Lecture 24
Jump to navigation
Jump to search


(Previous lecture) ... (Next lecture)
Interaction Databases used in Computational Biology
BCH441 Guest lecture:
- Francis Ouellette
- Associate Director, Informatics and Biocomputing and Senior Scientist, OICR
Links summary
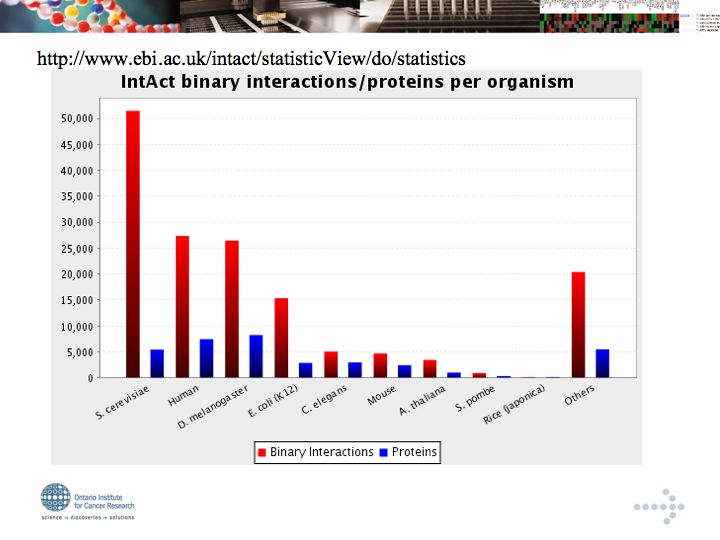
- IntAct - The EBI open access, archival interaction database
- BioGRID
- DIP (archival, not open access)
- MINT Topical, curatedl interaction sets
- Mpact
- BIND (non-commercial component not curated since 2003)
- HPRD
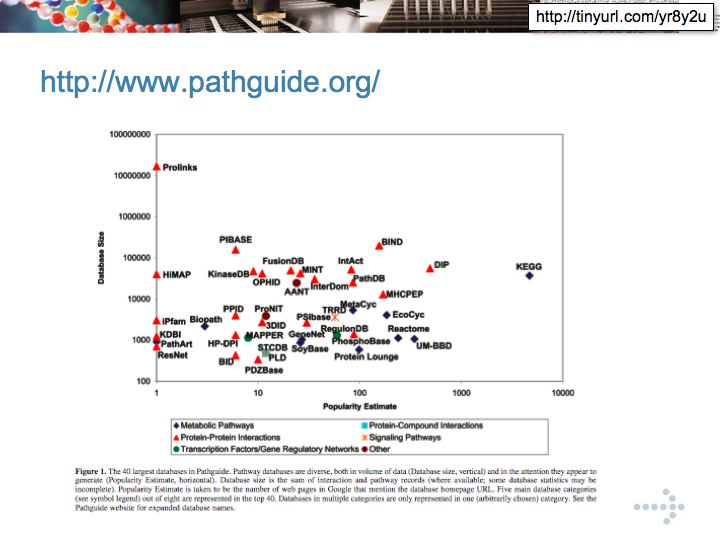
- Gary Bader's pathguide database
- Ulysses: Projection of Protein Networks across Species
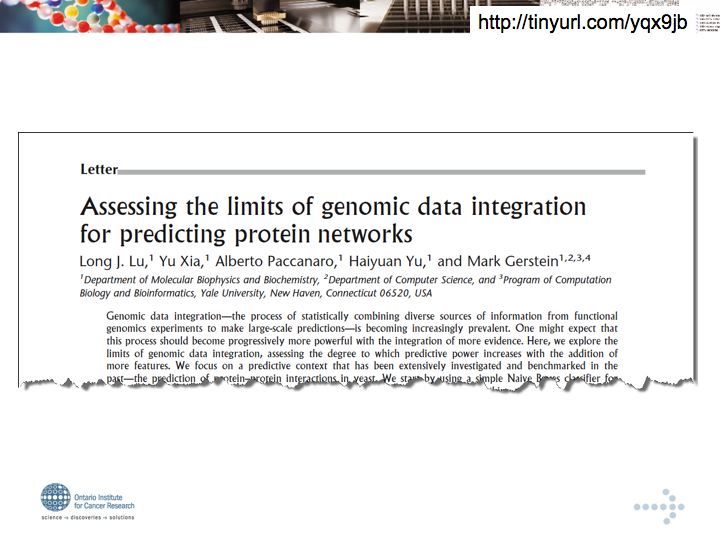
- Lu et al. (2005) Assessing the limits of genomic data integration for predicting protein networks.
- YPL240c
- Creative Commons License: atribution, share alike.
- Open Access overview
- Open Access Now
- DOAJ
- CIHR Open Access policy
Lecture slides
Introduction
Slide 001

Lecture 24, Slide 001
These lecture notes are provided under a Creative Commons License: atribution, share alike. (You can also use the "tinyurl" service that compresses long links and automatically redirects your browesr to the right page to access this link through the shorthand http://tinyurl.com/3cw4ql .
These lecture notes are provided under a Creative Commons License: atribution, share alike. (You can also use the "tinyurl" service that compresses long links and automatically redirects your browesr to the right page to access this link through the shorthand http://tinyurl.com/3cw4ql .
Slide 002
Slide 004
Slide 005
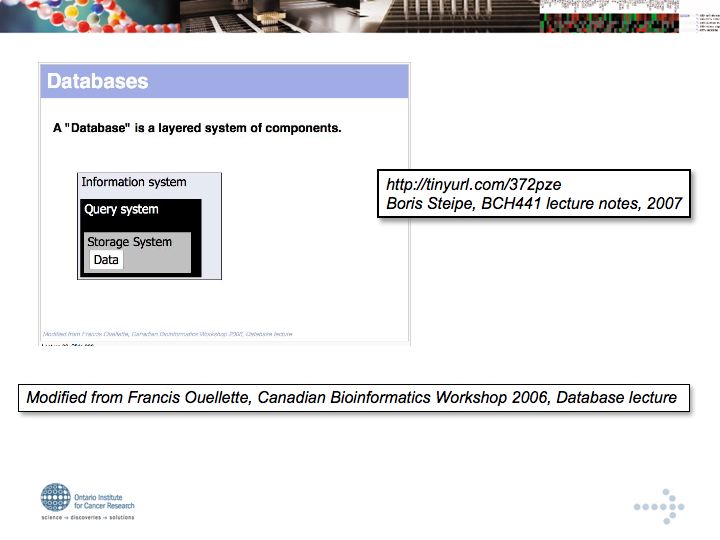
Bioinformatics and Databases
Slide 007
Slide 008
Slide 009
Slide 010
Slide 011
Slide 012
Slide 013
Slide 014
Slide 015
Slide 016
Slide 017
Slide 018
Slide 019
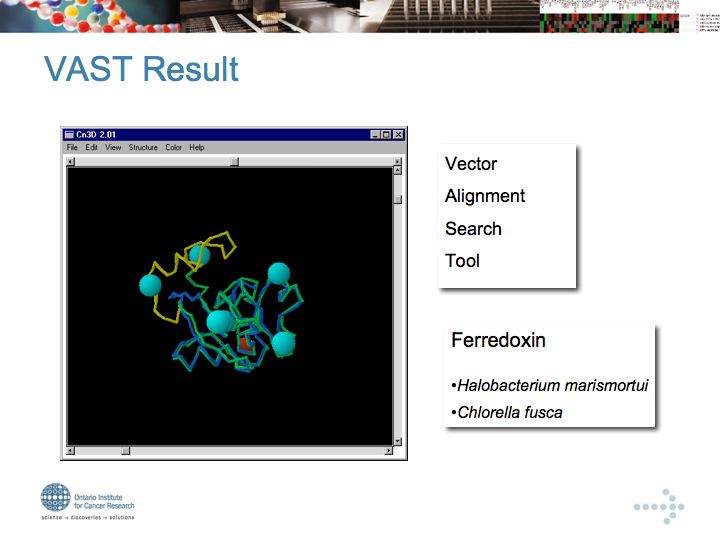
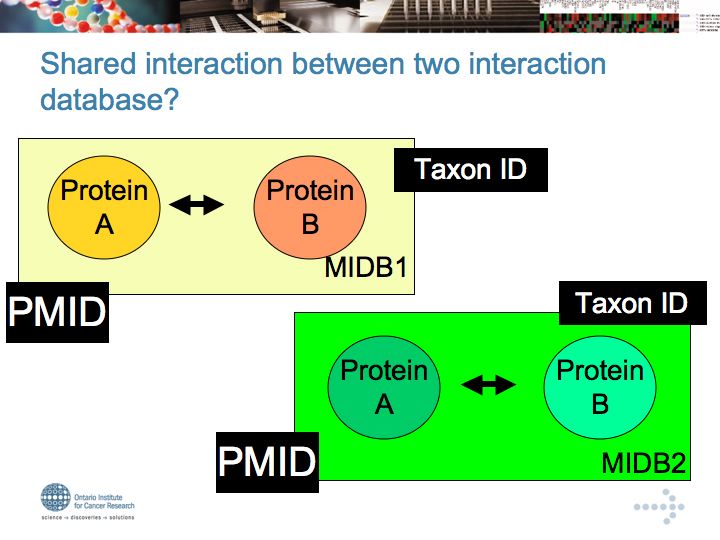
Interaction Databases
Slide 020
Slide 021

Lecture 24, Slide 021
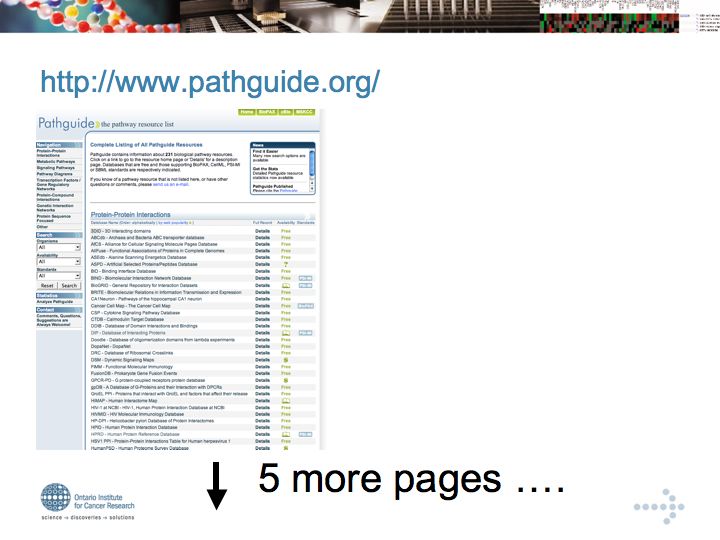
Gary Bader's pathguide database
Gary Bader's pathguide database
Slide 022
Slide 023
Slide 024
[[Image:24_slide024.jpg|frame|none|Lecture 24, Slide 024
Slide 025
Slide 026
Slide 027
Slide 028

Lecture 24, Slide 028
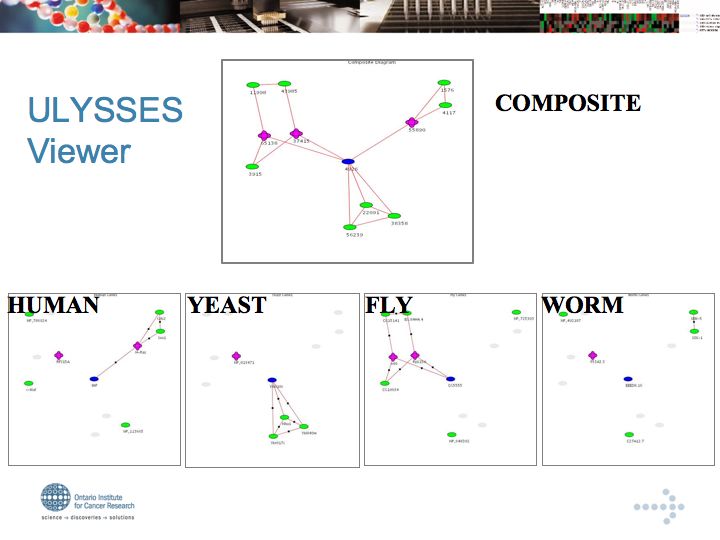
Ulysses: Projection of Protein Networks across Species
Ulysses: Projection of Protein Networks across Species
Slide 029
Slide 030
Slide 031
Slide 032
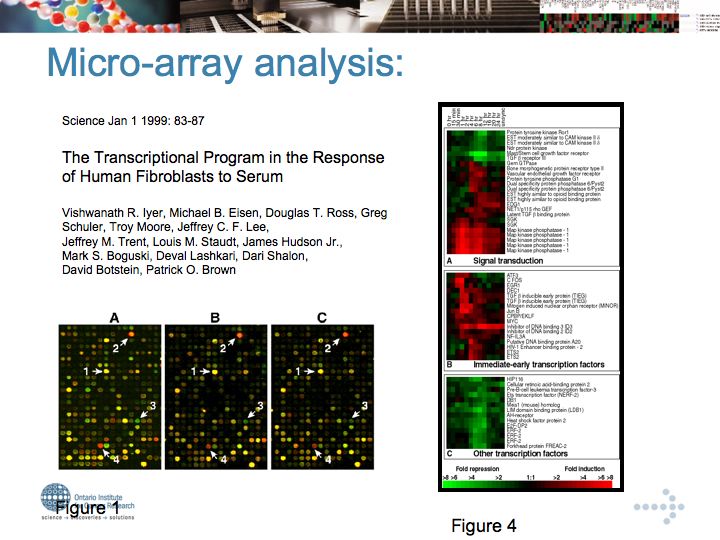
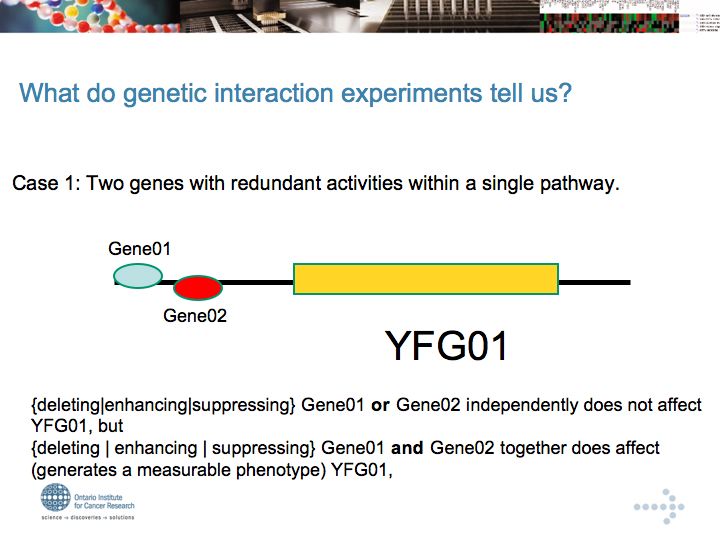
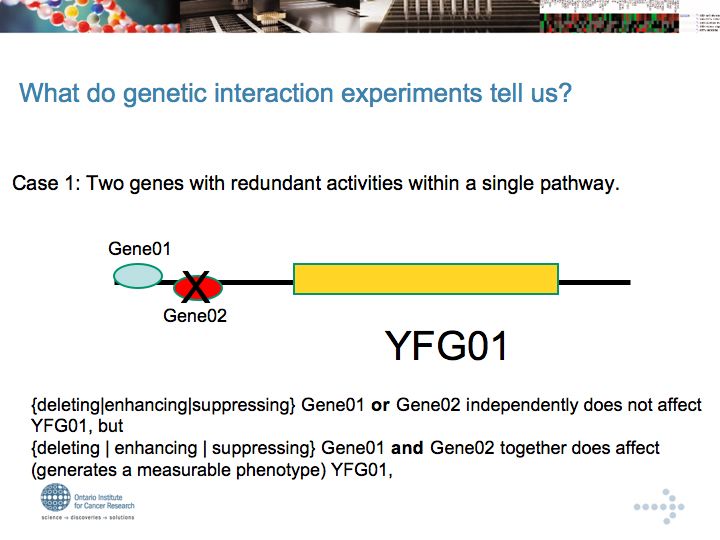
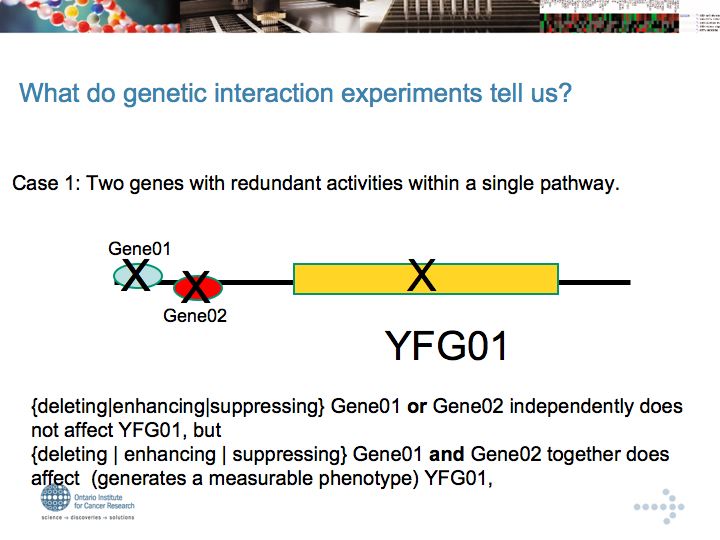
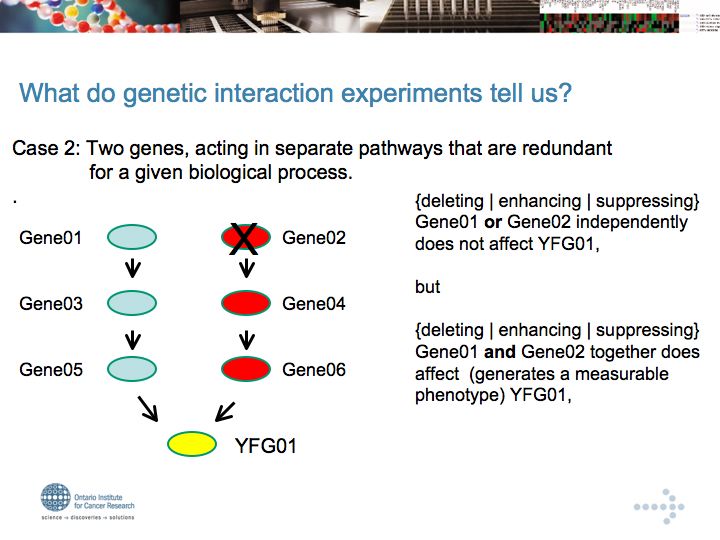
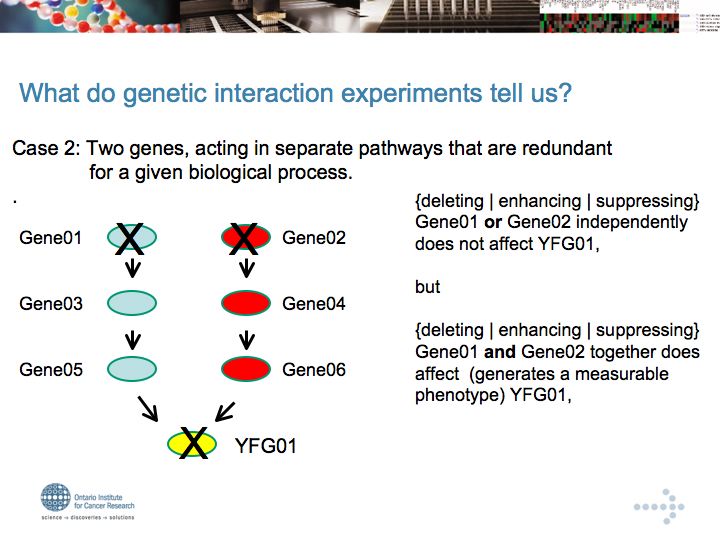
Genetic Interaction Networks
Slide 033
Slide 034
Slide 035
Slide 036
Slide 037
Slide 038
Slide 039
Slide 040
Slide 041
Slide 042
Slide 043
Slide 044
Slide 045

Lecture 24, Slide 045
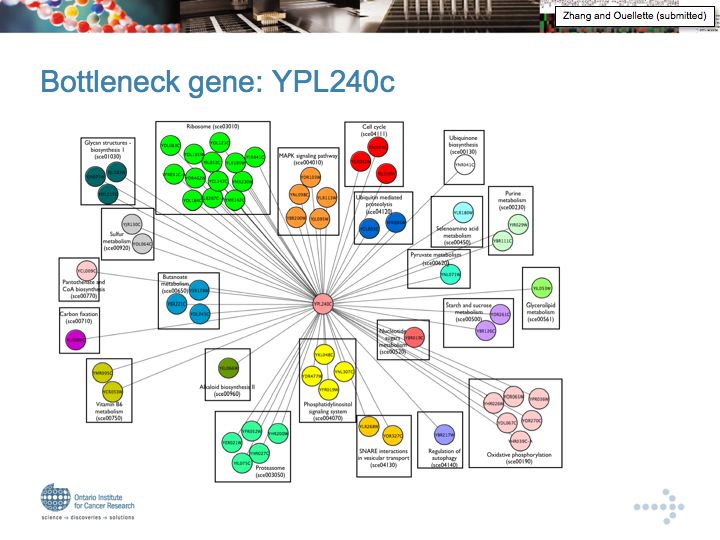
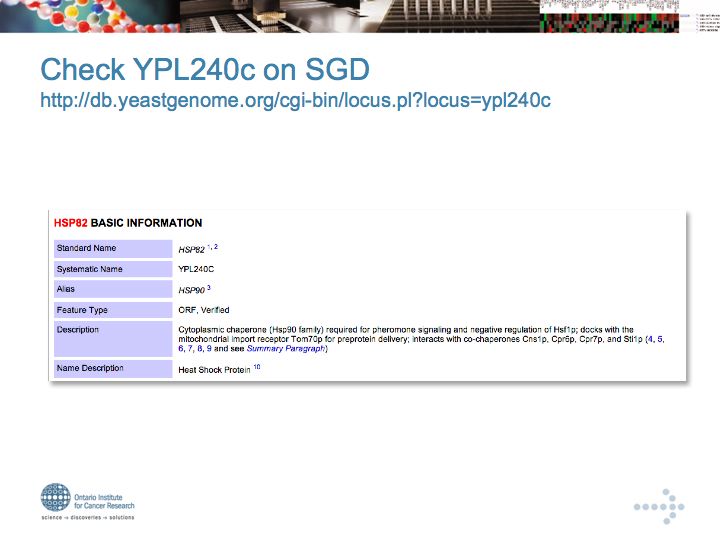
YPL240c
YPL240c
Open Access
Slide 046
Slide 047
Slide 048
Slide 049
Slide 050

Lecture 24, Slide 050
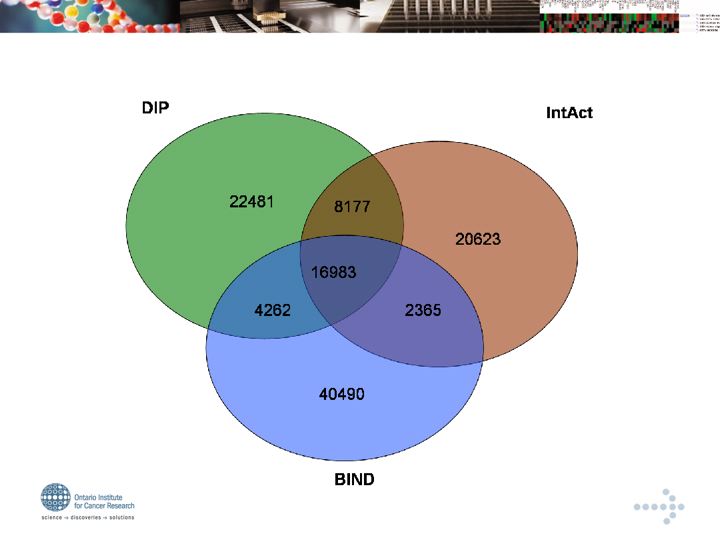
*DIP (archival, not open access)
*IntAct - The EBI open access, archival interaction database
*MINT Topical, curatedl interaction sets
*Mpact
*BioGRID
*BIND (non-commercial component not curated since 2003)
*HPRD
*Open Access overview
*Open Access Now
*DOAJ
*CIHR Open Access policy
*DIP (archival, not open access)
*IntAct - The EBI open access, archival interaction database
*MINT Topical, curatedl interaction sets
*Mpact
*BioGRID
*BIND (non-commercial component not curated since 2003)
*HPRD
*Open Access overview
*Open Access Now
*DOAJ
*CIHR Open Access policy