Difference between revisions of "User:Boris/Temp/APB"
| Line 1: | Line 1: | ||
| − | <!-- | + | <!-- {{Template:Active}} --> |
| − | + | {{Template:Inactive}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 13: | Line 8: | ||
<div style="padding: 5px; background: #A6AFD0; border:solid 1px #AAAAAA; font-size:200%;font-weight:bold;"> | <div style="padding: 5px; background: #A6AFD0; border:solid 1px #AAAAAA; font-size:200%;font-weight:bold;"> | ||
| − | Assignment | + | Assignment 4 - Homology modeling |
</div> | </div> | ||
| − | < | + | <div style="padding: 15px; background: #F0F1F7; border:solid 1px #AAAAAA; font-size:125%;color:#444444"> |
| − | + | ;How could the search for ultimate truth have revealed so hideous and visceral-looking an object? | |
| − | '' | + | ::''<small>Max Perutz (on his first glimpse of the Hemoglobin structure)</small>'' |
| + | </div> | ||
| + | | ||
| | ||
| + | Where is the hidden beauty in structure, and where, the "ultimate truth"? In the previous assignments we have studied sequence conservation in APSES family domains and looked at how these domains have evolved over time. We have seen that this is an ancient family, that had several members already in the cenancestor of all fungi, an organism that lived in the [http://www.ucmp.berkeley.edu/fungi/fungifr.html vendian period] of the proterozoic era of precambrian times, more than 600,000,000 years ago. | ||
| − | + | In order to understand how particular residues in the sequence contribute to the putative function of the protein, and why and how they are conserved throughout evolution, we would need to consider an explicit molecular model of an APSES domain protein, bound to its cognate DNA sequence. In particular, it would be interesting to correlate the conservation patterns we have observed in the MSAs with specific DNA binding interactions. Unfortunately, the 1MB1 structure does not have DNA bound and the evidence we have considered in Assignment 2 ([http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=10747782 Taylor ''et al.'', 2000]) is not sufficient to define the details of how a DNA double helix might be bound. These details would require the structure of a complex that contains protein as well as DNA. No such complex of an APSES domain has yet been crystallized. | |
| − | + | ||
| + | ''In this assignment you will construct a molecular model of the Mbp1 orthologue in your assigned organism, identify similar structures of distantly related domains for which protein-DNA complexes are known, define whether the available evidence allows you to distinguish between different modes of ligand binding, and assemble a hypothetical complex structure.'' | ||
| + | |||
| + | For the following, please remember the following terminology: | ||
| + | |||
| + | ;Target | ||
| + | :The protein that you are planning to model. | ||
| + | ;Template | ||
| + | :The protein whose structure you are using as a guide to build the model. | ||
| + | ;Model | ||
| + | :The structure that results from the modeling process. It has the '''Target sequence''' and is similar to the '''Template structure'''. | ||
| | ||
| − | + | A brief overview article on the construction and use of homology models is linked to the resource section at the bottom of this page. That section also contains links to other sites and resources you might require. | |
| − | |||
| − | |||
| − | + | {{Template:Preparation| | |
| − | + | care=Be sure you have understood all parts of the assignment and cover all questions in your answers! Sadly, we see too many assignments which, arduously effected, nevertheless intimate nescience of elementary tenets of molecular biology. If the sentence above did not trigger an urge to open a dictionary, you are trying to guess, rather than confirm possibly important information.| | |
| − | + | num=4| | |
| − | + | ord=fourth| | |
| + | due = Monday, November 5 at 10:00 in the morning}} | ||
| − | |||
| − | + | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> | |
| + | ==(1) Preparation== | ||
| + | </div> | ||
| − | |||
| − | + | <!-- | |
| − | |||
| − | |||
| + | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | ||
| + | ===Choosing a template (1 marks)=== | ||
| + | </div> | ||
| + | <br> | ||
| + | Often more than one related structure can be found in the PDB. We have discussed principles of selecting template structures in the lecture. Interestingly the PDB itself cannot be searched for the contents of its holdings, by structural- or sequence similarity, but there is always BLAST since the NCBI conveniently allows you to search against all sequences in PDB files. | ||
| − | + | *Use BLAST to identify all PDB files that contain APSES domains that are clearly homologuous to your target. (Document that you have searched in the correct subsection of the Genbank holdings). For the hits you find, consider how these structures differ and which features would make each more or less suitable for your task. Comment briefly on what options you have, select one template and note why you have decided to use this particular structure as a template. Include aspects of sequence similarity, length of the sequence, presence or absence of ligands and their potential effect on the structure, and experimental method and quality in your reasoning. | |
| − | |||
| − | |||
| − | + | *Note which sequence is contained in the coordinate section of the PDB file; note if and how this implied sequence differs from the sequences ... | |
| − | + | :*listed in the seqres records; | |
| − | * | + | :*given in the FASTA sequence for the template that the PDB provides; |
| − | * | + | :*and that stored by the NCBI. |
| − | |||
| − | *the | ||
| − | + | * In a table, establish the correspondence of the coordinate sequence numbering (defined by the residue numbers/insertion codes in the atom records) with your target sequence numbering. | |
| − | |||
| − | |||
| − | |||
| − | + | * Retrieve the most suitable template structure coordinate file from the PDB. | |
| − | + | --> | |
| − | |||
| − | |||
| − | |||
| − | + | | |
| + | | ||
| − | + | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | |
| + | === The input alignment (1 marks)=== | ||
| + | </div> | ||
| + | <br> | ||
| − | + | The sequence alignment between target and template is the single most important factor that determines the quality of your model. | |
| − | + | No homology modeling process will repair an incorrect alignment and it is useful to consider a homology model rather like a three-dimensional map of a sequence alignment, rather than a structure in its own right. In a homology modeling project, typically the largest amount of time should be spent on preparing the best possible alignment. Even though automated servers like the SwissModel server will align sequences and select template structures for you, it would be unwise to use these just because they are convenient, rather than the more sophisticated methods and more informed procedures we have discussed. Detailed analysis of fallacious models rarely leads to good results. | |
| − | |||
| − | |||
| − | + | The best possible alignment is usually constructed from a multiple sequence alignment that includes at least the target and template sequence and other related sequences as well. The additional sequences are an important aid in identifying the correct placement of insertions and deletions. Typically such an alignment will also include additional optimization steps to move insertions or deletions between target and template out of the secondary structure elements of the template structure. | |
| − | + | Here is an excerpt from the T-coffee aligned Mbp1 sequences: it contains all the residues of the yeast sequence that are found in the 1MB1 crystal structure - the '''template''' sequence for our homology model - and it has been edited to remove the N-terminal gaps in the sequence. Thus the N-terminus is 21 amino acids longer than the definition of the APSES domain in CDD (which starts with <code>SIMKR...</code>), the C- terminus is slightly shorter. | |
| − | |||
| − | |||
| − | + | Since the sequences are very similar between each other, there is no ambiguity in the alignment and the construction of a homology model should be straightforward. Normally one would spend considerable some effort at this stage to consider which parts of the target sequence and the template sequence appear to correctly aligned and to edit the alignment manually. In our case, evolutionary pressure was so strong that essentially all have evolved without a single indel in their sequence. | |
| − | |||
| − | + | I have added to the alignment the APSES domain of [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=116197493&dopt=GenPept XP_001224558], the ''Chaetomium globosum'' Mbp1 orthologue (MBP1_CHAGL). This will serve as the reference and fallback sequence. | |
| − | |||
| − | |||
| − | & | ||
| − | |||
| − | + | 1MB1 NQIYSARYSGVDVYEFIHSTG---SIMKRKKDDWVNATHILKAANFAKAKRTRILEKEV | |
| + | MBP1_CANGL NQIYSAKYSGVDVYEFIHPTG---SIMKRKNDGWVNATHILKAANFAKAKRTRILEKEV | ||
| + | MBP1_EREGO TQIYSAKYSGVEVYEFLHPTG---SIMKRKADDWVNATHILKAAKFAKAKRTRILEKEV | ||
| + | MBP1_KLULA NQIYSAKYSGVDVYEFIHPTG---SIMKRKADNWVNATHILKAAKFPKAKRTRILEKEV | ||
| + | MBP1_CANAL SQIYSATYSNVPAFEFVTSEG---PIMRRKKDSWINATHILKIAKFPKAKRTRILEKDV | ||
| + | MBP1_DEBHA TQIYSATYSNVPVFEFVTLEG---PIMRRKLDSWINATHILKIAKFPKAKRTRILEKDV | ||
| + | MBP1_YARLI MSIYKATYSGVPVYEFQCKNV---AVMRRKSDGWVNATHILKVAGFDKPQRTRILEKEV | ||
| + | MBP1_SCHPO SAVHVAVYSGVEVYECFIKGV---SVMRRRRDSWLNATQILKVADFDKPQRTRVLERQV | ||
| + | MBP1_USTMA KTIFKATYSGVPVYECIINNV---AVMRRRSDDWLNATQILKVVGLDKPQRTRVLEREI | ||
| + | MBP1_ASPNI SNVYSATYSSVPVYEFKIGTD---SVMRRRSDDWINATHILKVAGFDKPARTRILEREV | ||
| + | MBP1_ASPTE SKIYSATYSSVPVYEFKIEGD---SVMRRRADDWINATHILKVAGFDKPARTRILEREV | ||
| + | MBP1_CRYNE PKVYASVYSGVPVFEAMIRGI---SVMRRASDSWVNATQILKVAGVHKSARTKILEKEV | ||
| + | MBP1_GIBZE G-IYSASYSGVDVYEMEVNNI---AVMRRRNDSWLNATQILKVAGVDKGKRTKILEKEI | ||
| + | MBP1_NEUCR IYSLQATYSGVGVYEMEVNNV---AVMRRQKDGWVNATQILKVANIDKGRRTKILEKEI | ||
| + | MBP1_MAGGR P-IYTAVYSNVEVYEFEVNGV---AVMKRIGDSKLNATQILKVAGVEKGKRTKILEKEI | ||
| + | MBP1_ASPFU PQIYKAVYSNVSVYEMEVNGV---AVMKRRSDSWLNATQILKVAGVVKARRTKTLEKEI | ||
| + | MBP1_CHAGL AGIYSATYSGIPVYEYQFGPDMKEHVMRRREDNWINATHILKAAGFDKPARTRILERDV | ||
| + | |||
| + | 1MB1 LKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF | ||
| + | MBP1_CANGL LKEMHEKVQGGFGKYQGTWVPLNIAINLAEKFDVYQDLKPLF | ||
| + | MBP1_EREGO IKDTHEKVQGGFGKYQGTWVPLDIARRLAQKFEVLEELRPLF | ||
| + | MBP1_KLULA ITDTHEKVQGGFGKYQGTWIPLELASKLAEKFEVLDELKPLF | ||
| + | MBP1_CANAL QTGIHEKVQGGYGKYQGTYVPLDLGAAIARNFGVYDVLKPIF | ||
| + | MBP1_DEBHA QTGVHEKVQGGYGKYQGTYVPLDLGADIAKNFGVFDSLRPIF | ||
| + | MBP1_YARLI QKGVHEKVQGGYGKYQGTWVPLERAREIATLYDVDSHLAPIF | ||
| + | MBP1_SCHPO QIGAHEKVQGGYGKYQGTWVPFQRGVDLATKYKVDGIMSPIL | ||
| + | MBP1_USTMA QKGIHEKVQGGYGKYQGTWIPLDVAIELAERYNIQGLLQPIT | ||
| + | MBP1_ASPNI QKGVHEKVQGGYGKYQGTWIPLQEGRQLAERNNILDKLLPIF | ||
| + | MBP1_ASPTE QKGVHEKVQGGYGKYQGTWIPLPEGRLLAERNNIIDKLRPIF | ||
| + | MBP1_CRYNE LNGIHEKIQGGYGKYQGTWVPLDRGRDLAEQYGVGSYLSSVF | ||
| + | MBP1_GIBZE QTGEHEKVQGGYGKYQGTWIKFERGLQVCRQYGVEELLRPLL | ||
| + | MBP1_NEUCR QIGEHEKVQGGYGKYQGTWIPFERGLEVCRQYGVEELLSKLL | ||
| + | MBP1_MAGGR QTGEHEKVQGGYGKYQGTWIKYERALEVCRQYGVEELLRPLL | ||
| + | MBP1_ASPFU AAGEHEKVQGGYGKYQGTWVNYQRGVELCREYHVEELLRPLL | ||
| + | MBP1_CHAGL QKDVHEKIQGGYGKYQGTWIPLEQGRALAQRNNIYDRLRPIF | ||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| − | + | It should be obvious to you by now how you can copy a string of amino acids from such an alignment and create a FASTA file. However we need to take a little detour: this detour brings us to the question of sequence numbers. | |
| − | + | It is not straightforward at all how to number sequence in such a project. The "natural" way would be to start a sequential numbering from the start-codon of the full length protein and go sequentially from there. However imagine what would happen if a curator would discover that one of the splice-sites for a gene has been missed in automatic annotation. All of a sudden a corrected sequence would have a different length than the one that may have been used for earlier studies. Unfortunatlety, there is no mechanism (''wouldn't it be nice!'') that automatically goes back through the literature and your lab-journal and updates the revised sequence numbering... But there are other possible complications, regarding sequence numbers. The first residue of the CDD-APSES domain is not Residue 1 of the Mbp1 protein. The first residue of the 1MB1 FASTA file ''is'' the first residue of Mbp1 protein, but the last five residues are an artifiical His tag. Is H125 of 1MB1 the equivalent residue to R125 in MBP1_SACCE? The N-terminus of the Mbp1 crystal structure is disordered. The first residue in the structure is ASN 3, whereas the SEQRES records start with MET ... and so on. The take-home message is that a sequence number is nothing absolute, but something that makes sense only in a particular context. To emphasize this, we will write a FASTA header for our '''target''' sequence that lists the residues of the source sequence it correspond to. In terms of actual sequence numbering, we will adopt the numbering of the 1MB1 protein throughout to be able to consistently label particular amino acids. | |
| − | + | ||
| − | + | Access the sequence of "your" organism's Mbp1 Orthologue at UniProt. (You can use the links I have provided in the table below). | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<table style="border-left:1px solid #AAAAAA; border-bottom:1px solid #AAAAAA;" cellpadding="10" cellspacing="0"> | <table style="border-left:1px solid #AAAAAA; border-bottom:1px solid #AAAAAA;" cellpadding="10" cellspacing="0"> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<tr style="background: #BDC3DC;"> | <tr style="background: #BDC3DC;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><b><i>Organism</i></b></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><b><i>Organism</i></b></td> | ||
| − | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><b>Uniprot Accession</b></td> | |
| − | |||
| − | |||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><b>Uniprot | ||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Aspergillus fumigatus</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Aspergillus fumigatus</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q4WGN2_ASPFU Q4WGN2]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Aspergillus nidulans</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Aspergillus nidulans</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q5B8H6_EMENI Q5B8H6]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Aspergillus terreus</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Aspergillus terreus</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q0CQJ5_ASPTE Q0CQJ5]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Candida albicans</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Candida albicans</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q5ANP5_CANAL Q5ANP5]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Candida glabrata</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Candida glabrata</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q6FWD6_CANGL Q6FWD6]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Cryptococcus neoformans</i></td> |
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q5KHS0_CRYNE Q5KHS0]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Debaryomyces hansenii</i></td> |
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q6BSN6_DEBHA Q6BSN6]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Eremothecium gossypii</i></td> |
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q752H3_ASHGO Q752H3]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Gibberella zeae</i></td> |
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q4IEY8_GIBZE Q4IEY8]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Kluyveromyces lactis</i></td> |
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=MBP1_KLULA P39679]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Magnaporthe grisea</i></td> |
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q3S405_MAGGR Q3S405]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Neurospora crassa</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Neurospora crassa</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q7SBG9_NEUCR Q7SBG9]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Saccharomyces cerevisiae</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Saccharomyces cerevisiae</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=MBP1_YEAST P39678]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Schizosaccharomyces pombe</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Schizosaccharomyces pombe</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=RES2_SCHPO P41412]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #FFFFFF;"> | <tr style="background: #FFFFFF;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Ustilago maydis</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Ustilago maydis</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q4P117_USTMA Q4P117]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
<tr style="background: #E9EBF3;"> | <tr style="background: #E9EBF3;"> | ||
<td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Yarrowia lipolytica</i></td> | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"><i>Yarrowia lipolytica</i></td> | ||
| − | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;"> | + | <td style="border-right:1px solid #AAAAAA; border-top:1px solid #AAAAAA;">[http://www.ebi.uniprot.org/uniprot-srv/uniProtView.do?proteinId=Q6CGF5_YARLI Q6CGF5]</td> |
| − | |||
| − | |||
| − | |||
| − | |||
</tr> | </tr> | ||
</table> | </table> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | <div style="padding: 5px; background: #EEEEEE;"> | ||
| + | *Copy your organism's Mbp1 sequence from the alignment above. Then define the start- and end- sequence numbers of the '''target''' sequence relative to the full-length protein. Prepare a FASTA formatted file for the '''target''' sequence in your organism, giving it an appropriate header and include the sequence numbers. Refer to the [[Assignment_5_fallback_data|'''Fallback data''']] file if you are not sure about the format. (1 mark) | ||
</div> | </div> | ||
<br> | <br> | ||
| − | |||
| − | + | Your FASTA sequence should look similar to this: | |
| − | |||
| − | |||
| − | |||
| − | + | >1MB1: Mbp1_SACCE 1..100 | |
| + | NQIYSARYSGVDVYEFIHSTG---SIMKRKKDDWVNATHILKAANFAKAKRTRILEKEV | ||
| + | LKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF | ||
| − | + | | |
| − | + | | |
| − | | ||
| − | + | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> | |
| − | + | ==(2) Homology model== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ==(2) | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
| | ||
<div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | ||
| − | ===(2.1) | + | === (2.1) SwissModel (1 mark)=== |
</div> | </div> | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | + | Access the Swissmodel server at [http://swissmodel.expasy.org '''http://swissmodel.expasy.org'''] . Navigate to the '''Alignment Interface'''. | |
| − | |||
| − | |||
| − | + | <br><div style="padding: 5px; background: #EEEEEE;"> | |
| − | + | *Copy from the alignment above the 1MB1 sequence and the sequence from your organism, and paste it into the form field. Refer to the [[Assignment_5_fallback_data|'''Fallback Data file''']] if you are not sure about the format. | |
| − | + | :(You have to choose the format, and, if e.g. you choose a CLUSTAL format, you have to include a header line and a blank line. Other common problems uploading your alignment may include uploading a file that has not been saved as "text only" and periods i.e. "." in sequence names. Underscores appear to be safe.) | |
| − | |||
| − | + | * Click '''submit''' and define your '''target''' and '''template''' sequence. For the '''template sequence''' define the coordinate file and chain. (In our case the coordinate file is <code>'''1MB1'''</code> and the chain is "<code>'''_'''</code>" i.e. none, since the PDB file does not contain more than one chain. | |
| + | *Click '''submit''' and request the construction of a homology model: Enter your e-mail address and check the button for '''Normal Mode''', not "Swiss-PDB Viewer mode. (Important, since there will be problems with the output otherwise). Click '''submit'''. You should receive four files files by e-mail within half an hour or so. (1 mark) | ||
| − | + | (You do not need to submit any coordinate files with your assignment.) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| + | <br> | ||
| + | In case you do not wish to submit the modelling job yourself, you can access the result files for the from the [[Assignment_5_fallback_data|'''Fallback Data file''']]. | ||
| − | |||
| − | |||
<div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> | ||
| − | ==(3) | + | ==(3) Model analysis== |
</div> | </div> | ||
| | ||
| | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | ||
| − | ===(3.1) | + | === (3.1) The PDB file (1 mark)=== |
</div> | </div> | ||
<br> | <br> | ||
| − | + | Open your '''model''' coordinates in a text-editor (make sure you view the PDB file in a fixed-width font) and consider the following questions: (Alternatively, view the coordinates linked to the [[Assignment_5_fallback_data|'''Fallback Data file''']].) | |
<br><div style="padding: 5px; background: #EEEEEE;"> | <br><div style="padding: 5px; background: #EEEEEE;"> | ||
| − | * | + | *What is the residue number of the first residue in the '''model'''? What should it be, based on the alignment? If the putative DNA binding region was reported to be residues 50-74 in the Mbp1 protein, which residues of the '''model''' correspond to that? (1 mark) |
</div> | </div> | ||
| − | + | ||
| + | <!-- discuss flagging of loops - setting of B-factor to 99.0 --> | ||
| | ||
| Line 527: | Line 292: | ||
<div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | ||
| − | + | ===(3.2) first visualization (3 marks)=== | |
| − | ===(3.2) | ||
</div> | </div> | ||
<br> | <br> | ||
| − | + | In assignment 2 you have already studied the 1MB1 coordinate file and compared it to your organism's Mbp1 APSES domain, Since a homology model inherits its structural details from the '''template''', the model should look very similar to the original structure but contain the sequence of the '''target'''. | |
<br><div style="padding: 5px; background: #EEEEEE;"> | <br><div style="padding: 5px; background: #EEEEEE;"> | ||
| − | * | + | *Save the attachment of your '''model''' coordinates to your harddisk and visualize it in RasMol. (Alternatively, copy and save the coordinates from the [[Assignment_5_fallback_data|'''Fallback Data file''']] to your harddisk.) Make an informative view, divergent stereo and paste it into your assignment. (3 marks) |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
<br> | <br> | ||
| − | |||
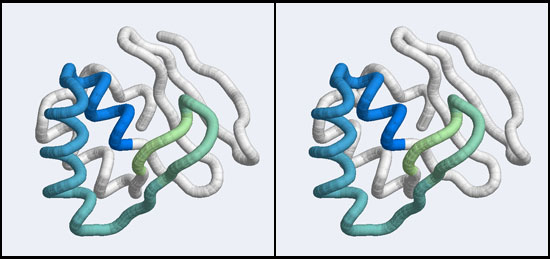
| − | + | [[Image:A5_Mbp1_subdomain.jpg|frame|none|Stereo-view of a subdomain within the 1MB1 structure that includes residues 36 to 76. The color gradient ramps from blue (36) to green (76).]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
| − | |||
| + | <div style="padding: 5px; background: #E9EBF3; border:solid 1px #AAAAAA;"> | ||
| − | + | ===(3.3) modeling a DNA ligand (4 marks)=== | |
| − | |||
| − | ==(4) | ||
</div> | </div> | ||
<br> | <br> | ||
| − | + | The really interesting question we could begin to address with our model is how sequence variation might be converted into changing DNA recognition sites, and then lead to changed cognate DNA binding sequences. But in order to address this, we would need to add a plausible model for a bound DNA molecule to our model. | |
| − | + | Since there is currently no software available that would accurately model such a complex from first principles, we will base this on homology modeling as well. This means we need to find a similar structure for which the complex structure is known. However, you may remember from the third assignment that the APSES domains in fungi seem to be a relatively small family. And there is no structure available of a protein-DNA complex. Now what? | |
| − | |||
| − | |||
| − | + | Remember that homologous sequences can have diverged to the point where their sequence similarity is no longer recognizable, however their structure may be quite well conserved. Thus if we could find similar structures in the PDB, these might provide us with some plausible hypotheses for how DNA is bound by APSES domains. We thus need a tool similar to BLAST, but not for the purpose of sequence alignment, but for structure alignment. A kind of BLAST for structures. | |
| − | |||
| − | |||
| − | + | However, very similar to BLAST, we might not want to search with the entire protein, if all we are interested in is a subdomain that binds to DNA. Attempting to match all structural elements in addition to the ones we are actually interested in is likely to make the search less specific - we would find false positives that are similar to some irrelevant part of our structure. However, defining too small of a subdomain would also lead to a loss of specificity: in the extreme it is easy to imagine that the search for e.g. a single helix would retrieve very many hits that would be quite meaningless. The arrangement of the residues from 50 to 74 that we have already discussed in Assignment 2 suggests that the compact subdomain from 36 to 76 (see the image above) might be a useful structure to search with: it contains the residues we are interested in and enough of connected secondary structure elements to be structurally meaningful. | |
| − | + | At the '''NCBI''', [http://www.ncbi.nlm.nih.gov/Structure/VAST/vast.shtml VAST] is a search tool for structural similarity search tool for this purpose. Unfortunately it does not seem to be able to handle a query with such a structural subdomain (the process did not finish after several days) but at least you can get a list of structural neighbors of the 1MB1 full-length template structure, by entering the PDB ID in a small form field on the VAST home page, and then clicking on the colored bar labeled "Chain" on the MMDB structure summary page. This precomputed page for the 1MB1 structure shows a number of diverse proteins matching to various helices and strands of the structure. | |
| − | + | At the '''EBI''' there are a number of very well designed structure analysis tools linked off the [http://www.ebi.ac.uk/Tools/structural.html '''Structural Analysis''' page]. As part of its MSD Services, the SSM (Secondary Structure Matching service) provides a well thought out interface for searching files from the PDB or uploading coordinates. | |
| − | + | After uploading the coordinates for residues 36 to 76 of the 1MB1 structure running the search and sorting the results by alignment length, the top hits include a number of nucleotide binding proteins such as a replication terminator (1F4K), the LexA repressor (1MVD) and a "Winged Helix" protein (1KQ8). These are all members of a much larger superfamily, the "winged helix" DNA binding domains ([http://cathwww.biochem.ucl.ac.uk/cgi-bin/cath/GotoCath.pl?cath=1.10.10.10 CATH 1.10.10.10]), of which hundreds of structures have been solved. They represent one branch of the tree of helix-turn-helix (HTH) DNA binding modules. (A recent review on HTH proteins is linked from the resources section at the bottom of this page). Winged Helix domains typically bind their cognate DNA with a "recognition helix" which precedes the beta hairpin and binds into the major groove; additional stabilizing interactions are provided by the edge of the beta strand binding into the minor groove. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | + | <!-- The other service the EBI structure links to is the DALI server. DALI was one of the first algorithms capable of large-scale protein structure searches; it was developed by Liisa Holm and is now hosted by her group in Helsinki. Submitting our search domain generates the e-mailed result linked to here. Both results (there are only two) are also found in the top 100 list of the SSM service. The winged helix domain 1DP7 merits some comment though: its structure shows a novel mode of binding for DNA. Here, it is the beta-wing, not the "recognition helix" that inserts into the major groove! We will consider this in more detail below. |
| − | + | First we shall explore some of the structures that SSM has returned. The SSM server presents its result details in Web pages, but it also allows to download the entire result set in an XML formatted file. This is a common method of data-interchange in bioinformatics but you would not want to actually read such a file and manually extract information (even though you could, in principle). Thus I have prepared a summary file of the alignment details of the SSM results. This should allow you to rapidly find the exact aligned residues in the matched domains. While I have derived this file from the output through a computer program I have written, you could easily have accessed the same information on the Web, had you run the query yourself. --> | |
| − | + | This is good news: once we have determined that the APSES domain is actually an example of a larger group of transcription factors, we can pick one of these for which a DNA complex structure is known. I have picked one such structure from the list of hits that were returned by SSM: it is the Elk-1 transcription factor. | |
| − | |||
| − | |||
| − | |||
| − | |||
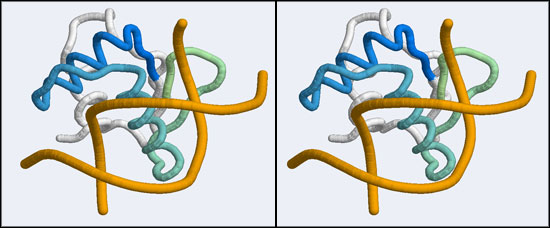
| − | + | [[Image:A5_canonical_wHTH.jpg|frame|none|Stereo-view of the canonical DNA binding mode of the Winged Helix domain family. Shown here is the Elk-1 transcription factor - an ETS DNA binding domain - in complex with a high-affinity binding site (pdb|1DUX). Note how the "recognition helix" inserts into the major groove of the DNA molecule. The color gradient ramps from blue (34) to green (84). Note how the first helix of the "helix-turn-helix" architecture serves only to position the recognition helix and makes few interactions by itself.]] | |
| − | + | Now all that is left to do is to bring the DNA molecule into the correct orientation for our '''model''' and then to combine the two files. We need to superimpose the Elk-1 protein/DNA complex onto our '''model'''. | |
| − | + | ;Structure superposition | |
| − | + | There are quite a number of superposition servers available on the Web, a remarkably comprehensive overview can be found in [http://en.wikipedia.org/wiki/Structural_alignment Wikipedia]. However, overengineering and black-box mentality makes our task more difficult than it need be: most tools do not allow users to specify particular alignment zones but attempt to automatically define the zones of residues to be supoerimposed according to some geometric target function. Almost none return the actual rotation matrix and translation vector that is used for the superposition. And almost none transform the coordinates of heteratoms such as solvent, ligands or DNA molecules along with the protein coordinates. An exception that I have found to be very useable is the [http://www.predictioncenter.org/local/lga/lga.html Local-Global Alignment server ('''LGA''')], written by Adam Zemla. The procedure is quite straightforward: | |
| − | |||
| − | |||
| − | |||
| − | + | *Define the structure to be rotated (1DUX in this case). This is a dimer, so download the file from the PDB and manually edit to contain only DNA chains A and B and protein chain C. | |
| + | *Define the structure to be held constant (1MB1 in this case). Download from PDB. | ||
| + | *Use the "browse" option to define both files as input on the LGA inpput form | ||
| + | *Use the option to have both coordinate sets included in your output: <code>-o2</code> | ||
| + | *Submit | ||
| − | + | The results arrive per e-mail. I have linked the resulting PDB file to the [[Assignment_5_fallback_data|'''Fallback Data page''']]. <small>If you run this analysis on your own, you may want to review the types of edits the edits I made to the PDB file to get it displayed correctly in Rasmol.</small> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br><div style="padding: 5px; background: #EEEEEE;"> | <br><div style="padding: 5px; background: #EEEEEE;"> | ||
| − | * | + | *Save the superimposed coordinates in a file, open and view in Rasmol and note how well the "recognition helix" and adjacent beta strands superimpose! (Alternatively, copy and save the coordinates from the c to your harddisk.) Make an informative view, divergent stereo and paste it into your assignment. (4 marks) |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> | <div style="padding: 5px; background: #BDC3DC; border:solid 1px #AAAAAA;"> | ||
| − | ==( | + | ==(4) Summary of Resources== |
</div> | </div> | ||
<br> | <br> | ||
;Links | ;Links | ||
| − | + | :* [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/restricted/Peitsch_2002_UseOfModels.pdf '''Review (PDF, restricted)''' Manuel Peitsch on Homology Modeling] | |
| − | :* [http:// | + | :* [http://biochemistry.utoronto.ca/undergraduates/courses/BCH441H/restricted/Aravind_2005_HTHdomains.pdf '''Review (PDF, restricted)''' Aravind ''et al.'' Helix-turn-helix domains] (background reading, not required reading) |
| − | + | :* [[Organism_list_2006|Assigned Organisms]] | |
| − | :* [http:// | + | :* [http://www.rcsb.org/pdb/file_formats/pdb/pdbguide2.2/guide2.2_frame.html '''PDB file format'''] |
| − | + | :* [http://en.wikipedia.org/wiki/Structural_alignment Wikipedia on '''Structural Superposition'''] <small>(although the article is called "Structural Alignment")</small> | |
| − | |||
| − | :* [ | ||
| − | :* [http://www. | ||
| − | |||
| − | :* [http:// | ||
| − | + | :* [[Assignment_5_fallback_data|'''Fallback Data page''']] | |
| − | :* [[ | ||
| − | |||
;Alignments | ;Alignments | ||
| − | |||
| − | |||
| − | |||
:* [[All_Mbp1_T-COFFEE|Mbp1 proteins '''T-Coffee''' aligned (text version)]] | :* [[All_Mbp1_T-COFFEE|Mbp1 proteins '''T-Coffee''' aligned (text version)]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | ||
Revision as of 15:28, 27 October 2007
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Contents
Assignment 4 - Homology modeling
- How could the search for ultimate truth have revealed so hideous and visceral-looking an object?
-
- Max Perutz (on his first glimpse of the Hemoglobin structure)
Where is the hidden beauty in structure, and where, the "ultimate truth"? In the previous assignments we have studied sequence conservation in APSES family domains and looked at how these domains have evolved over time. We have seen that this is an ancient family, that had several members already in the cenancestor of all fungi, an organism that lived in the vendian period of the proterozoic era of precambrian times, more than 600,000,000 years ago.
In order to understand how particular residues in the sequence contribute to the putative function of the protein, and why and how they are conserved throughout evolution, we would need to consider an explicit molecular model of an APSES domain protein, bound to its cognate DNA sequence. In particular, it would be interesting to correlate the conservation patterns we have observed in the MSAs with specific DNA binding interactions. Unfortunately, the 1MB1 structure does not have DNA bound and the evidence we have considered in Assignment 2 (Taylor et al., 2000) is not sufficient to define the details of how a DNA double helix might be bound. These details would require the structure of a complex that contains protein as well as DNA. No such complex of an APSES domain has yet been crystallized.
In this assignment you will construct a molecular model of the Mbp1 orthologue in your assigned organism, identify similar structures of distantly related domains for which protein-DNA complexes are known, define whether the available evidence allows you to distinguish between different modes of ligand binding, and assemble a hypothetical complex structure.
For the following, please remember the following terminology:
- Target
- The protein that you are planning to model.
- Template
- The protein whose structure you are using as a guide to build the model.
- Model
- The structure that results from the modeling process. It has the Target sequence and is similar to the Template structure.
A brief overview article on the construction and use of homology models is linked to the resource section at the bottom of this page. That section also contains links to other sites and resources you might require.
Preparation, submission and due date
- Read carefully.
- Be sure you have understood all parts of the assignment and cover all questions in your answers! Sadly, we see too many assignments which, arduously effected, nevertheless intimate nescience of elementary tenets of molecular biology. If the sentence above did not trigger an urge to open a dictionary, you are trying to guess, rather than confirm possibly important information.
Review the guidelines for preparation and submission of BCH441 assignments.
The due date for the assignment is Monday, November 5 at 10:00 in the morning.
(1) Preparation
The input alignment (1 marks)
The sequence alignment between target and template is the single most important factor that determines the quality of your model.
No homology modeling process will repair an incorrect alignment and it is useful to consider a homology model rather like a three-dimensional map of a sequence alignment, rather than a structure in its own right. In a homology modeling project, typically the largest amount of time should be spent on preparing the best possible alignment. Even though automated servers like the SwissModel server will align sequences and select template structures for you, it would be unwise to use these just because they are convenient, rather than the more sophisticated methods and more informed procedures we have discussed. Detailed analysis of fallacious models rarely leads to good results.
The best possible alignment is usually constructed from a multiple sequence alignment that includes at least the target and template sequence and other related sequences as well. The additional sequences are an important aid in identifying the correct placement of insertions and deletions. Typically such an alignment will also include additional optimization steps to move insertions or deletions between target and template out of the secondary structure elements of the template structure.
Here is an excerpt from the T-coffee aligned Mbp1 sequences: it contains all the residues of the yeast sequence that are found in the 1MB1 crystal structure - the template sequence for our homology model - and it has been edited to remove the N-terminal gaps in the sequence. Thus the N-terminus is 21 amino acids longer than the definition of the APSES domain in CDD (which starts with SIMKR...), the C- terminus is slightly shorter.
Since the sequences are very similar between each other, there is no ambiguity in the alignment and the construction of a homology model should be straightforward. Normally one would spend considerable some effort at this stage to consider which parts of the target sequence and the template sequence appear to correctly aligned and to edit the alignment manually. In our case, evolutionary pressure was so strong that essentially all have evolved without a single indel in their sequence.
I have added to the alignment the APSES domain of XP_001224558, the Chaetomium globosum Mbp1 orthologue (MBP1_CHAGL). This will serve as the reference and fallback sequence.
1MB1 NQIYSARYSGVDVYEFIHSTG---SIMKRKKDDWVNATHILKAANFAKAKRTRILEKEV MBP1_CANGL NQIYSAKYSGVDVYEFIHPTG---SIMKRKNDGWVNATHILKAANFAKAKRTRILEKEV MBP1_EREGO TQIYSAKYSGVEVYEFLHPTG---SIMKRKADDWVNATHILKAAKFAKAKRTRILEKEV MBP1_KLULA NQIYSAKYSGVDVYEFIHPTG---SIMKRKADNWVNATHILKAAKFPKAKRTRILEKEV MBP1_CANAL SQIYSATYSNVPAFEFVTSEG---PIMRRKKDSWINATHILKIAKFPKAKRTRILEKDV MBP1_DEBHA TQIYSATYSNVPVFEFVTLEG---PIMRRKLDSWINATHILKIAKFPKAKRTRILEKDV MBP1_YARLI MSIYKATYSGVPVYEFQCKNV---AVMRRKSDGWVNATHILKVAGFDKPQRTRILEKEV MBP1_SCHPO SAVHVAVYSGVEVYECFIKGV---SVMRRRRDSWLNATQILKVADFDKPQRTRVLERQV MBP1_USTMA KTIFKATYSGVPVYECIINNV---AVMRRRSDDWLNATQILKVVGLDKPQRTRVLEREI MBP1_ASPNI SNVYSATYSSVPVYEFKIGTD---SVMRRRSDDWINATHILKVAGFDKPARTRILEREV MBP1_ASPTE SKIYSATYSSVPVYEFKIEGD---SVMRRRADDWINATHILKVAGFDKPARTRILEREV MBP1_CRYNE PKVYASVYSGVPVFEAMIRGI---SVMRRASDSWVNATQILKVAGVHKSARTKILEKEV MBP1_GIBZE G-IYSASYSGVDVYEMEVNNI---AVMRRRNDSWLNATQILKVAGVDKGKRTKILEKEI MBP1_NEUCR IYSLQATYSGVGVYEMEVNNV---AVMRRQKDGWVNATQILKVANIDKGRRTKILEKEI MBP1_MAGGR P-IYTAVYSNVEVYEFEVNGV---AVMKRIGDSKLNATQILKVAGVEKGKRTKILEKEI MBP1_ASPFU PQIYKAVYSNVSVYEMEVNGV---AVMKRRSDSWLNATQILKVAGVVKARRTKTLEKEI MBP1_CHAGL AGIYSATYSGIPVYEYQFGPDMKEHVMRRREDNWINATHILKAAGFDKPARTRILERDV 1MB1 LKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF MBP1_CANGL LKEMHEKVQGGFGKYQGTWVPLNIAINLAEKFDVYQDLKPLF MBP1_EREGO IKDTHEKVQGGFGKYQGTWVPLDIARRLAQKFEVLEELRPLF MBP1_KLULA ITDTHEKVQGGFGKYQGTWIPLELASKLAEKFEVLDELKPLF MBP1_CANAL QTGIHEKVQGGYGKYQGTYVPLDLGAAIARNFGVYDVLKPIF MBP1_DEBHA QTGVHEKVQGGYGKYQGTYVPLDLGADIAKNFGVFDSLRPIF MBP1_YARLI QKGVHEKVQGGYGKYQGTWVPLERAREIATLYDVDSHLAPIF MBP1_SCHPO QIGAHEKVQGGYGKYQGTWVPFQRGVDLATKYKVDGIMSPIL MBP1_USTMA QKGIHEKVQGGYGKYQGTWIPLDVAIELAERYNIQGLLQPIT MBP1_ASPNI QKGVHEKVQGGYGKYQGTWIPLQEGRQLAERNNILDKLLPIF MBP1_ASPTE QKGVHEKVQGGYGKYQGTWIPLPEGRLLAERNNIIDKLRPIF MBP1_CRYNE LNGIHEKIQGGYGKYQGTWVPLDRGRDLAEQYGVGSYLSSVF MBP1_GIBZE QTGEHEKVQGGYGKYQGTWIKFERGLQVCRQYGVEELLRPLL MBP1_NEUCR QIGEHEKVQGGYGKYQGTWIPFERGLEVCRQYGVEELLSKLL MBP1_MAGGR QTGEHEKVQGGYGKYQGTWIKYERALEVCRQYGVEELLRPLL MBP1_ASPFU AAGEHEKVQGGYGKYQGTWVNYQRGVELCREYHVEELLRPLL MBP1_CHAGL QKDVHEKIQGGYGKYQGTWIPLEQGRALAQRNNIYDRLRPIF
It should be obvious to you by now how you can copy a string of amino acids from such an alignment and create a FASTA file. However we need to take a little detour: this detour brings us to the question of sequence numbers.
It is not straightforward at all how to number sequence in such a project. The "natural" way would be to start a sequential numbering from the start-codon of the full length protein and go sequentially from there. However imagine what would happen if a curator would discover that one of the splice-sites for a gene has been missed in automatic annotation. All of a sudden a corrected sequence would have a different length than the one that may have been used for earlier studies. Unfortunatlety, there is no mechanism (wouldn't it be nice!) that automatically goes back through the literature and your lab-journal and updates the revised sequence numbering... But there are other possible complications, regarding sequence numbers. The first residue of the CDD-APSES domain is not Residue 1 of the Mbp1 protein. The first residue of the 1MB1 FASTA file is the first residue of Mbp1 protein, but the last five residues are an artifiical His tag. Is H125 of 1MB1 the equivalent residue to R125 in MBP1_SACCE? The N-terminus of the Mbp1 crystal structure is disordered. The first residue in the structure is ASN 3, whereas the SEQRES records start with MET ... and so on. The take-home message is that a sequence number is nothing absolute, but something that makes sense only in a particular context. To emphasize this, we will write a FASTA header for our target sequence that lists the residues of the source sequence it correspond to. In terms of actual sequence numbering, we will adopt the numbering of the 1MB1 protein throughout to be able to consistently label particular amino acids.
Access the sequence of "your" organism's Mbp1 Orthologue at UniProt. (You can use the links I have provided in the table below).
| Organism | Uniprot Accession |
| Aspergillus fumigatus | Q4WGN2 |
| Aspergillus nidulans | Q5B8H6 |
| Aspergillus terreus | Q0CQJ5 |
| Candida albicans | Q5ANP5 |
| Candida glabrata | Q6FWD6 |
| Cryptococcus neoformans | Q5KHS0 |
| Debaryomyces hansenii | Q6BSN6 |
| Eremothecium gossypii | Q752H3 |
| Gibberella zeae | Q4IEY8 |
| Kluyveromyces lactis | P39679 |
| Magnaporthe grisea | Q3S405 |
| Neurospora crassa | Q7SBG9 |
| Saccharomyces cerevisiae | P39678 |
| Schizosaccharomyces pombe | P41412 |
| Ustilago maydis | Q4P117 |
| Yarrowia lipolytica | Q6CGF5 |
- Copy your organism's Mbp1 sequence from the alignment above. Then define the start- and end- sequence numbers of the target sequence relative to the full-length protein. Prepare a FASTA formatted file for the target sequence in your organism, giving it an appropriate header and include the sequence numbers. Refer to the Fallback data file if you are not sure about the format. (1 mark)
Your FASTA sequence should look similar to this:
>1MB1: Mbp1_SACCE 1..100 NQIYSARYSGVDVYEFIHSTG---SIMKRKKDDWVNATHILKAANFAKAKRTRILEKEV LKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLF
(2) Homology model
(2.1) SwissModel (1 mark)
Access the Swissmodel server at http://swissmodel.expasy.org . Navigate to the Alignment Interface.
- Copy from the alignment above the 1MB1 sequence and the sequence from your organism, and paste it into the form field. Refer to the Fallback Data file if you are not sure about the format.
- (You have to choose the format, and, if e.g. you choose a CLUSTAL format, you have to include a header line and a blank line. Other common problems uploading your alignment may include uploading a file that has not been saved as "text only" and periods i.e. "." in sequence names. Underscores appear to be safe.)
- Click submit and define your target and template sequence. For the template sequence define the coordinate file and chain. (In our case the coordinate file is
1MB1and the chain is "_" i.e. none, since the PDB file does not contain more than one chain.
- Click submit and request the construction of a homology model: Enter your e-mail address and check the button for Normal Mode, not "Swiss-PDB Viewer mode. (Important, since there will be problems with the output otherwise). Click submit. You should receive four files files by e-mail within half an hour or so. (1 mark)
(You do not need to submit any coordinate files with your assignment.)
In case you do not wish to submit the modelling job yourself, you can access the result files for the from the Fallback Data file.
(3) Model analysis
(3.1) The PDB file (1 mark)
Open your model coordinates in a text-editor (make sure you view the PDB file in a fixed-width font) and consider the following questions: (Alternatively, view the coordinates linked to the Fallback Data file.)
- What is the residue number of the first residue in the model? What should it be, based on the alignment? If the putative DNA binding region was reported to be residues 50-74 in the Mbp1 protein, which residues of the model correspond to that? (1 mark)
(3.2) first visualization (3 marks)
In assignment 2 you have already studied the 1MB1 coordinate file and compared it to your organism's Mbp1 APSES domain, Since a homology model inherits its structural details from the template, the model should look very similar to the original structure but contain the sequence of the target.
- Save the attachment of your model coordinates to your harddisk and visualize it in RasMol. (Alternatively, copy and save the coordinates from the Fallback Data file to your harddisk.) Make an informative view, divergent stereo and paste it into your assignment. (3 marks)
(3.3) modeling a DNA ligand (4 marks)
The really interesting question we could begin to address with our model is how sequence variation might be converted into changing DNA recognition sites, and then lead to changed cognate DNA binding sequences. But in order to address this, we would need to add a plausible model for a bound DNA molecule to our model.
Since there is currently no software available that would accurately model such a complex from first principles, we will base this on homology modeling as well. This means we need to find a similar structure for which the complex structure is known. However, you may remember from the third assignment that the APSES domains in fungi seem to be a relatively small family. And there is no structure available of a protein-DNA complex. Now what?
Remember that homologous sequences can have diverged to the point where their sequence similarity is no longer recognizable, however their structure may be quite well conserved. Thus if we could find similar structures in the PDB, these might provide us with some plausible hypotheses for how DNA is bound by APSES domains. We thus need a tool similar to BLAST, but not for the purpose of sequence alignment, but for structure alignment. A kind of BLAST for structures.
However, very similar to BLAST, we might not want to search with the entire protein, if all we are interested in is a subdomain that binds to DNA. Attempting to match all structural elements in addition to the ones we are actually interested in is likely to make the search less specific - we would find false positives that are similar to some irrelevant part of our structure. However, defining too small of a subdomain would also lead to a loss of specificity: in the extreme it is easy to imagine that the search for e.g. a single helix would retrieve very many hits that would be quite meaningless. The arrangement of the residues from 50 to 74 that we have already discussed in Assignment 2 suggests that the compact subdomain from 36 to 76 (see the image above) might be a useful structure to search with: it contains the residues we are interested in and enough of connected secondary structure elements to be structurally meaningful.
At the NCBI, VAST is a search tool for structural similarity search tool for this purpose. Unfortunately it does not seem to be able to handle a query with such a structural subdomain (the process did not finish after several days) but at least you can get a list of structural neighbors of the 1MB1 full-length template structure, by entering the PDB ID in a small form field on the VAST home page, and then clicking on the colored bar labeled "Chain" on the MMDB structure summary page. This precomputed page for the 1MB1 structure shows a number of diverse proteins matching to various helices and strands of the structure.
At the EBI there are a number of very well designed structure analysis tools linked off the Structural Analysis page. As part of its MSD Services, the SSM (Secondary Structure Matching service) provides a well thought out interface for searching files from the PDB or uploading coordinates.
After uploading the coordinates for residues 36 to 76 of the 1MB1 structure running the search and sorting the results by alignment length, the top hits include a number of nucleotide binding proteins such as a replication terminator (1F4K), the LexA repressor (1MVD) and a "Winged Helix" protein (1KQ8). These are all members of a much larger superfamily, the "winged helix" DNA binding domains (CATH 1.10.10.10), of which hundreds of structures have been solved. They represent one branch of the tree of helix-turn-helix (HTH) DNA binding modules. (A recent review on HTH proteins is linked from the resources section at the bottom of this page). Winged Helix domains typically bind their cognate DNA with a "recognition helix" which precedes the beta hairpin and binds into the major groove; additional stabilizing interactions are provided by the edge of the beta strand binding into the minor groove.
This is good news: once we have determined that the APSES domain is actually an example of a larger group of transcription factors, we can pick one of these for which a DNA complex structure is known. I have picked one such structure from the list of hits that were returned by SSM: it is the Elk-1 transcription factor.
Now all that is left to do is to bring the DNA molecule into the correct orientation for our model and then to combine the two files. We need to superimpose the Elk-1 protein/DNA complex onto our model.
- Structure superposition
There are quite a number of superposition servers available on the Web, a remarkably comprehensive overview can be found in Wikipedia. However, overengineering and black-box mentality makes our task more difficult than it need be: most tools do not allow users to specify particular alignment zones but attempt to automatically define the zones of residues to be supoerimposed according to some geometric target function. Almost none return the actual rotation matrix and translation vector that is used for the superposition. And almost none transform the coordinates of heteratoms such as solvent, ligands or DNA molecules along with the protein coordinates. An exception that I have found to be very useable is the Local-Global Alignment server (LGA), written by Adam Zemla. The procedure is quite straightforward:
- Define the structure to be rotated (1DUX in this case). This is a dimer, so download the file from the PDB and manually edit to contain only DNA chains A and B and protein chain C.
- Define the structure to be held constant (1MB1 in this case). Download from PDB.
- Use the "browse" option to define both files as input on the LGA inpput form
- Use the option to have both coordinate sets included in your output:
-o2 - Submit
The results arrive per e-mail. I have linked the resulting PDB file to the Fallback Data page. If you run this analysis on your own, you may want to review the types of edits the edits I made to the PDB file to get it displayed correctly in Rasmol.
- Save the superimposed coordinates in a file, open and view in Rasmol and note how well the "recognition helix" and adjacent beta strands superimpose! (Alternatively, copy and save the coordinates from the c to your harddisk.) Make an informative view, divergent stereo and paste it into your assignment. (4 marks)
(4) Summary of Resources
- Links
-
- Review (PDF, restricted) Manuel Peitsch on Homology Modeling
- Review (PDF, restricted) Aravind et al. Helix-turn-helix domains (background reading, not required reading)
- Assigned Organisms
- PDB file format
- Wikipedia on Structural Superposition (although the article is called "Structural Alignment")
[End of assignment]
If you have any questions at all, don't hesitate to mail me at boris.steipe@utoronto.ca or post your question to the Course Mailing List