Difference between revisions of "Lecture 02"
Jump to navigation
Jump to search




| Line 69: | Line 69: | ||
<tt> H = A C T (not '''G''')</tt><br> | <tt> H = A C T (not '''G''')</tt><br> | ||
<tt> V = G C A (not '''T''')</tt><br> | <tt> V = G C A (not '''T''')</tt><br> | ||
| − | <tt> N = A G C T (a'''n'''y) | + | <tt> N = A G C T (a'''n'''y)</tt> |
]] | ]] | ||
| + | |||
======Slide 009====== | ======Slide 009====== | ||
[[Image:L02_s009.jpg|frame|none|Lecture 02, Slide 009<br> | [[Image:L02_s009.jpg|frame|none|Lecture 02, Slide 009<br> | ||
Revision as of 13:50, 19 September 2007
Update Warning! This page has not been revised yet for the 2007 Fall term. Some of the slides may be reused, but please consider the page as a whole out of date as long as this warning appears here.
(Previous lecture) ... (Next lecture)
The Sequence Abstraction
...
Add:
- Exercises
- Summary points
Lecture Slides
Slide 001
Slide 002
Slide 003

Lecture 02, Slide 003
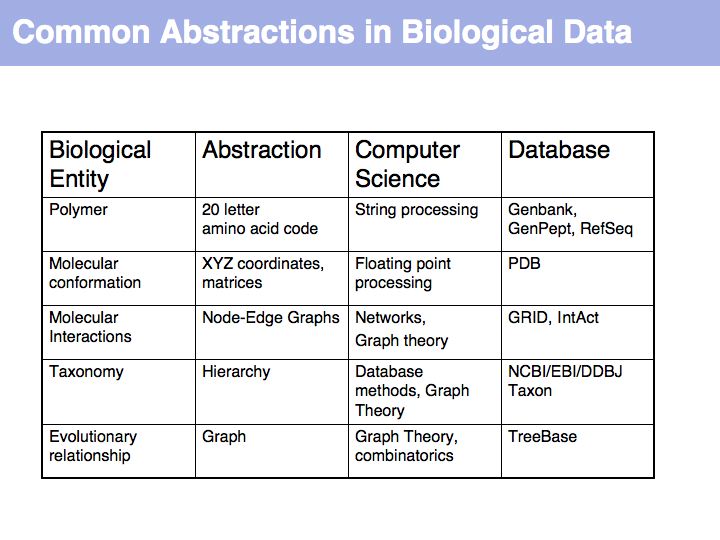
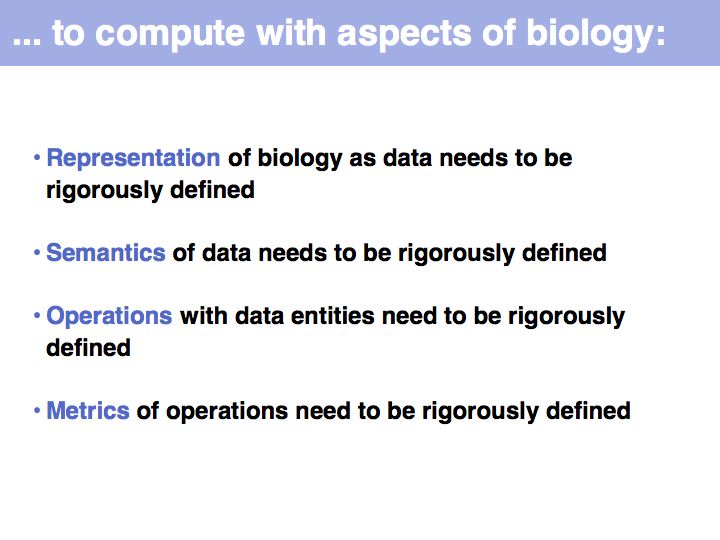
In order to make biology computable, we have to clarify the concepts we are working with. This is useful even beyond the requirements of bioinformatics. It is an exercise in clarifying the conceptual foundations of biology itself. In many instances, definitions in current, common use are deficient, either because our current state of knowledge has gone beyond the original ideas we were trying to subsume with a term (e.g. gene, or pathway), or because an inconsistent formal and colloquial meaning of terms lead to ambiguities (e.g. function), or because the technical meaning of terms is poorly understood and generally misused (e.g. homology).
In order to make biology computable, we have to clarify the concepts we are working with. This is useful even beyond the requirements of bioinformatics. It is an exercise in clarifying the conceptual foundations of biology itself. In many instances, definitions in current, common use are deficient, either because our current state of knowledge has gone beyond the original ideas we were trying to subsume with a term (e.g. gene, or pathway), or because an inconsistent formal and colloquial meaning of terms lead to ambiguities (e.g. function), or because the technical meaning of terms is poorly understood and generally misused (e.g. homology).
Slide 004
Slide 005

Lecture 02, Slide 005
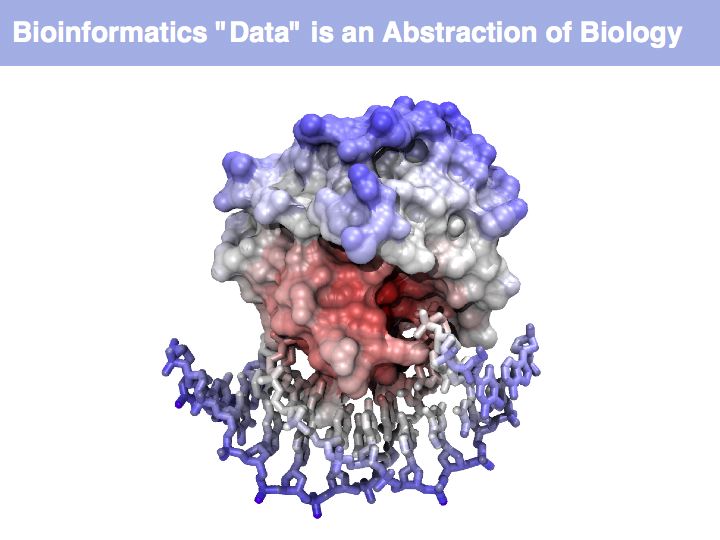
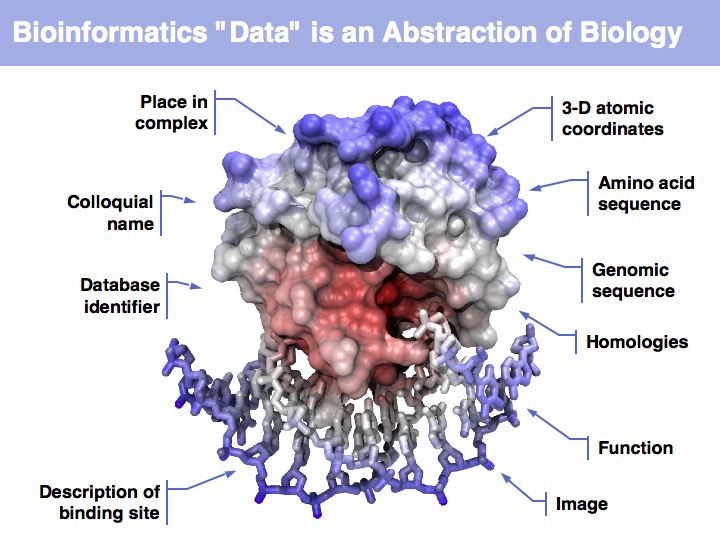
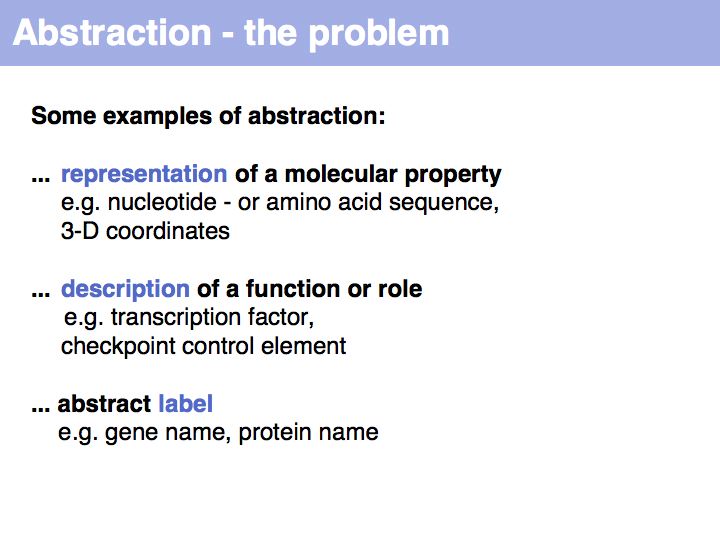
Asking about the best is an ill-posed question if the purpose is not specified. The best for what? There are many possible abstractions, each serving different purposes. Each of the abstractions above can serve as a representation of the biomolecule, each one emphasizes a different perspective and can serve a different purpose.
Asking about the best is an ill-posed question if the purpose is not specified. The best for what? There are many possible abstractions, each serving different purposes. Each of the abstractions above can serve as a representation of the biomolecule, each one emphasizes a different perspective and can serve a different purpose.
Slide 006

Lecture 02, Slide 006
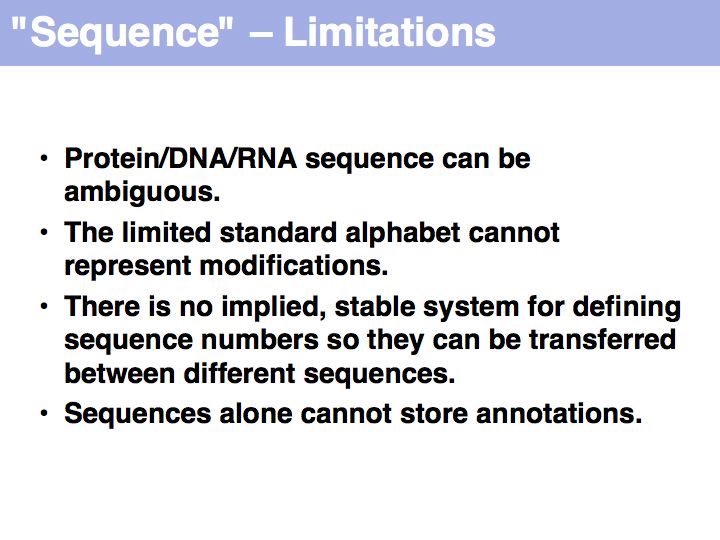
Working with abstractions implies we are no longer manipulating the biological entity, but it's representation. This distinction becomes crucial, when we start computing with representations to infer facts about the original entities! Inferences must be related back to biology! Common problems include that the abstraction may not be rich enough to capture the property we are investigating (e.g. one-letter sequence codes cannot represent amino acid modifications or sequence numbers), or the abstraction may be ambiguous (e.g. one protein may have more than one homologue in a related organism, thus the relationship between gene IDs is ambiguous) or the abstraction may not be unique (e.g. one protein may have more than one function, the same protein name may refer to unrelated proteins in different species).
Working with abstractions implies we are no longer manipulating the biological entity, but it's representation. This distinction becomes crucial, when we start computing with representations to infer facts about the original entities! Inferences must be related back to biology! Common problems include that the abstraction may not be rich enough to capture the property we are investigating (e.g. one-letter sequence codes cannot represent amino acid modifications or sequence numbers), or the abstraction may be ambiguous (e.g. one protein may have more than one homologue in a related organism, thus the relationship between gene IDs is ambiguous) or the abstraction may not be unique (e.g. one protein may have more than one function, the same protein name may refer to unrelated proteins in different species).
Slide 007
Slide 008

Lecture 02, Slide 008
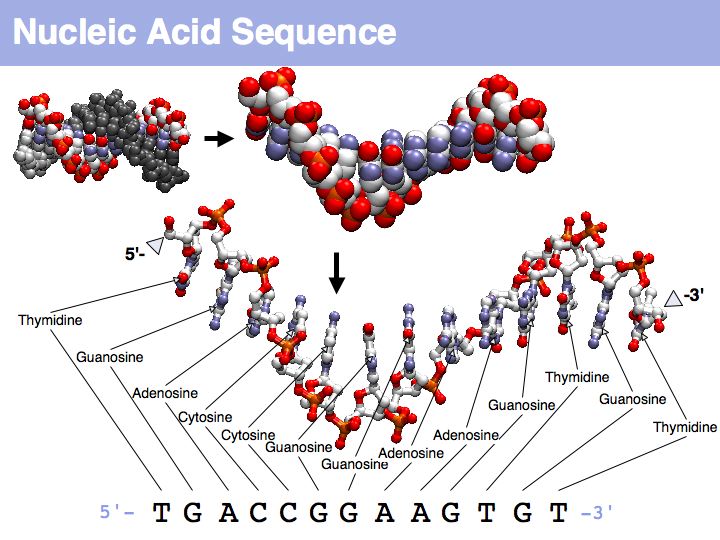
Nucleic acids can form heterocopolymers as DNA or RNA, thus their structural formula can be described (to a close approximation) simply by listing the nucleotide bases in a defined order. A one-letter code has been defined as a shorthand notation for this. By convention, DNA or RNA sequence is written in the direction from the bond with the 5' carbon of the ribose (or deoxyribose), to the bond with the 3'-carbon. Since the 5'-carbon carries a free 5'-phosphate at the terminus of the polynucleotide and the 3'-carbon carries a free OH, this direction happens to be the same direction that nucleic acid polymers are replicated by the DNA-polymerase: a phosphate of a single nucleotide is attached to the free -OH of the polynucleotide. This direction is also the direction of transcription of the RNA polymerase (for the same reason) and (incidentally) the direction of translation by the ribosome. Due to base-pair complementarity, only one strand of a double stranded sequence needs to be recorded since the sequence of the complementary strand is implied: it is simply the reverse complement. The one letter abbreviations are defined by the International Union of Pure and Applied Chemistry (IUPAC) as follows:
A = adenine
C = cytosine
G = guanine
T = thymine
R = G A (purine)
Y = T C (pyrimidine)
K = G T (keto)
M = A C (amino)
S = G C (strong bonds)
W = A T (weak bonds)
B = G T C (not A)
D = G A T (not C)
H = A C T (not G)
V = G C A (not T)
N = A G C T (any)
Nucleic acids can form heterocopolymers as DNA or RNA, thus their structural formula can be described (to a close approximation) simply by listing the nucleotide bases in a defined order. A one-letter code has been defined as a shorthand notation for this. By convention, DNA or RNA sequence is written in the direction from the bond with the 5' carbon of the ribose (or deoxyribose), to the bond with the 3'-carbon. Since the 5'-carbon carries a free 5'-phosphate at the terminus of the polynucleotide and the 3'-carbon carries a free OH, this direction happens to be the same direction that nucleic acid polymers are replicated by the DNA-polymerase: a phosphate of a single nucleotide is attached to the free -OH of the polynucleotide. This direction is also the direction of transcription of the RNA polymerase (for the same reason) and (incidentally) the direction of translation by the ribosome. Due to base-pair complementarity, only one strand of a double stranded sequence needs to be recorded since the sequence of the complementary strand is implied: it is simply the reverse complement. The one letter abbreviations are defined by the International Union of Pure and Applied Chemistry (IUPAC) as follows:
A = adenine
C = cytosine
G = guanine
T = thymine
R = G A (purine)
Y = T C (pyrimidine)
K = G T (keto)
M = A C (amino)
S = G C (strong bonds)
W = A T (weak bonds)
B = G T C (not A)
D = G A T (not C)
H = A C T (not G)
V = G C A (not T)
N = A G C T (any)
Slide 009

Lecture 02, Slide 009
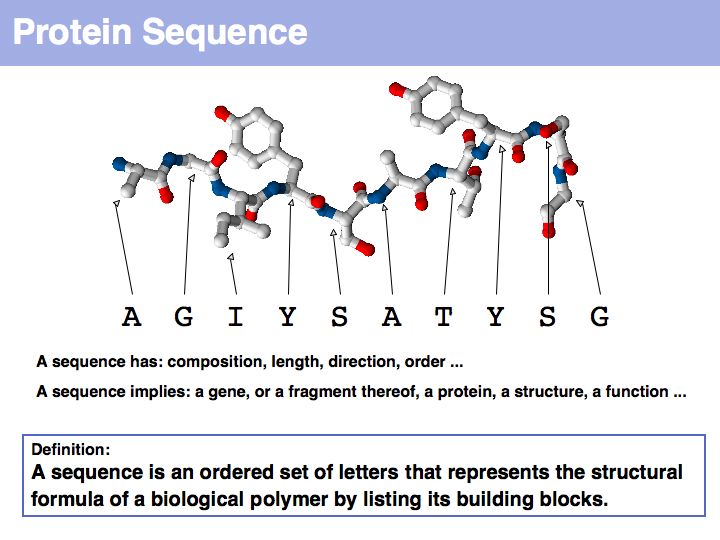
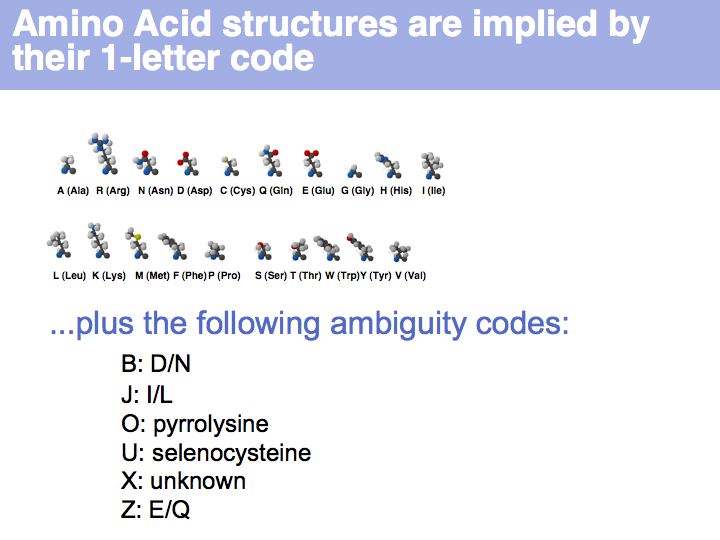
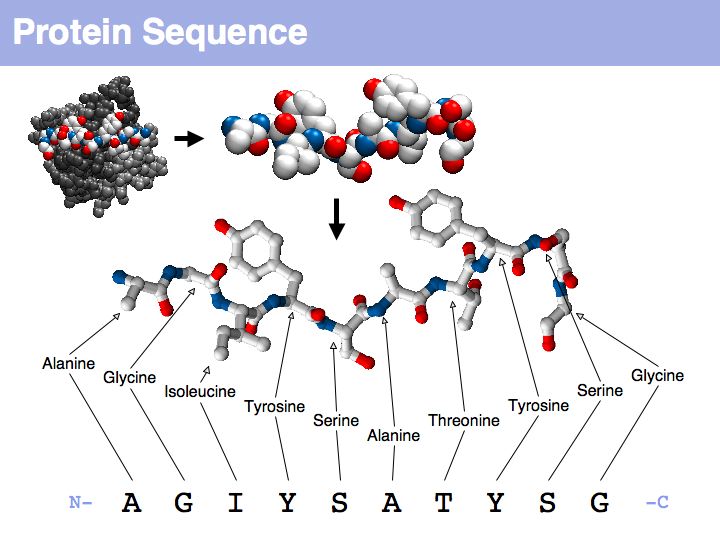
Proteins are amino acid heterocopolymers, thus their structural formula can be described (to a close approximation) simply by listing their constituent amino acid residues in a defined order. The one-letter code is a shorthand notation for this. By convention, protein sequence is written from aminoterminus (N-) to carboxyterminus (C-). This happens to be the same direction in which the protein is synthesized on the ribosome.
Proteins are amino acid heterocopolymers, thus their structural formula can be described (to a close approximation) simply by listing their constituent amino acid residues in a defined order. The one-letter code is a shorthand notation for this. By convention, protein sequence is written from aminoterminus (N-) to carboxyterminus (C-). This happens to be the same direction in which the protein is synthesized on the ribosome.
Slide 010
Slide 011
Slide 012

Lecture 02, Slide 012
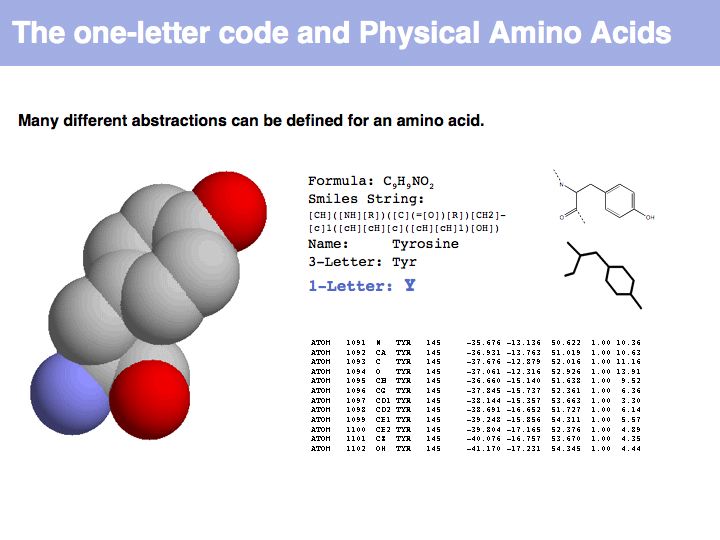
An amino acid is a molecule. A number of abstractions are in common use for this molecule: its chemical formula simply describes the elemental composition, a so called SMILES string (see also here) captures bonding topology as well, its information is equivalent to a chemical graph. A set of records of 3D coordinates can describe the three-dimensional conformation, this can in turn be displayed in a number of different image options - like a simple line drawing or a set of spheres, color coded by element, with relative sizes corresponding to the elements' Van der Waals radii.
An amino acid is a molecule. A number of abstractions are in common use for this molecule: its chemical formula simply describes the elemental composition, a so called SMILES string (see also here) captures bonding topology as well, its information is equivalent to a chemical graph. A set of records of 3D coordinates can describe the three-dimensional conformation, this can in turn be displayed in a number of different image options - like a simple line drawing or a set of spheres, color coded by element, with relative sizes corresponding to the elements' Van der Waals radii.
Slide 013
Slide 014
Slide 015
Slide 016

Lecture 02, Slide 016
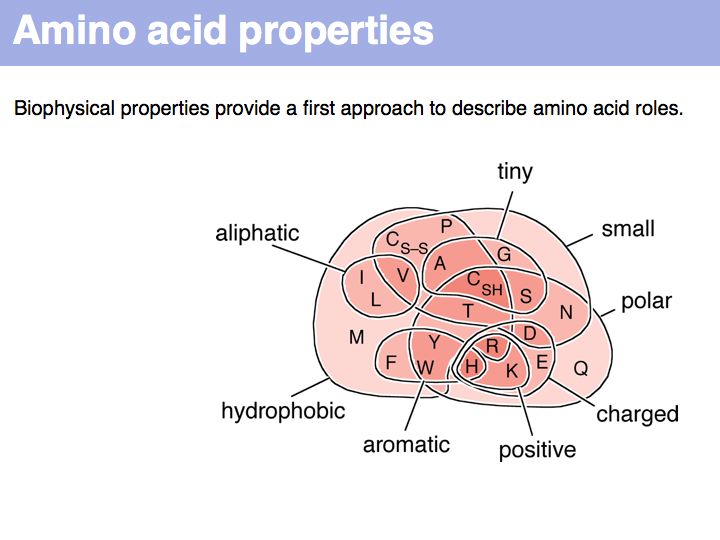
Hydrophobic amino acids - the group FAMILYVW - are found predominantly in the core of a protein, small amino acids such as GASC are often found in turns, charged amino acid sidechains - (+):KRH and (-)DE - are almost exclusively found on the surface; the energetic requirements for desolvation of the sidechain makes their incorporation into the core unfavourable.
Hydrophobic amino acids - the group FAMILYVW - are found predominantly in the core of a protein, small amino acids such as GASC are often found in turns, charged amino acid sidechains - (+):KRH and (-)DE - are almost exclusively found on the surface; the energetic requirements for desolvation of the sidechain makes their incorporation into the core unfavourable.
Slide 017

Lecture 02, Slide 017
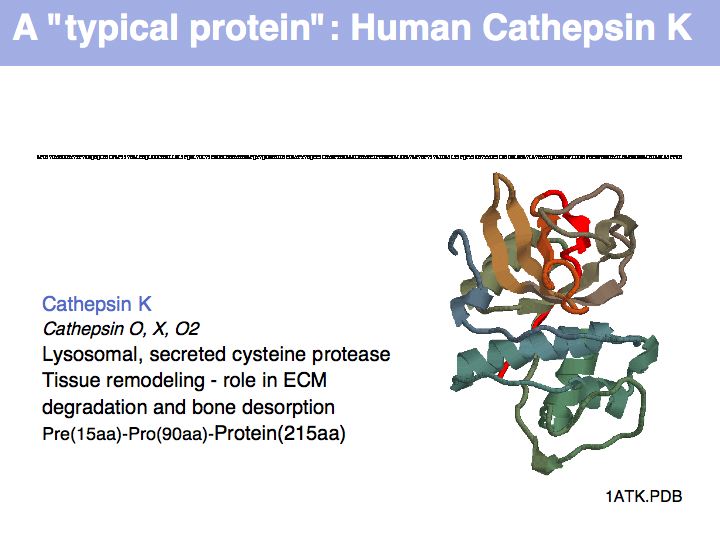
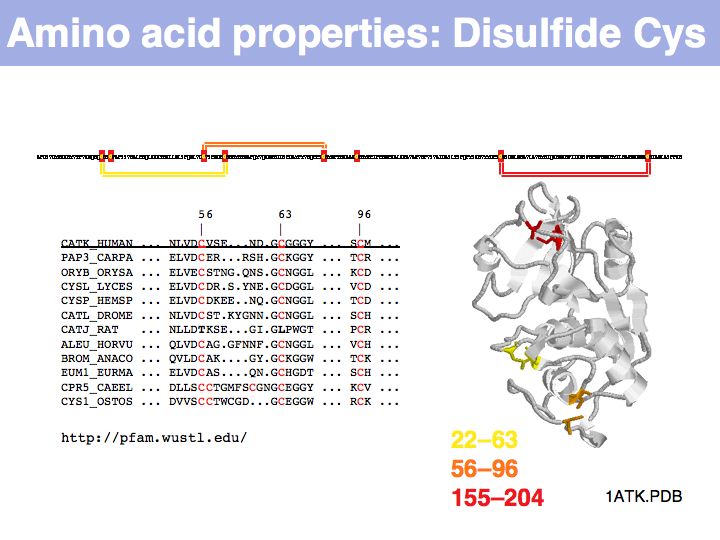
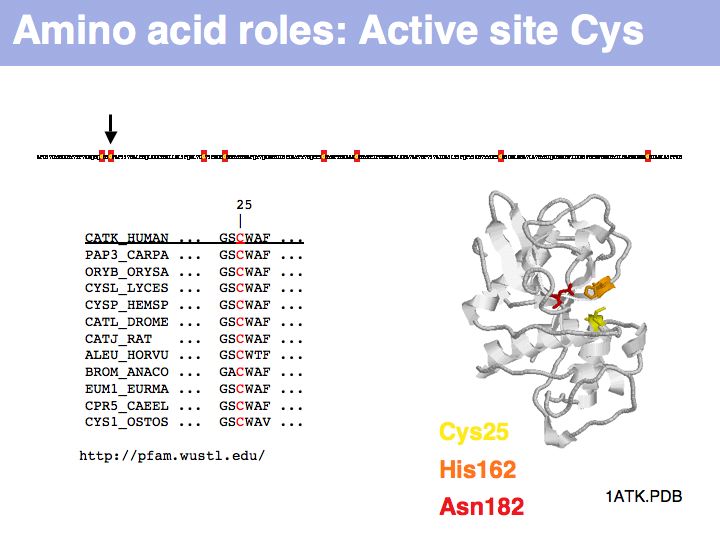
Cysteine can take on a number of different roles, depending on its context. Here, cysteine forms part of the active site, it is the nucleophile in the catalytic triad C-H-N; cathepsin is thuss an example of a cysteine-protease. This particular cysteine is absolutely conserved in related proteins.
Cysteine can take on a number of different roles, depending on its context. Here, cysteine forms part of the active site, it is the nucleophile in the catalytic triad C-H-N; cathepsin is thuss an example of a cysteine-protease. This particular cysteine is absolutely conserved in related proteins.
Slide 018
Slide 019

Lecture 02, Slide 019
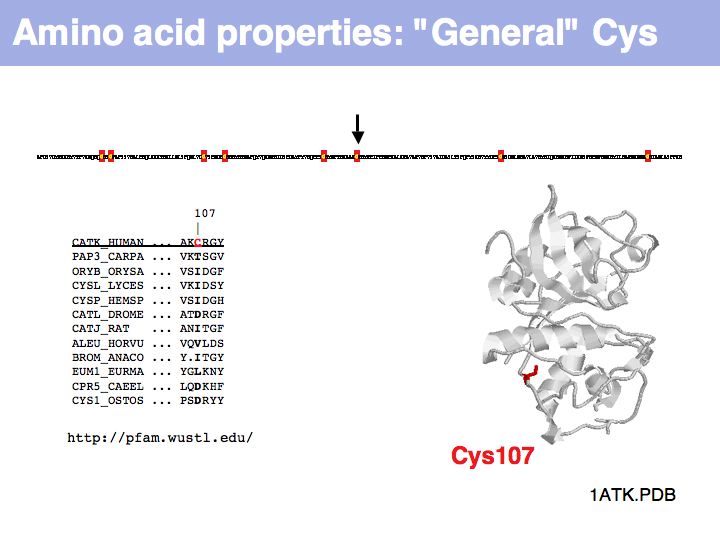
Cysteine can also be found in a very general role, simply as a somewhat polar, small residue. This general role is infrequent in secreted proteins since it can interfere with the formation of the correct disulfide topology and generally makes the protein sensitive to oxidation. Such cysteines are poorly conserved in related proteins.
Cysteine can also be found in a very general role, simply as a somewhat polar, small residue. This general role is infrequent in secreted proteins since it can interfere with the formation of the correct disulfide topology and generally makes the protein sensitive to oxidation. Such cysteines are poorly conserved in related proteins.
Slide 020

Lecture 02, Slide 020
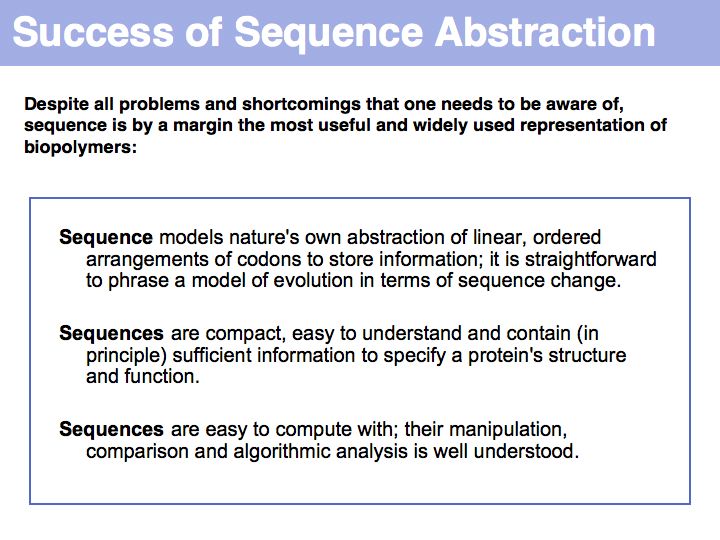
Sequence is the most important abstraction in biology; you need to know your amino acids in order to relate a sequence back to the biopolymer. Required knowledge is: the structural formula, the one- and three- letter codes and key properties (such as charge, relative size, polarity) for all 20 proteinogenic amino acids.
Sequence is the most important abstraction in biology; you need to know your amino acids in order to relate a sequence back to the biopolymer. Required knowledge is: the structural formula, the one- and three- letter codes and key properties (such as charge, relative size, polarity) for all 20 proteinogenic amino acids.
Slide 021

Lecture 02, Slide 021
In Shakespeare's classic tragedy of romantic love and family allegiance, Juliet encapsulates the play's central struggle in this phrase by claiming that Romeo's family name is an artificial and meaningless convention. Just like in the world of the sequence abstraction, this is only partially true: the problems are not just based in the fact that Romeo is called a Montague, but that he is in fact a member of that family. Pure, abstract labels, of course, can be arbitrarily replaced.
In Shakespeare's classic tragedy of romantic love and family allegiance, Juliet encapsulates the play's central struggle in this phrase by claiming that Romeo's family name is an artificial and meaningless convention. Just like in the world of the sequence abstraction, this is only partially true: the problems are not just based in the fact that Romeo is called a Montague, but that he is in fact a member of that family. Pure, abstract labels, of course, can be arbitrarily replaced.
Slide 022
Slide 023
Slide 024
Slide 025
Slide 026
Slide 027

Lecture 02, Slide 027
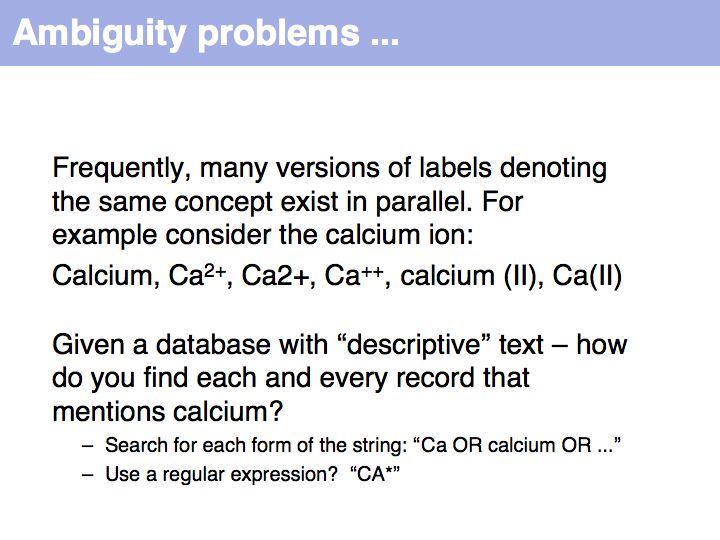
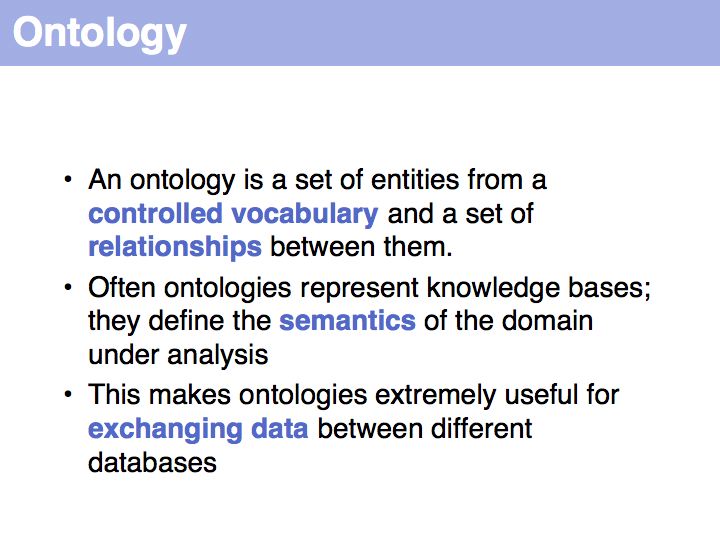
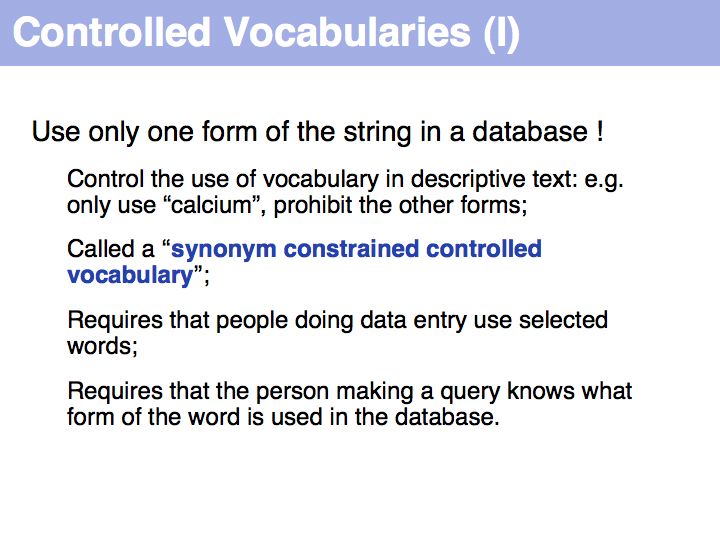
Often synonym constrained controlled vocabularies are presented as option lists on Web forms. If the CV is too long for this approach to be practical, defining the correct form becomes a challenge. In well engineered databases a lot of effort is spent on this task; typically a large dictionary of synonym mappings is employed in some form.
Often synonym constrained controlled vocabularies are presented as option lists on Web forms. If the CV is too long for this approach to be practical, defining the correct form becomes a challenge. In well engineered databases a lot of effort is spent on this task; typically a large dictionary of synonym mappings is employed in some form.
Slide 028

Lecture 02, Slide 028
NB. The situation that a unique property of an entity can be concisely described is the ideal case: in that case the identifier captures the most fundamental aspect of the molecule. For calcium, the element does not just have the atomic number 20, it is the element with 20 protons. Similarly oxytocin does not just have the sequence CYIQNCPLG, it is the peptide with that sequence. However, these are favourable exceptions and more commonly unique, abstract labels - such as identifiers - have to be defined.
NB. The situation that a unique property of an entity can be concisely described is the ideal case: in that case the identifier captures the most fundamental aspect of the molecule. For calcium, the element does not just have the atomic number 20, it is the element with 20 protons. Similarly oxytocin does not just have the sequence CYIQNCPLG, it is the peptide with that sequence. However, these are favourable exceptions and more commonly unique, abstract labels - such as identifiers - have to be defined.
Slide 029
Slide 030
Slide 031
Slide 032
Slide 033

Lecture 02, Slide 033
Read more about the FASTA format.
Read more about the FASTA format.
Slide 034
Slide 035
Slide 036
Slide 037
Slide 038
Slide 039
Slide 040
Slide 041
Slide 042
Slide 043
Slide 044
Slide 045
Slide 046
Slide 047
Slide 048
Slide 049
Slide 050
Slide 051
Slide 052
Slide 053
Slide 054
Slide 055

Lecture 02, Slide 055
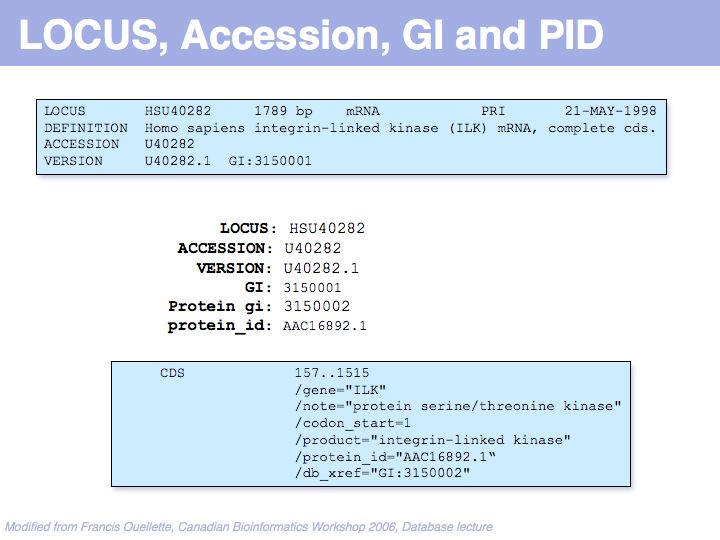
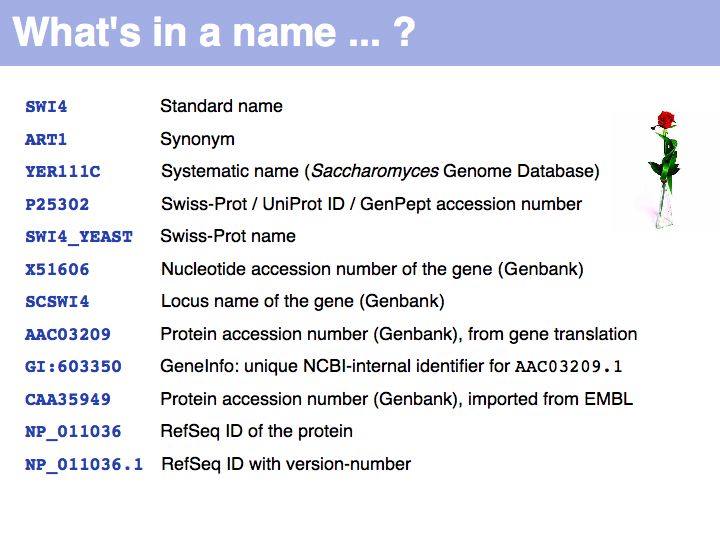
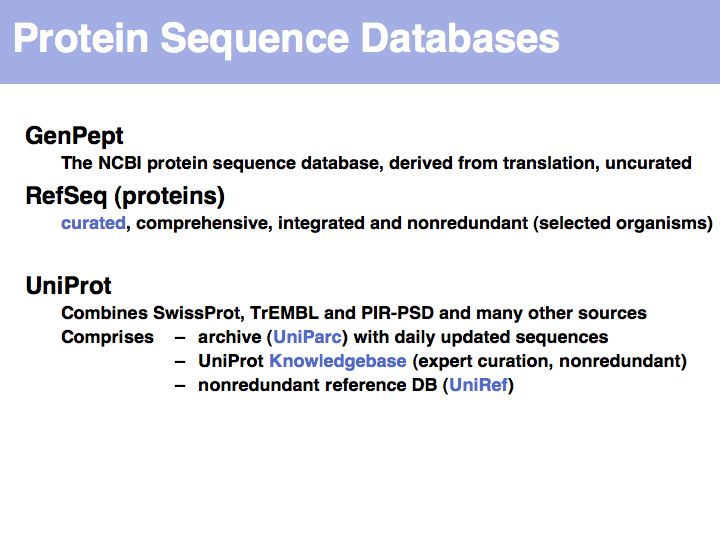
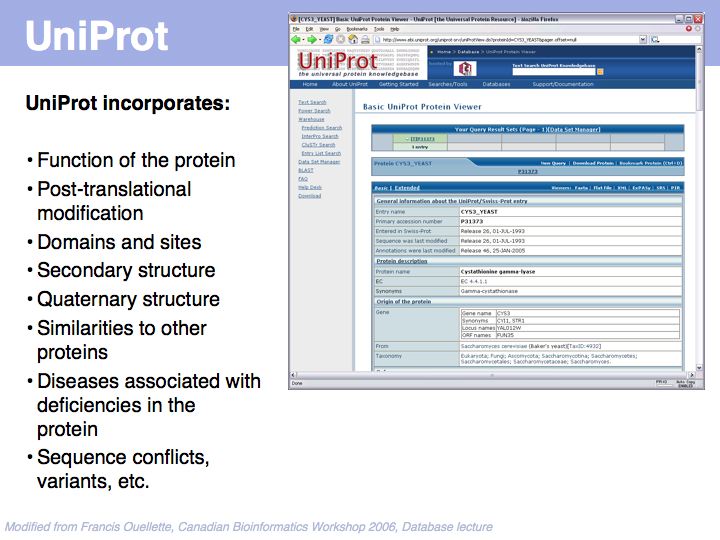
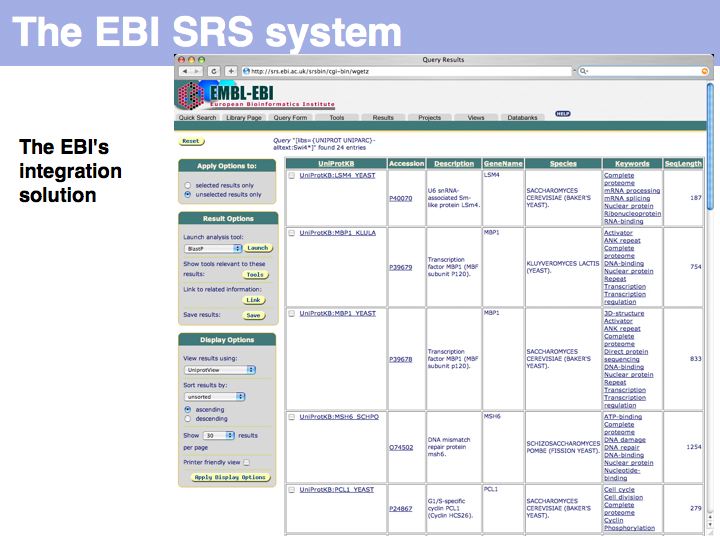
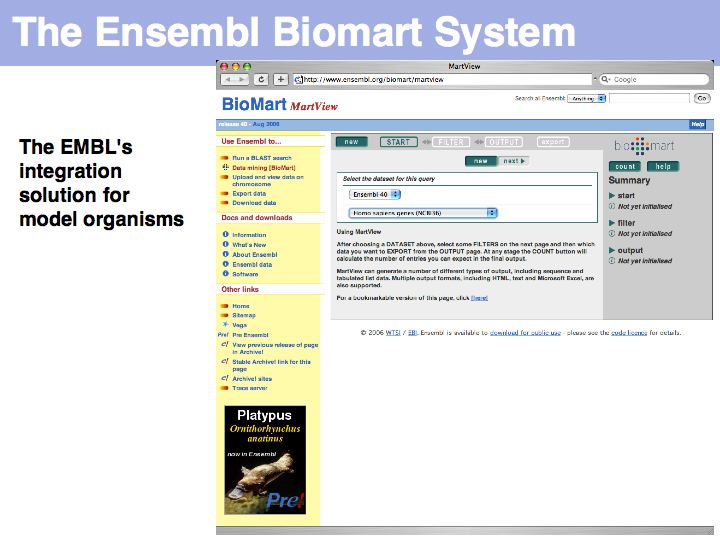
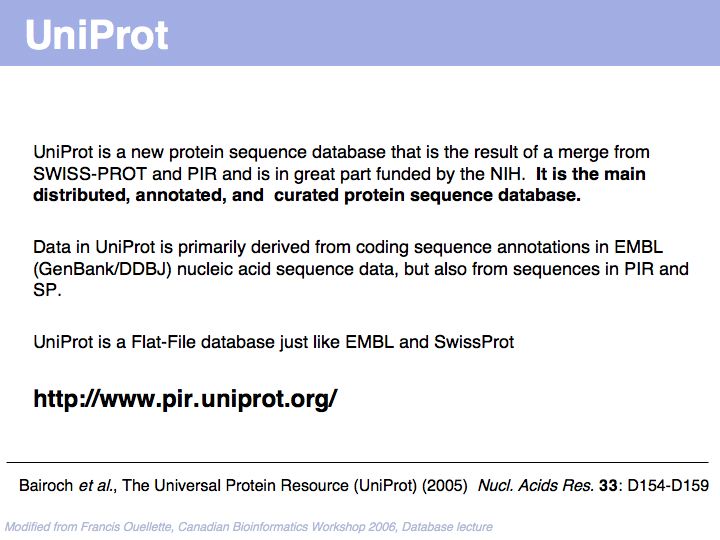
UniProt is arguably the more comprehensive resource, however it is not integrated with the GenBank world, although that would be reasonably straightforward to do. For example, neither does the NCBI record for the Swi4 protein contain a reference to the UniProt accession number, nor does the UniProt record for the same protein contain an NCBI accession number or a GI. National database politics are not always in the best interest of the worldwide scientific community.
UniProt is arguably the more comprehensive resource, however it is not integrated with the GenBank world, although that would be reasonably straightforward to do. For example, neither does the NCBI record for the Swi4 protein contain a reference to the UniProt accession number, nor does the UniProt record for the same protein contain an NCBI accession number or a GI. National database politics are not always in the best interest of the worldwide scientific community.