Difference between revisions of "Homology modeling fallback data"
Jump to navigation
Jump to search
m |
|||
| Line 34: | Line 34: | ||
Explanation of the output is found [http://swissmodel.expasy.org/workspace/index.php?func=special_help '''in the SwissModel Help page''']. | Explanation of the output is found [http://swissmodel.expasy.org/workspace/index.php?func=special_help '''in the SwissModel Help page''']. | ||
| − | [[Image: | + | [[Image:ModelledRange.png|frame|none|Graphical comparison of template and target sequence to emphasize which regions have been modelled.]] |
| − | [[Image: | + | |
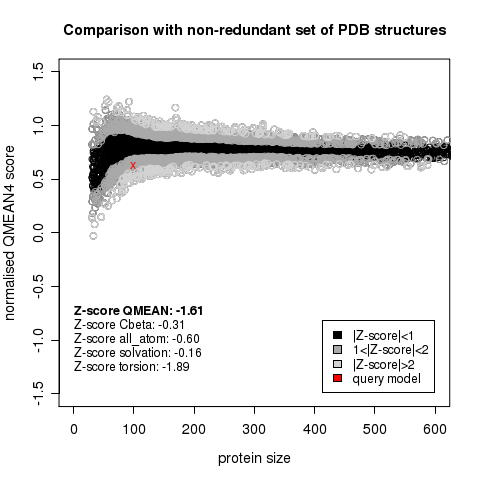
| + | [[Image:QMeanScore.png|frame|none|Model QMean score vs. expected QMean scores of protein structures, as a function of domain size.]] | ||
| + | |||
| + | |||
| + | |||
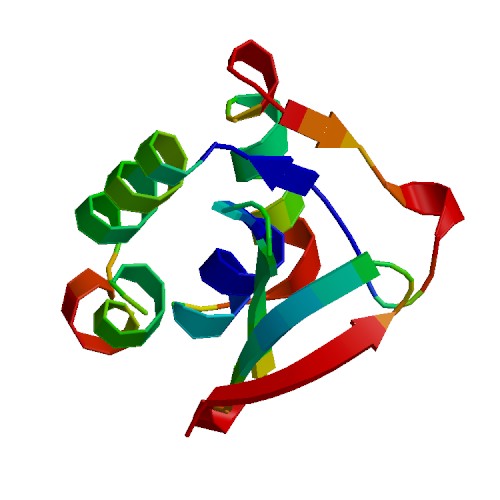
| + | [[Image:ColoringByScore.jpg|frame|none|Cartoon view of the model, colored by ANOLEA scores]] | ||
| + | |||
The target-template alignment: | The target-template alignment: | ||
| − | TARGET 1 | + | TARGET 1 IK -GVSVMRRRR DSWLNATQIL KVADFDKPQR |
| − | + | 1mb1A 3 nqiysarysg vdvyef--ih stgsimkrkk ddwvnathil kaanfakakr | |
| − | + | ||
TARGET ssshhhh hh hhhh | TARGET ssshhhh hh hhhh | ||
| − | + | 1mb1A sssssss ssssss ss sssssss ssshhhh hh hhhh | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | TARGET 32 TRVLERQVQI GAHEKVQGGY GKYQGTWVPF QRGVDLATK ---------- | ||
| + | 1mb1A 51 trilekevlk ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf | ||
| + | |||
| + | TARGET hhhhhhhhh sss ssss h hhhhhhhhh | ||
| + | 1mb1A hhhhh sss ssss h hhhhhhhhh hh hh | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
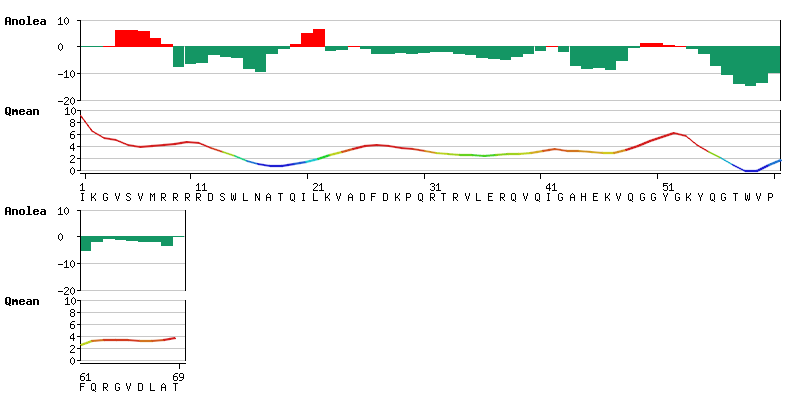
| − | [[Image: | + | [[Image:ModelQualityEstimation.png|frame|none|Local quality estimation: '''ANOLEA''' and QMean''' scores as graphical output]] |
==Model coordinates== | ==Model coordinates== | ||
* [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/2007/MBP1_CHAGL.txt MBP1_CHAGL.pdb] | * [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/2007/MBP1_CHAGL.txt MBP1_CHAGL.pdb] | ||
Revision as of 03:19, 30 November 2011
Please remember: if you use information from this page as a fallback for technical problems with the assignment, you must document this in your submission. State what you tried, what didn't work and only then use the data from here.
Contents
Target sequence
This is just one of the reference species' sequences - I chose Mbp1_SCHPO because it is the most distantly related to saccharomyces cerevisiae.
>MBP1_SCHPO NP_593032 23..96 IKGVSVMRRRRDSWLNATQILKVADFDKPQRTRVLERQVQIGAHEKVQGGYGKYQGTWVPFQRGVDLATK YKV
FASTA formatted target-template alignment
This uses the 1MB1 PDB file as template, the sequence was extracted from coordinates using the WhatIf server.
>1MB1_A 3..100 NQIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKET HEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF >MBP1_SCHPO NP_593032 23..96 ----------------IKGV-SVMRRRRDSWLNATQILKVADFDKPQRTRVLERQVQIGA HEKVQGGYGKYQGTWVPFQRGVDLATK-----------
SwissModel response
Explanation of the output is found in the SwissModel Help page.
The target-template alignment:
TARGET 1 IK -GVSVMRRRR DSWLNATQIL KVADFDKPQR
1mb1A 3 nqiysarysg vdvyef--ih stgsimkrkk ddwvnathil kaanfakakr
TARGET ssshhhh hh hhhh
1mb1A sssssss ssssss ss sssssss ssshhhh hh hhhh
TARGET 32 TRVLERQVQI GAHEKVQGGY GKYQGTWVPF QRGVDLATK ----------
1mb1A 51 trilekevlk ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf
TARGET hhhhhhhhh sss ssss h hhhhhhhhh
1mb1A hhhhh sss ssss h hhhhhhhhh hh hh