Gaps only to allow changing indels.
- (C) Pressing the spacebar once should insert a gap character before the selected column in all sequences. Insert as many gaps as you need to align the beginning of sequences with the corresponding residues of 1MB1
S I M .... - (D) Now insert as many gaps as you need into the structure sequence, to align it completely with the Mbp1_SACCE APSES domain sequence. (Simply select residues in the sequence and use the space bar to insert gaps. (Note: I have noticed a bug that sometimes prevents slider or keyboard input to the MultiSeq window; it fails to regain focus after operations in a different window. I don't know whether this is a Mac related problem or a more general bug in MultiSeq. When this happens I quit VMD and restore the session from a saved state. It is a bit annoying but not mission-critical.)
- (E) When you are done, it may be prudent to save the state of your alignment. Use File → Save Session...
- (5) Color by similarity
- (A) Use the View → Coloring → Sequence similarity → BLOSUM30 option to color the residues in the alignment and structure. This clearly shows you where conserved and variable residues are located and allows to analyze their structural context.
- (B) You can adjust the color scale in the usual way by navigating to VMD main → Graphics → Colors..., choosing the Color Scale tab and adjusting the scale midpoint (0.75 works well for me).
- (C) Navigate to the Representations window and apply the color scheme to your tube-and-sidechain representation: double-click on the NewCartoon representation to hide it and use User coloring of your Tube and Licorice representations to apply the sequence similarity color gradient that MultiSeq has calculated. The example below shows in principle what you could expect to see (without sidechains).
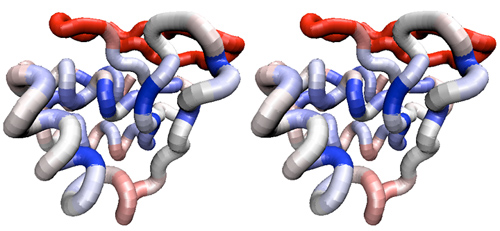
Stereo view of a tube representation of an APSES domain structure, colored according to residue similarity of all fungal APSES domains as defined in this assignment. A BLOSUM30 similarity matrix was applied and a gradient midpoint of 0.75. The domain is oriented with the putative recognition helix towards the front, left and the "wing" on the right.
- (D) Now delete all non-Mbp1 sequences from the alignment and recalculate the similarity coloring using only the Mbp1 orthologues. You may want to shift the gradient midpoint to 0.9 or so since overall conservation is much higher. Again study the conservation patterns.
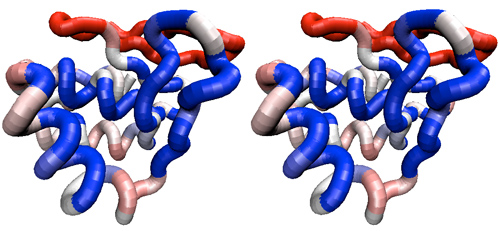
Stereo view of a tube representation of an APSES domain structure, colored according to residue similarity of all Mbp1 orthologue APSES domains, as defined in this assignment. A BLOSUM50 similarity matrix was applied and a gradient midpoint of 0.90. The domain is oriented with the putative recognition helix towards the front, left and the "wing" on the right.
- Analysis (1 mark)
- Generate two parallel stereo views that shows the APSES domain backbone and selected sidechains as described above. One should be colored by sequence similarity among all APSES domains, the other by similarity among only the Mbp1 orthologues. Scale and rotate the structure so that the putative DNA binding domain is easily visible. Paste both views into your assignment in a compressed format, as was explained for Assignment 2.
- Briefly discuss what you see (with reference to specific residues and sidechains) and what you conclude about residue conservation in the alignment of all APSES domains. Are the patterns of sequence variation for structurally conserved residues compatible with the notion that all APSES domains have a common fold?
- Briefly discuss how the situation changes when you compare only Mbp1 orthologues with each other. Never mind that overall conservation is higher: does the distribution of conserved residues in the context of the domain change, and if so, how? Are the patterns of sequence variation for functionally conserved residues compatible with the notion that all Mbp1 orthologues have a similar function?
NB: These are not yes/no questions but require reference to specific residues and observations.
(6) Summary of Resources
- Links
- Sequences
- Alignments
- Mbp1 proteins:
- APSES domains:
- Further reading
[End of assignment]
If you have any questions at all, don't hesitate to mail me at boris.steipe@utoronto.ca or post your question to the Course Mailing List