Difference between revisions of "UCSF Chimera"
m |
|||
| Line 8: | Line 8: | ||
| − | [[Image:Mbp1_Model_complex.jpg|frame|none|'''A complex between a | + | [[Image:Mbp1_Model_complex.jpg|frame|none|'''A complex between a transcription factor and its cognate DNA binding site'''<br> |
| − | The | + | The coordinates of [http://pdb.org/pdb/explore/explore.do?structureId=1PUE '''1PUE'''] were downloaded from the PDB; separate models for protein and DNA were defined in Chimera and a solvent-accessible surface was calculated for the protein component of the complex. Rainbow coloring by sequence number was applied to the residues. The protein surface is transparent and the protein is displayed in "pipe-and-loops" mode. The scene was exported from Chimera to the POVray raytracer and rendered with standard parameters. ]] |
Revision as of 18:11, 10 September 2014
UCSF Chimera
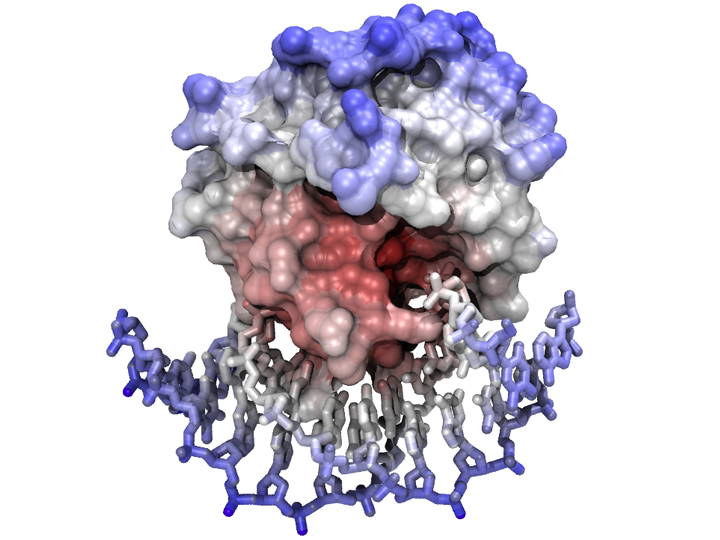
The coordinates of 1PUE were downloaded from the PDB; separate models for protein and DNA were defined in Chimera and a solvent-accessible surface was calculated for the protein component of the complex. Rainbow coloring by sequence number was applied to the residues. The protein surface is transparent and the protein is displayed in "pipe-and-loops" mode. The scene was exported from Chimera to the POVray raytracer and rendered with standard parameters.
UCSF Chimera is a powerful, well engineered, molecular graphics package with a large number of functions that is under ongoing development and maintenance. It is one of a number of freely available tools to visualize and manipulate three-dimensional molecular datasets, but it stands out due to its rich features, the versatility of its use and the ease with which common tasks can be accomplished and the fact that it is being actively developed and maintained. You should learn to work with it in this course, but you should also be aware that there are alternatives, each with different strengths and weaknesses. Some of the more widely used free systems are:
A comprehensive list of molecular graphics systems can be found here.
Installing Chimera
Task:
- Access the Chimera homepage and navigate to the Download section.
- Find the the newest version for your platform in the table and click on the file to download it.
- Follow the instructions to install Chimera.
Chimera tutorials
The Chimera User Guide site has a set of associated tutorials and there are more tutorials linked from here.