Introduction
Jump to navigation
Jump to search










I N T R O D U C T I O N
Objectives
- Understand that in molecular biology, the amount of knowledge based on computational inference is steadily surpassing that derived from experimental observation.
- Be aware of rapid change in the filed of bioinformatics. This relates to databases, as well as procedures and requires scientists to continuously update themselves with new data and approaches.
- Understand that the goal of the course is not primarily the transfer of bits of knowledge, but to acquire the skill to devise novel problem-solving strategies wherever your own work requires it.
Links
- NCBI (National Center for Biotechnology Information)
- PDB (Protein structure DataBase)
- KEGG (the Kyoto Encyclopedia of Genes and Genomes)
- CGDN bioinformatics portal
- Bioinformatics.org
- Genome Canada Bioinformatics HelpDesk
- The International Society for Computational Biology
- VMD
Slides
What is Bioinformatics?
Slide 0007

Introduction, slide 0007
From its beginning, it was recognized that molecular biology is an information science, just as much as a molecular science. Scientists have documented an unbroken flow of information: from the storage of information in the non-random sequence of nucleotide heterocopolymers, to the self-organized acquisition of structure and function in proteins that provides a selective advantage for evolution. The abstractions and models that focus on inheritable information, rather than on the details of its representation, have proven to be remarkably powerful in explaining the basic features of life, such as robust self-organization and the process of evolution.
From its beginning, it was recognized that molecular biology is an information science, just as much as a molecular science. Scientists have documented an unbroken flow of information: from the storage of information in the non-random sequence of nucleotide heterocopolymers, to the self-organized acquisition of structure and function in proteins that provides a selective advantage for evolution. The abstractions and models that focus on inheritable information, rather than on the details of its representation, have proven to be remarkably powerful in explaining the basic features of life, such as robust self-organization and the process of evolution.
Slide 0008

Introduction, slide 0008
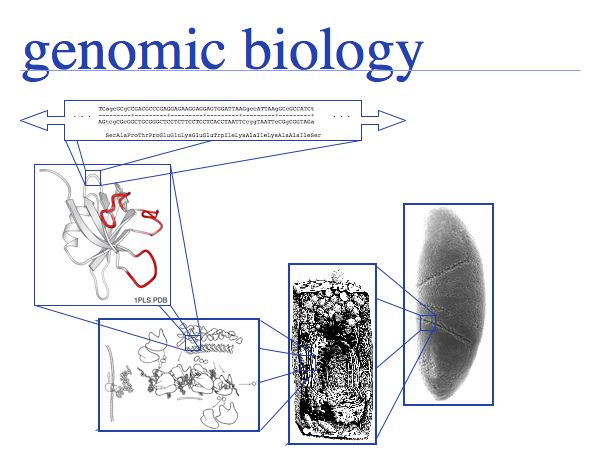
In the genomic - or post-genomic - era, the relationship of information and molecule becomes even more pertinent. In principle, all information that is required to specify an organism is contained in its genome. The genome can be fully sequenced therefore the information is accessible to us. However, the expression of the information is organized in a hierarchical fashion, in complex, interacting subsystems. Knowledge of a DNA sequence does not yet allow us to predict the protein's structure. Knowledge of a protein's structure does not yet allow us to predict its interactions and assembly to molecular "machines". Knowledge of these complexes does not yet allow us to piece together their functional connections, as they build up the complex metabolic or regulatory systems, or the structural framework of a cell. At each level, incomplete information prevents us from predicting the next-higher level of organization from its components. The sheer volume of data is a comparatively minor obstacle.
In the genomic - or post-genomic - era, the relationship of information and molecule becomes even more pertinent. In principle, all information that is required to specify an organism is contained in its genome. The genome can be fully sequenced therefore the information is accessible to us. However, the expression of the information is organized in a hierarchical fashion, in complex, interacting subsystems. Knowledge of a DNA sequence does not yet allow us to predict the protein's structure. Knowledge of a protein's structure does not yet allow us to predict its interactions and assembly to molecular "machines". Knowledge of these complexes does not yet allow us to piece together their functional connections, as they build up the complex metabolic or regulatory systems, or the structural framework of a cell. At each level, incomplete information prevents us from predicting the next-higher level of organization from its components. The sheer volume of data is a comparatively minor obstacle.
Slide 0009

Introduction, slide 0009
The current emphasis on -omic sciences creates novel challenges both in the quantity as well as the quality of scientific enquiry. The scale has become larger; molecular components are analyzed not in isolation but in their associations;comparison between genes within and across species is a major source of new insight and the absence of particular components and features is just as informative as their presence. However the availability of technology has led at times to a purely methods-driven agenda.
The current emphasis on -omic sciences creates novel challenges both in the quantity as well as the quality of scientific enquiry. The scale has become larger; molecular components are analyzed not in isolation but in their associations;comparison between genes within and across species is a major source of new insight and the absence of particular components and features is just as informative as their presence. However the availability of technology has led at times to a purely methods-driven agenda.
Slide 0010

Introduction, slide 0010
The US National Center of Biotechnology Information is one of the world's major centres for molecular data.
The US National Center of Biotechnology Information is one of the world's major centres for molecular data.
Slide 0011

Introduction, slide 0011
The PDB (Protein structure DataBase) is the world's central repository for 3D structural data of proteins and nucleic acids.
The PDB (Protein structure DataBase) is the world's central repository for 3D structural data of proteins and nucleic acids.
Slide 0012

Introduction, slide 0012
KEGG' (the Kyoto Encyclopedia of Genes and Genomes) is one of a group of data resources that focus on the functional relationships of the components of biological systems. Note that sequences, structures and functions are complementary aspects of the same molecular entities. Cross-referencing between databases and ensuring consistency is a major challenge and task of biological data management.
KEGG' (the Kyoto Encyclopedia of Genes and Genomes) is one of a group of data resources that focus on the functional relationships of the components of biological systems. Note that sequences, structures and functions are complementary aspects of the same molecular entities. Cross-referencing between databases and ensuring consistency is a major challenge and task of biological data management.
Slide 0013

Introduction, slide 0013
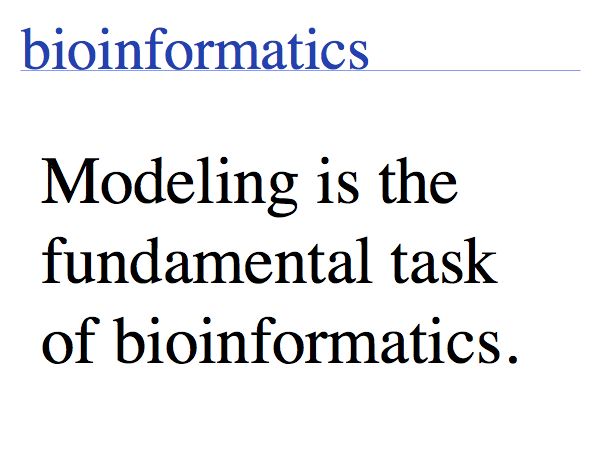
Bioinformatics can be viewed as the science that develops between the two poles data management and computational modeling of life. On one hand, if we look at the practice of bioinformatics, we can conclude that biological data management is what bioinformatics is all about. On the other hand, bioinformatics as a science is a way to study biology. And this aspect - which I like to refer to as "Computational Biology" - is not well described by data management. It has a lot more to do with modeling, and the question of understanding biology.
Bioinformatics can be viewed as the science that develops between the two poles data management and computational modeling of life. On one hand, if we look at the practice of bioinformatics, we can conclude that biological data management is what bioinformatics is all about. On the other hand, bioinformatics as a science is a way to study biology. And this aspect - which I like to refer to as "Computational Biology" - is not well described by data management. It has a lot more to do with modeling, and the question of understanding biology.
Slide 0014
Slide 0015
Learning "Bioinformatics"
Slide 0017
Course resources and supporting sites
Slide 0024
Slide 0027
Slide 0028

Introduction, slide 0028
CGDN bioinformatics portal - home of the Canadian Bioinformatics Workshops
CGDN bioinformatics portal - home of the Canadian Bioinformatics Workshops
Slide 0029

Introduction, slide 0029
Bioinformatics.org
Bioinformatics.org
Browse the archives of the BioBB mailing list - it may be quite useful to subscribe to get a better idea of what's going on in the field.
Slide 0030

Introduction, slide 0030
Genome Canada Bioinformatics HelpDesk
Genome Canada Bioinformatics HelpDesk
Slide 0031

Introduction, slide 0031
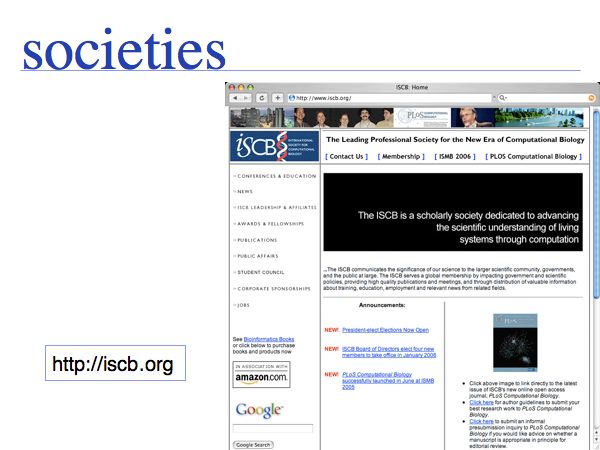
The International Society for Computational Biology (among other activities) host ISMB - the world's largest bioinformatics conference.
The International Society for Computational Biology (among other activities) host ISMB - the world's largest bioinformatics conference.
Slide 0032
Slide 0033

Introduction, slide 0033
VMD is a free, widely used, richly featured and well supported molecular viewer that we will be using throughout the course. On the homepage, you can download the program, find tutorials and handbooks and subscribe to the support mailing list or simply browse the list archives.
VMD is a free, widely used, richly featured and well supported molecular viewer that we will be using throughout the course. On the homepage, you can download the program, find tutorials and handbooks and subscribe to the support mailing list or simply browse the list archives.