BIO Assignment Week 2
Assignment for Week 2
Scenario, Labnotes, R-functions, Databases, Data Modeling
Note! This assignment is currently active. All significant changes will be announced on the mailing list.
- Parts labelled as "TBC" are in progress and will be made available as they are being completed.
Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz.
Contents
The Scenario
I have introduced the concept of "cargo cult science" in class. The "cargo" in Bioinformatics is to understand biology. This includes understanding how things came to be the way they are, and how they work. Both relate to the concept of function of biomolecules, and the systems they contribute to. But "function" is a rather poorly defined concept and exploring ways to make it rigorous and computable will be the major objective of this course. The realm of bioinformatics contains many kingdoms and duchies and shires and hidden glades. To find out how they contribute to the whole, we will proceed on a quest. We will take a relatively well-characterized protein that is part of a relatively well-characterized process, and ask what its function is. We will examine the protein's sequence, its structure, its domain composition, its relationship to and interactions with other proteins, and through that paint a picture of a "system" that it contributes to.
Our quest will revolve around a transcription factor that plays an important role in the regulation of the cell cycle: Mbp1 is a key component of the MBF complex (Mbp1/Swi6) in yeast. This complex regulates gene expression at the crucial G1/S-phase transition of the mitotic cell cycle and has been shown to bind to the regulatory regions of more than a hundred target genes. It is therefore a DNA binding protein that acts as a control switch for a key cellular process.
We will start our quest with information about the Mbp1 protein of Baker's yeast, Saccharomyces cerevisiae, one of the most important model organisms. Baker's yeast is a eukaryote that has been studied genetically and biochemically in great detail for many decades, and it is easily manipulated with high-throughput experimental methods. But each of you will use this information to study not Baker's yeast, but a related organism. You will explore the function of the Mbp1 protein in some other species from the kingdom of fungi, whose genome has been completely sequenced; thus our quest is also an exercise in model-organism reasoning: the transfer of knowledge from one, well-studied organism to others.
It's reasonable to hypothesize that such central control machinery is conserved in most if not all fungi. But we don't know. Many of the species that we will be working with have not been characterized in great detail, and some of them are new to our class this year. And while we know a fair bit about Mbp1, we probably don't know very much at all about the related genes in other organisms: whether they exist, whether they have similar functional features and whether they might contribute to the G1/S checkpoint system in a similar way. Thus we might discover things that are new and interesting. This is a quest of discovery.
Here are the steps of the assignment for this week:
- We'll need to explore what data is available for the Mbp1 protein.
- We'll need to pick a species to adopt for exploration.
- We'll need to define what data we want to store and design a datamodel.
However, before we head off into the Internet: have you thought about how to document such a "quest"? How will you keep notes? Obviously, computational research proceeds with the same best-practice principles as any wet-lab experiment. We have to keep notes, ensure our work is reproducible, and that our conclusions are supported by data. I think it's pretty obvious that paper notes are not very useful for bioinformatics work. Ideally, you should be able to save results, and link to files and Webpages.
Keeping Labnotes
Consider it a part of your assignment to document your activities in electronic form. Here are some applications you might think of - but (!) disclaimer, I myself don't use any of these (yet) (except the Wiki of course).
- Evernote - a web hosted, automatically syncing e-notebook.
- Nevernote - the Open Source alternative to Evernote.
- Google Keep - if you have a Gmail account, you can simply log in here. Grid-based. Seems a bit awkward for longer notes. But of course you can also use Google Docs.
- Microsoft OneNote - this sounds interesting and even though I have had my share of problems with Microsoft products, I'll probably give this a try. Syncing across platforms, being able to format contents and organize it sounds great.
- The Student Wiki - of course. You can keep your course notes with your User pages.
Are you aware of any other solutions? Let us know!
Keeping such a journal will be helpful, because the assignments are integrated over the entire term, and later assignments will make use of earlier results. But it is also excellent practice for "real" research. Expand the section below for details - written from a Wiki perspective but generally applicable.
Data Sources
SGD - a Yeast Model Organism Database
Yeast happens to have a very well maintained model organism database - a Web resource dedicated to Saccharomyces cerevisiae. Where such resources are available, they are very useful for the community. For the general case however, we need to work with one of the large, general data providers - the NCBI and the EBI. But in order to get a sense of the type of data that is available, let's visit the SGD database first.
Task:
Access the information page on Mbp1 at the Saccharomyces Genome Database.
- Browse through the Summary page and note the available information: you should see:
- information about the gene and the protein;
- Information about it's roles in the cell curated at the Gene Ontology database;
- Information about knock-out phenotypes; (Amazing. Would you have imagined that this is a non-essential gene?)
- Information about protein-protein interactions;
- Regulation and expression;
- A curators' summary of our understanding of the protein. Mandatory reading.
- And key references.
- Access the Protein tab and note the much more detailed information.
- Domains and their classification;
- Sequence;
- Shared domains;
- and much more...
You will notice that some of this information relates to the molecule itself, and some of it relates to its relationship with other molecules. Some of it is stored at SGD, and some of it is cross-referenced from other databases. And we have textual data, numeric data, and images.
How would you store such data to use it in your project? We will work on this question at the end of the assignment.
If we were working on yeast, most data we need is right here: curated, kept current and consistent, referenced to the literature and ready to use. But you'll be working on a different species and we'll explore the much, much larger databases at the NCBI for this. The upside is that most of the information like this is available for your species. The downside is that we'll have to integrate information from many different sources "by hand".
NCBI databases
The NCBI (National Center for Biotechnology Information) is the largest international provider of data for genomics and molecular biology. With its annual budget of several hundred million dollars, it organizes a challenging program of data management at the largest scale, it makes its data freely and openly available over the Internet, worldwide, and it runs significant in-house research projects.
Let us explore some of the offerings of the NCBI that can contribute to our objective of studying a particular gene in an organism of interest.
Entrez
Task:
Remember to document your activities.
- Access the NCBI website at http://www.ncbi.nlm.nih.gov/ [1]
- In the search bar, enter
mbp1and click Search. - On the resulting page, look for the Protein section and click on the link. What do you find?
The result page of your search in "All Databases" is the "Global Query Result Page" of the Entrez system. If you follow the "Protein" link, you get taken to the more than 450 sequences in the NCBI Protein database that contain the keyword "mbp1". But when you look more closely at the results, you see that the result is quite non-specific: searching only by keyword retrieves an Arabidopsis protein, bacterial proteins, a Saccharomyces protein (perhaps one that we are actually interested in), Maltose Binding Proteins - and much more. There must be a more specific way to search, and indeed there is. Time to read up on the Entrez system.
Task:
- Navigate to the Entrez Help Page and read about the Entrez system, especially about:
- Boolean operators,
- wildcards,
- limits, and
- filters.
- You should minimally understand:
- How to search by keyword;
- How to search by gene or protein name;
- How to restrict a search to a particular organism.
Don't skip this part, you don't need to know the options by heart, but you should know they exist and how to find them. It would be great to have a synopsis of the important fields for reference, wouldn't it?Why don't you go and create one: I have put a template page on the Student Wiki (A synopsis of Entrez codes). Editors welcome!
Keyword and organism searches are pretty universal, but apart from that, each NCBI database has its own set of specific fields. You can access the keywords via the Advanced Search interface of any of the database pages.
Protein Sequence
Task:
With this knowledge we can restrict the search to proteins called "Mbp1" that occur in Baker's Yeast. Return to the Global Search page and in the search field, type:
Mbp1[protein name] AND
"Saccharomyces cerevisiae"[organism]
This should find one and only one protein. Follow the link into the protein database: since this is only one record, the link takes you directly to the result: CAA98618.1—a data record in Genbank Flat File (GFF) format[2]. The database identifier CAA98618.1 tells you that this is a record in the GenPept database. There are actually several, identical versions of this sequence in the NCBI's holdings. A link to "Identical Proteins" near the top of the record shows you what these are:
Some of the sequences represent duplicate entries of the same gene (Mbp1) in the same strain (S288c) of the same species (S. cerevisiae). In particular:
- there are three GenBank; records (
CAA52271.1,CAA98618.1andDAA11800.1); these are archival entries, submitted by independent yeast genome research projects;
- there is an entry in the RefSeq database:
NP_010227.1. This is the preferred entry for us to work with. RefSeq is a curated, non-redundant database which solves a number of problems of archival databases. You can recognize RefSeq identifiers – they always look like NP_12345.1, NM_12345.1, XP_12345.1, NC_12345.1 etc. This reflects whether the sequence is protein, mRNA or genomic, and inferred or obtained through experimental evidence. The RefSeq IDNP_010227.1actually appears twice, once linked to its genomic sequence, and once to its mRNA.
- there is a SwissProt sequence
P39678.1[3]. This link is kind of a big deal. It's a cross-reference into UniProt, the huge protein sequence database maintained by the EBI (European Bioinformatics Institute), which is the NCBI's counterpart in Europe. SwissProt entries have the highest annotation standard overall and are expertly curated. Many Webservices that we will encounter, work with UniProt ID's (e.g.P39678.1), rather than RefSeq. But it used to be until recently that the two databases did not link to each other, mostly for reasons of funding politics. It's great to see that this divide has now been overcome.
- Finally, there are four entries of the same sequence in different yeast strains. These don't have to be identical, they just happen to be. Sometimes we find identical sequences in quite divergent species. Therefore I would not actually consider
EIW11153.1,AJU86440.1,AJU58508.1, andAJU61971.1to be identical proteins, although they have the same sequence.
Note all the .1 suffixes of the sequence identifiers. These are version numbers. Two observations:
- It's great that version numbers are now used throughout the NCBI database. This is good database engineering practice because it's really important for reproducible research that updates to database records are possible, but recognizable. When working with data you always must provide for the possibility of updates, and manage the changes transparently and explicitly. Proper versioning should be a part of all datamodels.
- When searching, or for general use, you should omit the version number, i.e. use
NP_010227orP39678notNP_010227.1resp.P39678.1. This way the database system will resolve the identifier to the most current, highest version number (unless you want the older one, of course).
Task:
- Note down the RefSeq ID and the UniProt (SwissProt) ID in your journal.
- Follow the link to the RefSeq entry
NP_010227.1. - Explore the page and follow these links (note the contents in your journal):
- Under "Analyze this Sequence": Identify Conserved Domains
- Under "Protein 3D Structure": See all 3 structures...
- Under "Pathways for the MBP1 gene": Cell cycle - yeast
- Under "Related information" Proteins with Similar Sequence
As we see, this is a good start page to explore all kinds of databases at the NCBI via cross-references.
PubMed
Arguably one of the most important databases in the life sciences is PubMed and this is a good time to look at PubMed in a bit more detail.
Task:
- Return back to the MBP1 RefSeq record.
- Find the PubMed links under Related information in the right-hand margin and explore them.
- The first one (PubMed) will take you to records that cite the sequence record;
- The second one (PubMed (RefSeq)) will take you to articles that relate to the Mbp1 gene or protein;
- The third one (PubMed (Weighted)) applies a weighting algorithm to find broadly relevant information - an example of literature data mining. PubMed(weighted) appears to give a pretty good overview of systems-biology type, cross-sectional and functional information.
But neither of the searches finds all Mbp1 related literature.
- On any of the PubMed pages open the Advanced query page and study the keywords that apply to PubMed searches. These are actually quite important and useful to remember. Make yourself familiar with the section on Search field descriptions and tags in the PubMed help document, (in particular [DP], [AU], [TI], and [TA]), how you use the History to combine searches, and the use of AND, OR, NOT and brackets. Understand how you can restrict a search to reviews only, and what the link to Related citations... is useful for.
- Now find publications with Mbp1 in the title. In the result list, follow the links for the two Biochemistry papers, by Taylor et al. (2000) and by Deleeuw et al. (2008). Download the PDFs, we will need them later.
Now, we were actually trying to find related proteins in a different species. Our next task is therefore to decide what that species should be.
Choosing YFO (Your Favourite Organism)
In this section we create a lottery to assign species at (pseudo) random to students. We'll try the following procedure.
- First, I create a list of suitable species.
- Then, we put this list into the body of an R function.
- The function picks one of the species at random - but to make sure this process is reproducible, we'll set a seed for the random number generator. Obviously, everyone has to use a different seed, or else everyone would end up getting the same species assigned.
- Thus we'll use your Student Number as the seed. This is an integer, so it can be used, and it's unique to each of you. The choice is then random, reproducible and unique.
You may notice that this process does not guarantee that everyone gets a different species, and that all species are chosen at least once. I don't think doing that is possible in a "stateless" way (i.e. I don't want to have to remember who chose what species), given that I don't know all of your student numbers. But if anyone can think of a better solution, that would be neat.
Is it possible that all of you end up working on the same species anyway? Indeed. That's the problem with randomness. But it is not very likely.
What about the "suitable species" though? Where do they come from? For the purposes of the course "quest", we need species
- that actually have transcription factors that are related to Mbp1;
- whose genomes have been sequenced; and
- for which the sequences have been deposited in the RefSeq database, NCBI's unique sequence collection.
Task:
- Access the R tutorial and work through the section on Writing your own functions. It is short, and crucial for your work.
Here is R code to assign the species:
Task:
- Read, try to understand and then execute the following R-code.
pickSpecies <- function(ID) {
# this function randomly picks a fungal species
# from a list. It is seeded by a student ID. Therefore
# the pick is random, but reproducible.
# first, define a list of species:
Species <- c(
"Agaricus bisporus (AGABI)",
"Arthrobotrys oligospora (ARTOL)",
"Arthroderma benhamiae (ARTBE)",
"Aureobasidium subglaciale (AURSU)",
"Auricularia delicata (AURDE)",
"Batrachochytrium dendrobatidis (BATDE)",
"Baudoinia panamericana (BAUPA)",
"Beauveria bassiana (BEABA)",
"Bipolaris sorokiniana (BIPSO)",
"Blastomyces dermatitidis (BLADE)",
"Botrytis cinerea (BOTCI)",
"Capronia epimyces (CAPEP)",
"Chaetomium thermophilum (CHATH)",
"Cladophialophora yegresii (CLAYE)",
"Clavispora lusitaniae (CLALU)",
"Coccidioides immitis (COCIM)",
"Colletotrichum graminicola (COLGR)",
"Coniophora puteana (CONPU)",
"Coniosporium apollinis (CONAP)",
"Coprinopsis cinerea (COPCI)",
"Cordyceps militaris (CORMI)",
"Cryptococcus neoformans (CRYNE)",
"Cyphellophora europaea (CYPEU)",
"Dactylellina haptotyla (DACHA)",
"Debaryomyces hansenii (DEBHA)",
"Dichomitus squalens (DICSQ)",
"Endocarpon pusillum (ENDPU)",
"Eremothecium gossypii (EREGO)",
"Eutypa lata (EUTLA)",
"Exophiala aquamarina (EXOAQ)",
"Fibroporia radiculosa (FIBRA)",
"Fomitiporia mediterranea (FOMME)",
"Fonsecaea pedrosoi (FONPE)",
"Fusarium pseudograminearum (FUSPS)",
"Gaeumannomyces graminis (GAEGR)",
"Glarea lozoyensis (GLALO)",
"Gloeophyllum trabeum (GLOTR)",
"Heterobasidion irregulare (HETIR)",
"Histoplasma capsulatum (HISCA)",
"Kazachstania africana (KAZAF)",
"Kluyveromyces lactis (KLULA)",
"Komagataella pastoris (KOMPA)",
"Laccaria bicolor (LACBI)",
"Lachancea thermotolerans (LACTH)",
"Leptosphaeria maculans (LEPMA)",
"Lodderomyces elongisporus (LODEL)",
"Magnaporthe oryzae (MAGOR)",
"Malassezia globosa (MALGL)",
"Marssonina brunnea (MARBR)",
"Metarhizium robertsii (METRO)",
"Meyerozyma guilliermondii (MEYGU)",
"Microsporum gypseum (MICGY)",
"Millerozyma farinosa (MILFA)",
"Moniliophthora roreri (MONRO)",
"Myceliophthora thermophila (MYCTH)",
"Naumovozyma dairenensis (NAUDA)",
"Nectria haematococca (NECHA)",
"Neofusicoccum parvum (NEOPA)",
"Neosartorya fischeri (NEOFI)",
"Ogataea parapolymorpha (OGAPA)",
"Paracoccidioides brasiliensis (PARBR)",
"Penicillium rubens (PENRU)",
"Pestalotiopsis fici (PESFI)",
"Phanerochaete carnosa (PHACA)",
"Pneumocystis murina (PNEMU)",
"Podospora anserina (PODAN)",
"Postia placenta (POSPL)",
"Pseudocercospora fijiensis (PSEFI)",
"Pseudogymnoascus destructans (PSEDE)",
"Pseudozyma hubeiensis (PSEHU)",
"Puccinia graminis (PUCGR)",
"Punctularia strigosozonata (PUNST)",
"Pyrenophora teres (PYRTE)",
"Rasamsonia emersonii (RASEM)",
"Rhinocladiella mackenziei (RHIMA)",
"Scheffersomyces stipitis (SCHST)",
"Schizophyllum commune (SCHCO)",
"Sclerotinia sclerotiorum (SCLSC)",
"Serpula lacrymans (SERLA)",
"Setosphaeria turcica (SETTU)",
"Sordaria macrospora (SORMA)",
"Spathaspora passalidarum (SPAPA)",
"Stereum hirsutum (STEHI)",
"Talaromyces marneffei (TALMA)",
"Tetrapisispora phaffii (TETPH)",
"Thielavia terrestris (THITE)",
"Tilletiaria anomala (TILAN)",
"Togninia minima (TOGMI)",
"Torulaspora delbrueckii (TORDE)",
"Trametes versicolor (TRAVE)",
"Tremella mesenterica (TREME)",
"Trichoderma virens (TRIVI)",
"Trichophyton rubrum (TRIRU)",
"Tuber melanosporum (TUBME)",
"Uncinocarpus reesii (UNCRE)",
"Vanderwaltozyma polyspora (VANPO)",
"Verticillium alfalfae (VERAL)",
"Wallemia mellicola (WALME)",
"Wickerhamomyces ciferrii (WICCI)",
"Yarrowia lipolytica (YARLI)",
"Zygosaccharomyces rouxii (ZYGRO)",
"Zymoseptoria tritici (ZYMTR)"
)
set.seed(ID) # seed the random number generator
choice <- sample(Species, 1) # pick a random element
return(choice)
}- Execute the function
pickSpecies()with your student ID as its argument. Example:
> pickSpecies(991234567)
[1] "Coccidioides immitis (COCIM)"- Note down the species name and its five letter label on your Student Wiki user page. Use this species whenever this or future assignments refer to YFO.
Selecting "your" gene
Task:
- Back at the Mbp1 protein page follow the link to Run BLAST... under "Analyze this sequence".
- This allows you to perform a sequence similarity search. You need to set two parameters:
- As Database, select Reference proteins (refseq_protein) from the drop down menu;
- In the Organism field, type the species you have selected as YFO and select the corresponding taxonomy ID.
- Click on Run BLAST to start the search.
This should find a handful of genes, all of them in YFO. If you find none, or hundreds, or they are not all in the same species, you did something wrong. Ask on the mailing list and make sure to fix the problem.
- Note the results in your Journal.
Data Storage
Now that we have a better sense of what our data is, we need to consider ways to model and store it. Let's talk about storage first.
Any software project requires modelling on many levels - data-flow models, logic models, user interaction models and more. But all of these ultimately rely on a data model that defines how the world is going to be represented in the computer for the project's purpose. The process of abstraction of data entities and defining their relationships can (and should) take up a major part of the project definition, often taking several iterations until you get it right. Whether your data can be completely described, consistently stored and efficiently retrieved is determined to a large part by your data model.
Databases can take many forms, from memories in your brain, to shoe-cartons under your bed, to software applications on your computer, or warehouse-sized data centres. Fundamentally, these all do the same thing: collect information and make it available.
Let us consider collecting information on APSES-domain transcription factors in various fungi, with the goal of being able to compare them. Let's specify this as follows:
- cross reference the source databases;
- study if they have the same features (e.g. domains);
- and compare the features.
The underlying information can easily be retrieved for a protein from its RefSeq or UniProt entry.
Text files
A first attempt to organize the data might be simply to write it down in a large text file:
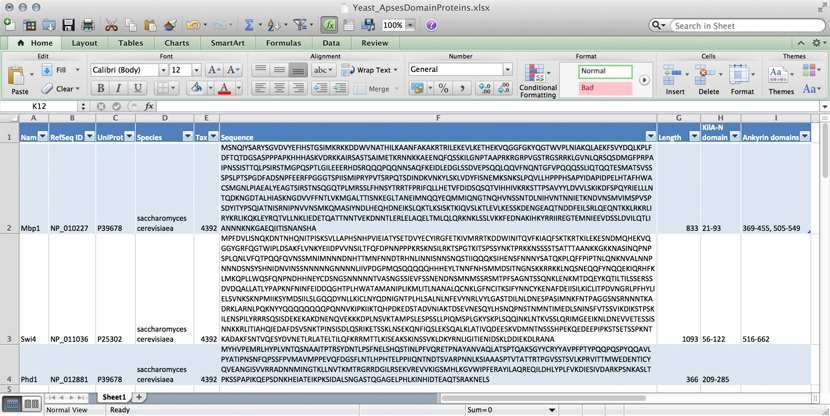
name: Mbp1 refseq ID: NP_010227 uniprot ID: P39678 species: Saccharomyces cerevisiae taxonomy ID: 4392 sequence: MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKR TRILEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF DFTQTDGSASPPPAPKHHHASKVDRKKAIRSASTSAIMETKRNNKKAEEN QFQSSKILGNPTAAPRKRGRPVGSTRGSRRKLGVNLQRSQSDMGFPRPAI PNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQPQQNNSAQFKEI DLEDGLSSDVEPSQQLQQVFNQNTGFVPQQQSSLIQTQQTESMATSVSSS PSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDINDKV NKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPYIDAPIDPELHTAFH WACSMGNLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRI FQLLHETVFDIDSQSQTVIHHIVKRKSTTPSAVYYLDVVLSKIKDFSPQY RIELLLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTAN EIMNQQYEQMMIQNGTNQHVNSSNTDLNIHVNTNNIETKNDVNSMVIMSP VSPSDYITYPSQIATNISRNIPNVVNSMKQMASIYNDLHEQHDNEIKSLQ KTLKSISKTKIQVSLKTLEVLKESSKDENGEAQTNDDFEILSRLQEQNTK KLRKRLIRYKRLIKQKLEYRQTVLLNKLIEDETQATTNNTVEKDNNTLER LELAQELTMLQLQRKNKLSSLVKKFEDNAKIHKYRRIIREGTEMNIEEVD SSLDVILQTLIANNNKNKGAEQIITISNANSHA length: 833 Kila-N domain: 21-93 Ankyrin domains: 369-455, 505-549 ...
... and save it all in one large text file and whenever you need to look something up, you just open the file, look for e.g. the name of the protein and read what's there. Or - for a more structured approach, you could put this into several files in a folder.[4] This is a perfectly valid approach and for some applications it might not be worth the effort to think more deeply about how to structure the data, and store it in a way that it is robust and scales easily to large datasets. Alas, small projects have a tendency to grow into large projects and if you work in this way, it's almost guaranteed that you will end up doing many things by hand that could easily be automated. Imagine asking questions like:
- How many proteins do I have?
- What's the sequence of the Kila-N domain?
- What percentage of my proteins have an Ankyrin domain?
- Or two ...?
Answering these questions "by hand" is possible, but tedious.
Spreadsheets
Many serious researchers keep their project data in spreadsheets. Often they use Excel, or an alternative like the free OpenOffice Calc, or Google Sheets, both of which are compatible with Excel and have some interesting advantages. Here, all your data is in one place, easy to edit. You can even do simple calculations - although you should never use Excel for statistics[5]. You could answer What percentage of my proteins have an Ankyrin domain? quite easily[6].
There are two major downsides to spreadsheets. For one, complex queries need programming. There is no way around this. You can program inside Excel with Visual Basic. But you might as well export your data so you can work on it with a "real" programming language. The other thing is that Excel does not scale very well. Once you have more than a hundred proteins in your spreadsheet, you can see how finding anything can become tedious.
However, just because it was built for business applications, and designed for use by office assistants, does not mean it is intrinsically unsuitable for our domain. It's important to be pragmatic, not dogmatic, when choosing tools: choose according to your real requirements. Sometimes "quick and dirty" is just fine, because quick.
R
R can keep complex data in data frames and lists. If we do data analysis with R, we have to load the data first. We can use any of the read.table() functions for structured data, read lines of raw text with readLines(), or slurp in entire files with scan(). But we could also keep the data in an R object in the first place that we can read from disk, analyze, modify, and write back. In this case, R becomes our database engine.
# Sample construction of an R database table as a dataframe
# Data for the Mbp1 protein
proteins <- data.frame(
name = "Mbp1",
refSeq = "NP_010227",
uniProt = "P39678",
species = "Saccharomyces cerevisiae",
taxId = "4392",
sequence = paste(
"MSNQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKR",
"TRILEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF",
"DFTQTDGSASPPPAPKHHHASKVDRKKAIRSASTSAIMETKRNNKKAEEN",
"QFQSSKILGNPTAAPRKRGRPVGSTRGSRRKLGVNLQRSQSDMGFPRPAI",
"PNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQPQQNNSAQFKEI",
"DLEDGLSSDVEPSQQLQQVFNQNTGFVPQQQSSLIQTQQTESMATSVSSS",
"PSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDINDKV",
"NKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPYIDAPIDPELHTAFH",
"WACSMGNLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRI",
"FQLLHETVFDIDSQSQTVIHHIVKRKSTTPSAVYYLDVVLSKIKDFSPQY",
"RIELLLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTAN",
"EIMNQQYEQMMIQNGTNQHVNSSNTDLNIHVNTNNIETKNDVNSMVIMSP",
"VSPSDYITYPSQIATNISRNIPNVVNSMKQMASIYNDLHEQHDNEIKSLQ",
"KTLKSISKTKIQVSLKTLEVLKESSKDENGEAQTNDDFEILSRLQEQNTK",
"KLRKRLIRYKRLIKQKLEYRQTVLLNKLIEDETQATTNNTVEKDNNTLER",
"LELAQELTMLQLQRKNKLSSLVKKFEDNAKIHKYRRIIREGTEMNIEEVD",
"SSLDVILQTLIANNNKNKGAEQIITISNANSHA",
sep=""),
seqLen = 833,
KilAN = "21-93",
Ankyrin = "369-455, 505-549",
stringsAsFactors = FALSE)
# add data for the Swi4 protein
proteins <- rbind(proteins,
data.frame(
name = "Swi4",
refSeq = "NP_011036",
uniProt = "P25302",
species = "Saccharomyces cerevisiae",
taxId = "4392",
sequence = paste(
"MPFDVLISNQKDNTNHQNITPISKSVLLAPHSNHPVIEIATYSETDVYEC",
"YIRGFETKIVMRRTKDDWINITQVFKIAQFSKTKRTKILEKESNDMQHEK",
"VQGGYGRFQGTWIPLDSAKFLVNKYEIIDPVVNSILTFQFDPNNPPPKRS",
"KNSILRKTSPGTKITSPSSYNKTPRKKNSSSSTSATTTAANKKGKKNASI",
"NQPNPSPLQNLVFQTPQQFQVNSSMNIMNNNDNHTTMNFNNDTRHNLINN",
"ISNNSNQSTIIQQQKSIHENSFNNNYSATQKPLQFFPIPTNLQNKNVALN",
"NPNNNDSNSYSHNIDNVINSSNNNNNGNNNNLIIVPDGPMQSQQQQQHHH",
"EYLTNNFNHSMMDSITNGNSKKRRKKLNQSNEQQFYNQQEKIQRHFKLMK",
"QPLLWQSFQNPNDHHNEYCDSNGSNNNNNTVASNGSSIEVFSSNENDNSM",
"NMSSRSMTPFSAGNTSSQNKLENKMTDQEYKQTILTILSSERSSDVDQAL",
"LATLYPAPKNFNINFEIDDQGHTPLHWATAMANIPLIKMLITLNANALQC",
"NKLGFNCITKSIFYNNCYKENAFDEIISILKICLITPDVNGRLPFHYLIE",
"LSVNKSKNPMIIKSYMDSIILSLGQQDYNLLKICLNYQDNIGNTPLHLSA",
"LNLNFEVYNRLVYLGASTDILNLDNESPASIMNKFNTPAGGSNSRNNNTK",
"ADRKLARNLPQKNYYQQQQQQQQPQNNVKIPKIIKTQHPDKEDSTADVNI",
"AKTDSEVNESQYLHSNQPNSTNMNTIMEDLSNINSFVTSSVIKDIKSTPS",
"KILENSPILYRRRSQSISDEKEKAKDNENQVEKKKDPLNSVKTAMPSLES",
"PSSLLPIQMSPLGKYSKPLSQQINKLNTKVSSLQRIMGEEIKNLDNEVVE",
"TESSISNNKKRLITIAHQIEDAFDSVSNKTPINSISDLQSRIKETSSKLN",
"SEKQNFIQSLEKSQALKLATIVQDEESKVDMNTNSSSHPEKQEDEEPIPK",
"STSETSSPKNTKADAKFSNTVQESYDVNETLRLATELTILQFKRRMTTLK",
"ISEAKSKINSSVKLDKYRNLIGITIENIDSKLDDIEKDLRANA",
sep=""),
seqLen = 1093,
KilAN = "56-122",
Ankyrin = "516-662",
stringsAsFactors = FALSE)
)
# how many proteins?
nrow(proteins)
#what are their names?
proteins[,"name"]
# how many do not have an Ankyrin domain?
sum(proteins[,"Ankyrin"] == "")
# save it to file
save(proteins, file="proteinData.Rda")
# delete it from memory
rm(proteins)
# check...
proteins # ... yes, it's gone
# read it back in:
load("proteinData.Rda")
# did this work?
sum(proteins[,"seqLen"]) # 1926 amino acids
# add another protein: Phd1
proteins <- rbind(proteins,
data.frame(
name = "Phd1",
refSeq = "NP_012881",
uniProt = "P39678",
species = "Saccharomyces cerevisiae",
taxId = "4392",
sequence = paste(
"MPFDVLISNQKDNTNHQNITPISKSVLLAPHSNHPVIEIATYSETDVYEC",
"MYHVPEMRLHYPLVNTQSNAAITPTRSYDNTLPSFNELSHQSTINLPFVQ",
"RETPNAYANVAQLATSPTQAKSGYYCRYYAVPFPTYPQQPQSPYQQAVLP",
"YATIPNSNFQPSSFPVMAVMPPEVQFDGSFLNTLHPHTELPPIIQNTNDT",
"SVARPNNLKSIAAASPTVTATTRTPGVSSTSVLKPRVITTMWEDENTICY",
"QVEANGISVVRRADNNMINGTKLLNVTKMTRGRRDGILRSEKVREVVKIG",
"SMHLKGVWIPFERAYILAQREQILDHLYPLFVKDIESIVDARKPSNKASL",
"TPKSSPAPIKQEPSDNKHEIATEIKPKSIDALSNGASTQGAGELPHLKIN",
"HIDTEAQTSRAKNELS",
sep=""),
seqLen = 366,
KilAN = "209-285",
Ankyrin = "", # No ankyrin domains annotated here
stringsAsFactors = FALSE)
)
# check:
proteins[,"name"] #"Mbp1" "Swi4" "Phd1"
sum(proteins[,"Ankyrin"] == "") # Now there is one...
sum(proteins[,"seqLen"]) # 2292 amino acids
# [END]
The third way to use R for data is to connect it to a "real" database:
- a relational database like mySQL, MariaDB, or PostgreSQL;
- an object/document database like {{WP|MongoDB};
- or even a graph-database like Neo4j.
R "drivers" are available for all of these. However all of these require installing extra software on your computer: the actual database, which runs as an independent application. If you need a rock-solid database with guaranteed integrity, industry standard performance, and scalability to even very large datasets and hordes of concurrent users, don't think of rolling your own solution. One of the above is the way to go.
MySQL and friends
MySQL is a free, open relational database that powers some of the largest corporations as well as some of the smallest laboratories. It is based on a client-server model. The database engine runs as a daemon in the background and waits for connection attempts. When a connection is established, the server process establishes a communication session with the client. The client sends requests, and the server responds. One can do this interactively, by running the client program /usr/local/mysql/bin/mysql (on Unix systems). Or, when you are using a program such as R, Python, Perl, etc. you use the appropriate method calls or functions—the driver—to establish the connection.
These types of databases use their own language to describe actions: SQL - which handles data definition, data manipulation, and data control.
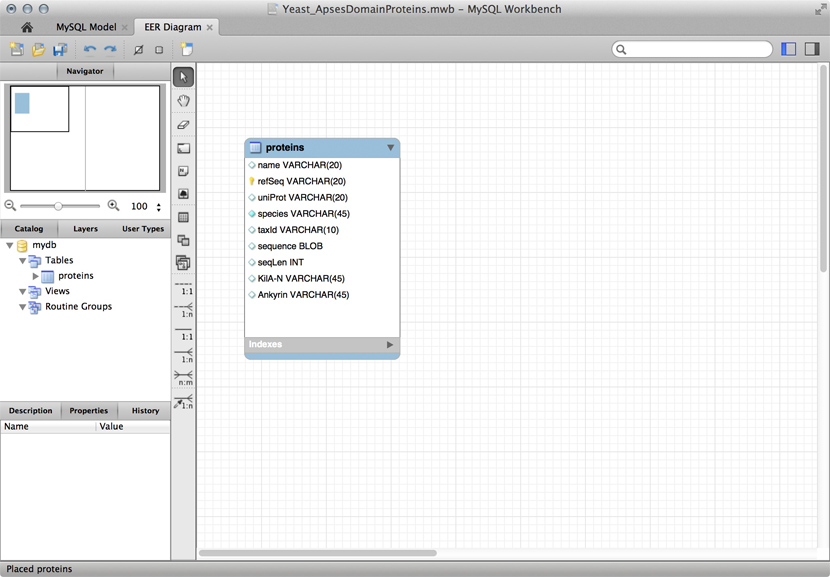
Just for illustration, the Figure above shows a table for our APSES domain protein data, built as a table in the MySQL workbench application and presented as an Entity Relationship Diagram (ERD). There is only one entity though - the protein "table". The application can generate the actual code that implements this model on a SQL compliant database:
CREATE TABLE IF NOT EXISTS `mydb`.`proteins` (
`name` VARCHAR(20) NULL,
`refSeq` VARCHAR(20) NOT NULL,
`uniProt` VARCHAR(20) NULL,
`species` VARCHAR(45) NOT NULL COMMENT ' ',
`taxId` VARCHAR(10) NULL,
`sequence` BLOB NULL,
`seqLen` INT NULL,
`KilA-N` VARCHAR(45) NULL,
`Ankyrin` VARCHAR(45) NULL,
PRIMARY KEY (`refSeq`))
ENGINE = InnoDB
This looks at least as complicated as putting the model into R in the first place. Why then would we do this, if we need to load it into R for analysis anyway. There are several important reasons.
- Scalability: these systems are built to work with very large datasets and optimized for performance. In theory R has very good performance with large data objects, but not so when the data becomes larger than what the computer can keep in memory all at once.
- Concurrency: when several users need to access the data potentially at the same time, you must use a "real" database system. Handling problems of concurrent access is what they are made for.
- ACID compliance. ACID describes four aspects that make a database robust, these are crucial for situations in which you have only partial control over your system or its input, and they would be quite laborious to implement for your hand built R data model:
- Atomicity: Atomicity requires that each transaction is handled "indivisibly": it either succeeds fully, with all requested elements, or not at all.
- Consistency: Consistency requires that any transaction will bring the database from one valid state to another. In particular any data-validation rules have to be enforced.
- Isolation: Isolation ensures that any concurrent execution of transactions results in the exact same database state as if transactions would have been executed serially, one after the other.
- Durability: The Durability requirement ensures that a committed transaction remains permanently committed, even in the event that the database crashes or later errors occur. You can think of this like an "autosave" function on every operation.
All the database systems I have mentioned above are ACID compliant[7].
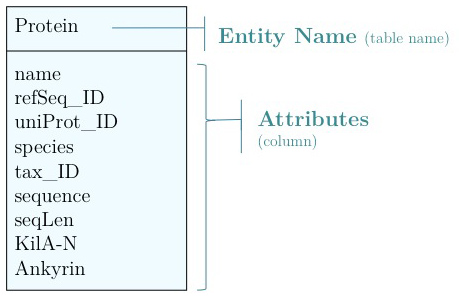
Data modelling
|
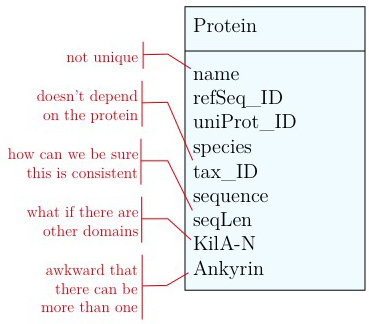
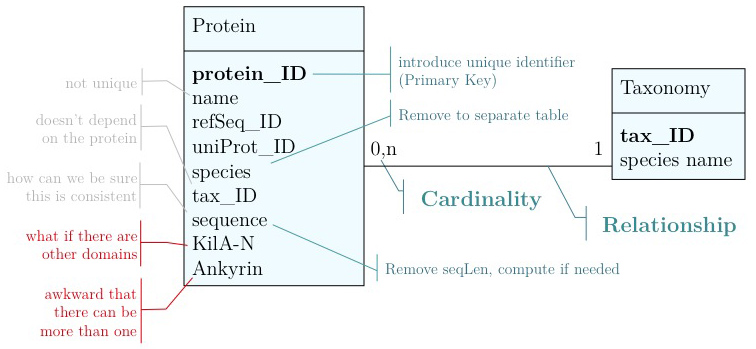
As you have seen above, the actual specification of a data model in R or as a sequence of SQL statements is quite technical and not well suited to obtain an overview for the model's main features that we would need for its design. We'll thus introduce a modelling convention: the Entity-Relationship Diagram (ERD). These are semi-formal diagrams that show the key features of the model. Currently we have only a single table defined, with a number of attributes. If we think a bit about our model and its intended use, it should become clear that there are a number of problems. They have to do with efficiency, and internal consistency. Problems include:
|
- Unique identifier
- Every entity in our data model should have its own, unique identifier. Typically this will simply be an integer that we should automatically increment with every new entry. Automatically. We have to be sure we don't make a mistake.
- Move species/tax_id to separate table
- If the relationship between two attributes does not actually depend on our protein, we move them to their own table. One identifier remains in our protein table. We call this a "foreign key". The relationship between the two tables is drawn as a line, and the cardinalities of the relationship are identified. "Cardinalities" means: how many entities of one table can be associated with one entity of the other table. Here,
0, non the left side means: a giventax_IDdoes not have to actually occur in the protein table i.e. we can put species in the table for which we actually have no proteins. There could also be many ("n") proteins for one species in our database. On the right hand side1means: there is exactly one species annotated for each protein. No more, no less.
- Remove redundant data
This is almost always a good idea. It's usually better just to compute seqLen or similar from the data. The exception is if something is expensive to compute and/or used often. Then we may store the reult in our datamodel, while making our procedures watertight so we store the correct values.
These are relatively easy repairs. Treating the domain annotations correctly requires a bit more surgery.
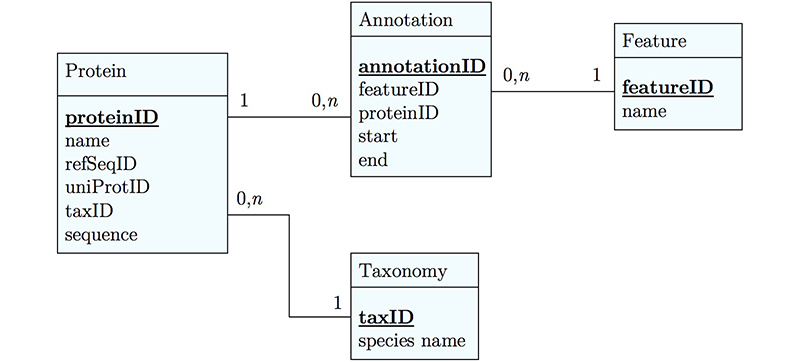
It's already awkward to work with a string like "21-93" when we need integer start and end values. We can parse them out, but it would be much more convenient if we can store them directly. But something like "369-455, 505-549" is really terrible. First of all it becomes an effort to tell how many domains there are in the first place, and secondly, the parsing code becomes quite involved. And that creates opportunities for errors in our logic and bugs in our code. And finally, what about if we have more features that we want to annotate? Should we have attributes like Kil-A N start, Kil-A N end, Ankyrin 01 start, Ankyrin 01 end, Ankyrin 02 start, Ankyrin 02 end, Ankyrin 03 start, Ankyrin 30 end... No, that would be absurdly complicated and error prone. There is a much better approach that solves all three problems at the same time. Just like with our species, we create a table that describes features. We can put any number of features there, even slightly different representations of canonical features from different data sources. Then we create a table that stores every feature occurrence in every protein. We call this a junction table and this is an extremely common pattern in data models. Each entry in this table links exactly one protein with exactly one feature. Each protein can have 0, n features. And each feature can be found in 0, n proteins.
With this simple schematic, we obtain an excellent overview about the logical structure of our data and how to represent it in code. Such models are essential for the design and documentation of any software project.
Time to put this into practice: design your own data model.
Task:
- Use your imagination about what kind of data you would like to store to study the "function" of a protein, specifically the transcription factors in YFO that are related to Mbp1.
- Write down what you would like to store.
- Sketch a relational data model for your data. Put it on paper, or print it out. Bring it to class for Tuesday's quiz.
- That is all.
Links and resources
Footnotes and references
- ↑ If you find this URL hard to remember, consider the acronyms:
- ncbi.nlm.nih.gov
- NCBI: National Center for Biotechnology Information
- NLM: National Library of Medicine
- NIH: National Institutes of Health
- GOV: the US GOVernment top-level domain
- ↑ If there would have been more than one match, you would have gotten a list of results, as before.
- ↑ Actually the "real" SwissProt identifier would be patterned like
MBP1_YEAST.P39678is the corresponding UniProt identifier. - ↑ Your operating system can help you keep the files organized. The "file system" is a database.
- ↑ For real: Excel is miserable and often wrong on statistics, and it makes horrible, ugly plots. See here and here why Excel problems are not merely cosmetic.
- ↑ At the bottom of the window there is a menu that says "sum = ..." by default. This provides simple calculations on the selected range. Set the choice to "count", select all Ankyrin domain entries, and the count shows you how many cells actually have a value.
- ↑ For a list of relational Database Management Systems, see here.
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.