BIO Assignment 5 2011
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Assignment 4 - Phylogenetic Analysis
Please note: This assignment is currently inactive. Unannounced changes may be made at any time.
Introduction
- Nothing in Biology makes sense except in the light of evolution.
- Theodosius Dobzhansky
... but does evolution make sense in the light of biology?
As we have seen in the previous assignments, the Mbp1 transcription factor has homologues in all other fungi, yet - looking at orthologues - this is not always a clear one-to-one mapping of related genes to each other. It appears that various systems of APSES domain transcription factors have evolved independently. Of course this bears directly on our notion of function - what it means to say that two genes in different organisms have the "same" function. In case two organisms both have an orthologous gene for the same, distinct function, this may be warranted. But what if that gene has duplicated in one of them, and the two paralogues now perform different, related functions in one organism? In order to be able to even ask such questions, we need to understand how we can make the evolutionary history of gene families explicit. This is the domain of phylogenetic analysis. We can ask questions like: how many paralogues did the cenancestor of a clade possess? Which of these underwent additional duplications in the phylogenesis of the organism I am studying? Did any genes get lost? And - adding additional biological insight to the picture - did the observed duplications lead to the "invention" of new biological systems? When was that? And how did the species benefit from this event?
We will develop some of this kind of analysis in this assignment. In the previous assignment you have established which genes are the reciprocally most closely related orthologues to Mbp1 in yeast. In this assignment, we will analyse their evolutionary relationship and compare it to the evolutionary relationship of all fungal APSES domains. The goal is to define families of related transcription factors and their evolutionary history.
A number of good tools for phylogenetic analysis exist; general purpose packages include the (free) PHYLIP package and the (commercial) PAUP package. Specialized tools for tree-building include Treepuzzle or Mr. Bayes. This assignment is conctructed around programs that are availble in PHYLIP, however you are welcome to use other tools that fulfil a similar purpose if you wish. In this field, researchers consider trees that have been built with ML (maximum likelihood) methods to be more reliable than trees that are built with parsimony methods, or distance methods such as NJ (Neighbor Joining). However ML methods are also much more compute-intensive. Just like with multiple sequence alignments, some algorithms will come closer to guessing the truth and others will not and usually it is hard to tell, which is the more trustworthy of two diverging results. The prudent researcher tries out alternatives and forms her own opinion. Specifically, we may usually assume results that converge, independent of the algorithm, to be more reliable than those that depend strongly on a particular algorithm or details of input data.
But regarding algorithm and rersources: we will take two shortcuts in this assignment (and both shortcuts are things you should not do in real life):
One: we will use an efficient tree-building algorithm, not the best-available one. This is an algorithm which is available on the Web, without the need for you to install software on your own machine. In real life you would of course use the most accurate algortihm you can lay your hands on, regardless of the resources this requires, since it makes no sense to waste your time on a careful analysis of inaccurate trees. Your supervisor would want it so as well. And if not she, the reviewers of your manuscript.
Two: we will assume the tree the algorithm constructs is correct. In real life you would establish its reliability with a bootstrap procedure: repeat the tree-building a hundred times with partial data and see which branches and groupings are robust and which depend on the details of the data. But we should still acknowledge that bifurcations that are very close to each other have not been" resolved". Any conscientious reviewer would flag such leniency and send your results back to you for a bootstrapping exercise at the computer. In phylogenetic analysis, not all lines that the program draws are equally trustworthy. Dont take the trees as a given fact just because a program suggests this. Look at the evidence, use your reasoning, and analyse them critically.
In case you want to review concept of trees, clades, LCAs OTUs and the like, I have linked an excellent and very understandable introduction-level article on phylogenetic analysis to the resource section at the bottom of this page.
Preparation, submission and due date
Read carefully. Be sure you have understood all parts of the assignment and cover all questions in your answers! Sadly, we always get assignments back in which important aspects have simply overlooked marks unnecessarily. If you did not notice that the above did not make sense, you are reading what you expect, not what is written.
Prepare a Microsoft Word document with a title page that contains:
- your full name
- your Student ID
- your e-mail address
- the organism name you have been assigned
Follow the steps outlined below. You are encouraged to write your answers in short answer form or point form, like you would document an analysis in a laboratory notebook. However, you must
- document what you have done,
- note what Web sites and tools you have used,
- paste important data sequences, alignments, information etc.
If you do not document the process of your work, we will deduct marks. Try to be concise, not wordy! Use your judgement: are you giving us enough information so we could exactly reproduce what you have done? If not, we will deduct marks. Avoid RTF and unnecessary formating. Do not paste screendumps. Keep the size of your submission below 1.5 MB.
Write your answers into separate paragraphs and give each its title. Save your document with a filename of:
A3_family name.given name.doc
(for example my fourth assignment would be named: A4_steipe.boris.doc - and don't switch the order of your given name and familyname please!)
Finally e-mail the document to [boris.steipe@utoronto.ca] before the due date.
Your document must not contain macros. Please turn off and/or remove all macros from your Word document; we will disable macros, since they pose a security risk.
With the number of students in the course, we have to economize on processing the assignments. Thus we will not accept assignments that are not prepared as described above. If you have technical difficulties, contact me.
The due date for the assignment is Thursday, December 7. at 24:00 (last day of class). In case you need more time since the assignment was posted late, an extension is automatically granted to Tuesday, December 19. at 10:00 in the morning.
Grading
Don't wait until the last day to find out there are problems! Assignments that are received past the due date will have one mark deducted and an additional mark for every full twelve hour period past the due date. Assignments received more than 5 days past the due date will not be assessed. If you need an extension, you must arrange this beforehand.
Marks are noted below in the section headings for of the tasks. A total of 10 marks will be awarded, if your assignment answers all of the questions. A total of 2 bonus marks (up to a maximum of 10 overall) can be awarded for particularily interesting findings, or insightful comments. A total of 2 marks can be subtracted for lack of form or for glaring errors. The marks you receive will
- count directly towards your final marks at the end of term, for BCH441 (undergraduates), or
- be divided by two for BCH1441 (graduates).
(1) Preparations
(1.1) Preparing Input Files (2 marks)
Introduction: Task
For this assignment, we start from the multiple sequence alignments we have constructed in the last assignment. We will edit alignments to make them suitable for phylogenetic analysis. We will construct a tree and we will discuss tree analysis in the end.
Introduction: Principle
In order to use molecular sequences for the construction of phylogenetic trees, you have to build a multiple alignment first, then edit it. This is important: all sequences have to be edited to contain the exact same number of characters and to hold aligned characters in corresponding positions. Phylogeny programs are not meant to revise an alignment but to analyse evolutionary relationships, given the alignment.
The result of the tree estimation is a decision about likely relationships. Fundamentally all the programs do is to decide which sequences had common ancestors. Distance based phylogeny programs have a way to convert sequence comparisons into evolutionary distances (applying a model of evolution such as a mutation data matrix, calculating one number for each pair of sequences and using that to estimate a tree by grouping together the most highly related sequences (e.g. with an NJ, Neigbor Joining, algorithm). Alternatively one can find trees that are most compatible with the observed sequences and the specific model of evolutionary change through point-mutations (either or by minimizing the number of mutation events over the tree (Parsimony) or by finding the tree for which the observed sequences would be the most likely (ML, Maximum Likelihood)). Clearly, in order for any of this to work, one must not include fragments of sequence which have evolved under a totally different evolutionary model, such as domain fusion, or insertion/deletion of residues. The goal is not to be as comprehensive and complete as possible but to input the columns of aligned residues that will best represent the phylogenetic relationships between the sequences.
Introduction: Problems
Gaps are a real problem here, as usual. Strictly speaking, the similarity score of an alignment program as well as the distance score of a phylogeny program are not calculated for an ordered sequence, but for a sum of independent values, one for each aligned columns of characters. The order of the columns does not change the score. Hoever for the alignment, this is no longer true, once gaps are introduced: we treat an insertion gap-character differently from an elongation character. Most alignment programs use a model with a constant gap insertion penalty and a linear gap extension penalty. This is not rigourously justified from biology, but parametrized (or you could say "tweaked") to correspond to our observations. However, most phylogeny programs, (such as the programs in PHYLIP) do not work in this way. PHYLIP strictly operates on columns of characters and treats a gap character like a residue with the one letter code "-". This underestimates the distance between gapped sequences, since any evolutionary model should reflect the fact that gaps are much less likely than point mutations. If the gap is very long though, all events are counted individually as many single substitutions (rather than one lengthy one) and this overestimates the distance. And it gets worse: long stretches of gaps can make sequences appear similar in a way that is not justified, just because they are identical in the "-" character. When there are unambiguous gaps, one might be tempted to fudge the alignment by inserting matching characters into sequences that are ungapped (e.g. five "A"s each into the ungapped sequences and five "-" each into the gapped sequences), however, I would caution against this approach since it possibly introduces even more non-obvious implicit assumptions and potential for error. It is therefore common to remove most, if not all gaps.
Introduction: Practice
In practice, follow the fundamental principle that all characters in a column should be related by homology. This implies the following rules of thumb:
- Remove all stretches of residues in which the alignment appears ambiguous.
- Remove all frayed N- and C- termini, especially regions in which not all sequences that are being compared appear homologous.
- Remove all but approximately one column from gapped regions, and all residues N- and C- terminal of the gap in which the alignment appears questionable. ( I would keep one gapped column as a placeholder for a rare and very distinct evolutionary event, rather than simply deleting them all, some researchers remove all gaps).
- Also, consider that neither residues that are completely different between all species, nor residues that are completely conserved are informative for relationship distances.
- If your sequences are too long, you may run out of memory. 60-80 aligned residues should be plenty and if the sequences fit on a single line you will save yourself potential trouble with block-wise vs. interleaved input.
- (A very useful trick with Microsoft Word is that you can select blocks of text and entire columns in the document with your mouse: hold the "ALT" key depressed while you click and drag your mouse to select. This will greatly facilitate the preparation of sequences. You can treat that selection as any other selected text, color characters, or delete them. Importantly, you can also cut and paste entire columns! Of course, this will only work as expected if you use a fixed-width font such as Courier. )
The preparation of the input file of aligned residues, used by the PHYLIP package is straightforward in principle; just carefully follow the instructions in PHYLIP's well written documentation. If you plan to use an outgroup for your tree, it is a good idea to move that to the first line of your alignment, since this is where PHYLIP will look for it by default.
Some notes on how to avoid common editing troubles. Copy the sequences frrom the link provided below. Paste them into a document, using the Word "Edit -> Paste special -> Unformatted text". Set the page-setup to "landscape", the font-size to something small, then you can put every sequence into one line. You can replace all paragraph marks ("^p") with (nothing) to remove them, then replace the FASTA header line character ">" with paragraphs ("^p") to separate them by line again. Take special note that your files must not include tab characters. You can use Word to globally replace all tabs (specified as "^t") with a blank, to make sure. Spaces count, so display your alignment in a fixed-width font, such as Courier ("Courier New" on Windows), not a proportional-width font such as Times, Arial, or Helvetica, and ensure all characters in your alignments align as they should. As always, make sure you save your input files as "Text Only".
- A note if you are working on a Mac and saving input on disk, to run with a locally installe PHYLIP version: here MS Word will play one of its usual shenanigans on you since it writes text files with the old-style OS 9 Carriage Return characters
(\r; ASCII 13; hex 0D; CR). Just by looking at the file, this is quite invisible but such "Carriage returns" are not going to be recognized by PHYLIP and most other self-respecting UNIX based programs. It may not make a difference when you paste your sequences to a Web server; but if you compute things locally it will appear to the program as though everything were in one line). And this can (and did) lead to head-banging rounds of frustration. You need to replace them with Linefeed resp. Newline characters(\n; ASCII 10; hex 0A; LF)and you can't even do that within Word(!). Open a UNIX terminal window and navigate to the directory where your files reside. Then type:
- tr "\r" "\n" < infile > outfile
- ... where outfile is different from infile (careful: if a file by the name of outfile already exists, tr will cheerfully overwrite it.) Alternatively you could type the following perl one-liner :
- perl -e 'while(<>){tr/\r/\n/;print}' < infile > outfile
In your assignment submission, clearly identify the source sequences you are using, as well as the alignment method you have used. Paste your unaltered source alignment into your document, clearly highlight or otherwise color the columns that you have selected, annotate why you have selected them and paste your result as well. Here is an example of what this might look like:
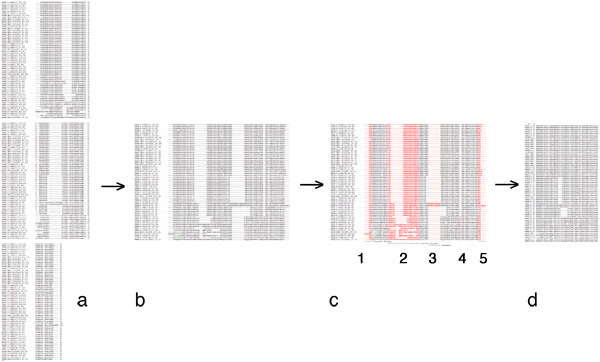
Introduction: Web Service and data
You have two choices for completing the assignment: either to peruse one of the on-line Webservices that generously provide a compute-intensive task such as PHYLIP, or to download and install the program at home. If you choose the former, one of your options is the PHYLIP service at the Institut Pasteur. I have tried it, and it works - however not entirely without problems. Uninformative errors will occur when your input is too large for the system's memory (like: "sequences not aligned" ... "out of memories" and such) but what is worse, after submitting a number of jobs, the system locked me out, asking me to what an unspecified time until results would be sent by e-mail (three days later, that hasn't happened). Regrettably, this is not documented. If you can live with that, the integration of their services in a logical sequence of steps is good and some of their services are a bit more advanced than plain out of the box PHYLIP. If you rateher decide to install PHYLIP, good for you. That is easy to do, well documented, there are much less limitations on memory - but if you don't read and understand the instructions carefully, you may be in for a spell of frustration.
Either way, I have posted typical input files and result files here, to allow you to bail out in case technical problems become overwhelming. If you use the data posted here instead of your own, you must document that fact and explain what you have tried, and why that has failed. If you fail to do that, we will deduct marks - the posted data is a fallback, not a shortcut.
In this assignment, we will use a simple distance based tree construction method. This represent a reasonable compromise between accurracy and speed, especially when applied to moderately dissimilar sequences. In genereal, distance methods include two steps: (1) calculate a pairwise-distance matrix between sequences, (2) construct a tree, based on the matrix. Thus all the information in the alignment bewtween two pairs of sequences is collapsed into a single number: their pairwise distance. Alternative approaches, parsimony as well as ML based algorithms take individual columns into account. Parsimony based methods construct inaccurate trees when they can't make good estimates for the required number of sequence changes, if the sequences become too dissimilar. ML based methods are considered the most accurate for dissimilar sequences, however they are also very compute intensive and the full-length APSES domain alignment for 74 species run about half a day on my workstation. Thus we will use distance based methods here, specifically the UPGMA variant of the neighbor joining algorithm.
Prepare an input file that is representative of the whole protein, and one that represents only the APSES domains.
- Access one of the MSAs for Mbp1 proteins, linked from the resources section at the bottom of the page. Choose an MSA that you have determined in your third assignment to be "reliable" and (briefly) justify your choice. Prepare a PHYLIP formatted input file from this MSA, restricting the number of characters to no more than 60. Follow the considerations dicussed above. In particular you should choose some residues from each of the three aligned regions (The APSES domains, the Ankyrin domains and the C-terminal aligned region), to represent the diversity between these proteins. Document this as described above. (See the fallback data in case you get stuck) (1 mark)
- Prepare a second PHYLIP formatted input file from this MSA, that contains only the APSES domains. (See the fallback data in case you get stuck) (1 mark)
(1.2) Calculating Trees (3 marks)
- Using the protdist program of PHYLIP, calculate a distance matrix for both files. (See the fallback data in case you get stuck) (1 mark)
- If you use the PHYLIP Webserver, do the following: use a neighbor joining algorithm (bionj on the PHYLIP server), construct a tree for both input files (on the server: run ... "on outfile" ) When the program is done, select the option drawgram and click Run the selected program on treefile. Choose a cladogram tree-style and a suitable output format (e.g. postscript). Paste the trees into your assignment.
- If you use a locally installed version of PHYLIP use neighbor with the UPGMA method to construct a tree for both input files. Open the file outfile in a text-editor, copy and paste the trees into your assignment.
- (1 mark) for either of the above
- Briefly discuss whether the trees are fundamentally similar or whether there are important differences (i.e. differences in topology). If there are differences in topology, which branch(es) would have to be moved to make the trees congruent? (1 mark)
(2) Analysis
It is surprisingly difficult to find a comprehensive phylogenetic analysis of the fungal species for which the genomes have been sequenced, although one would assume this to be data of considerable utility for the community. I have constructed a cladogram for the species we are analysing, based on data published for 1551 fungal ribosomal sequences. Such rRNA trees are a standard method of phylogenetic analysis, supported by the assumption that rRNA sequences are monophyletic and have evolved under comparable selective pressure in all species.
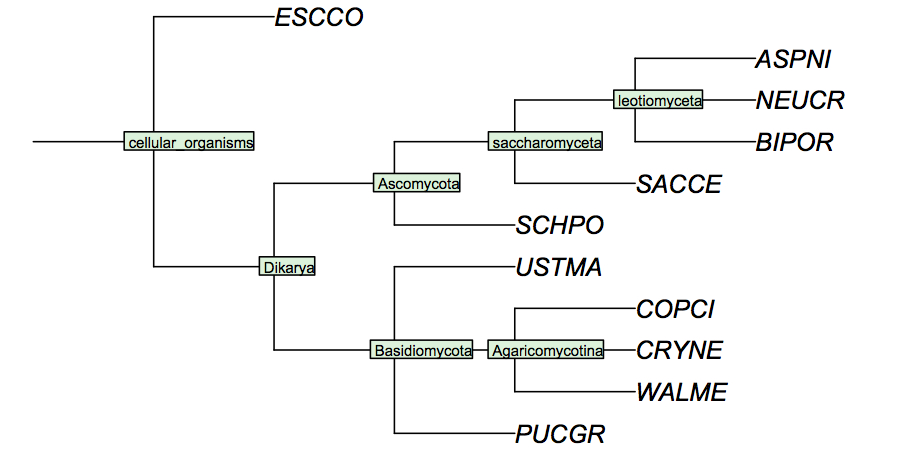
(2.1) Correspondence of Gene trees and Phylogenetic Tree (1 mark)
Both your Mbp1 trees should of course correspond to the reference cladogram of fungi, after all, we have chosen the orthologues of Mbp1 for this task. However, in practice this is usually not exactly the case. The treest are likely going to be different in parts. Note that phylogenetic trees that differ only in rotations around branchpoints are considerd identical ! Differences in phylogenetic trees only concern the topology, i.e. the branching order.
- Briefly compare one of the phylogenetic trees with the reference cladogram. Are there species that are not in the expected position? Does this mean the cladogram is not correct? (1 mark)
(2.2) Evolutionary History of the APSES Domain (2 marks)
In order to study the evolutionary history of the entire gene family, we have to prepare a tree of all 74 APSES genes. I have done this with the program promlk of the PHYLIP package. You can access the APSES domains reference tree here.
This is a complicated tree, and it can look impenetrably confusing at first. Here are two principles that will help you make sense of the tree.
A: A gene that is present in an ancestral species, is inherited in all descendent species. The gene has to be observed in all OTUs, unless its has been lost (which is a rare event). This means, if a gene is present in two widely divergent species, but in none other of the descendants of the LCA, it is possible that there is some problem with the tree (long branch attraction maybe), or the sequence has been acquired through horizontal gene transfer.
B: Paralogous genes in an ancestral species should give rise to monophyletic subtrees for each of the genes, in all descendants; this means: if the LCA of a branch has e.g. three genes, we would expect three copies of the species cladogram below this branchpoint, one for each of these genes. Each of these subtrees should recapitulate the reference phylogenetic tree of the OTUs, up to the branchpoint of their LCA.
With these two simple principles (you should draw them out on a piece of paper if they do not seem obvious to you), you can probably pry the reference tree of all APSES domains apart quite nicely. A few colored pencils and a printout of the tree will help.
- Identify at least two (more or less) monophyletic subtrees of the tree that recapitulate (more or less) the species tree. Write down their branch-point numbers. In one of these subtrees, define the correspondence between the letters in the species tree and the branchpoints in the gene tree as far as possible. (The correspondence is not going to be exact. ) (1 mark)
- Discuss briefly if there are features of the gene tree that are systematically inconsistent with the reference cladogram: choose one species from the gene tree that violates the expectation from the species tree and check what it's close neighbors are for the other parts of the tree. Do these sequences always appear more closely or more distantly related than they should be? (1 mark)
(2.3) Unraveling the Mbp1 clade (2 marks)
If we consider the Mbp1 clade (the clade containing all 16 of our Mbp1 orthologues), descending from branchpoint 115, we note that it contains 16 additional genes, one from each of the species we have studied, however both subtrees are polyphyletic within the clade. (In what follows I will refer to subgroups of the tree as Clades and label them with their branch number in this reference tree.) It is thus tempting to speculate that this subtree, Clade 115 is actually a mix-up of two ancestral genes, one of them an orthologue to Mbp1, the other to a different gene - perhaps Swi4. If this were true, there would be three possible explanations for the polyphyletic tree: (i) either we have mislabeled the orthologues, or (ii) we have constructed an incorrect tree, or (iii) our model of how these genes have evolved is wrong. Actually, it is rather obvious when we compare the clades that branch off at 117 and 128, why this is not a simple case of mislabeling the genes ...
- Explain briefly why it is unlikely that the clades at branchpoints 117 and 128 are each descendants of a single cenancestral gene. (1 mark)
Could this point to a problem with the columns we have selected for the tree building? Have we biased our input data and then constructed an incorrect tree? To test this, a revised tree was constructed, taking the genes from Clade 115, realigning them and constructing a new tree. You can have a look at the revised Mbp1 APSES domain tree, and the procedure - or you can just take my word for it, the trees are substantially similar. Apparently the precise choice of columns has a minor effect on the results, as it should have. We should note in general however that branch points that are close together in time may be poorly resolvable, while branch points that are separated by longer evolutionary distances are more reliable. In summary, save for some inaccuracies, we have no reason to believe the tree is fundamentally incorrect.
So what about or model of evolution? What do we actually expect?
As we had written above, we expect to see anumber of more or less faithful copies of the species tree, for each of gene in the cenancestor (or LCA for later duplications).
And as you see, this is not exactly what we find in Clade 115. What we do find however is that Clade 115 reflects our expectations in part. Clade 123 pretty well recapitulates the Saccharomycotina, as does Clade 118, and when we consider Clade 128, apart from some "noise" we see both the Dikaryomycota (in Clade 142) reasonably grouped together, as well as the Euascomycotina (in Clade 129") - in fact the most striking departure from our expected tree for Mbp1 is the placement of Clade 123, which we would expect attached below branch 128, not above it where we have found it.
This may be due to insufficient resolution of the trees. Whenever we have long, evolutionary distances with a few OTUs clustered at the end, we can be reasonbly certain of the branch. When branching events occur at similar times, the order is uncertain and any specific ordering would probably not be well supported in bootstrapping.
What is striking however is the parallelism between Clade 123 and Clade 118. Apparently branchpoint 117 marks a duplication event of the ancestral Mbp1 gene in the Saccharomycotina. Here our expectation appears to be borne out. And given the uncertainty in the precise ordering between branchpoints 117 and 128, the Clade overall is thus actually quite consistent with the species tree.
- Finally, find one example of a gene duplication, below branchpoint 128.
(3) Summary of Resources
- Links
- Sequences
- Alignments
- Mbp1 proteins:
- APSES domains:
[End of assignment]
If you have any questions at all, don't hesitate to mail me at boris.steipe@utoronto.ca or post your question to the Course Mailing List