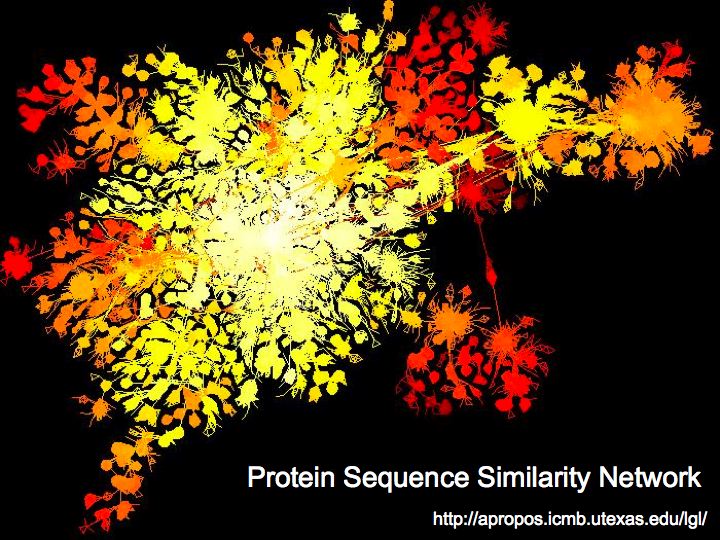
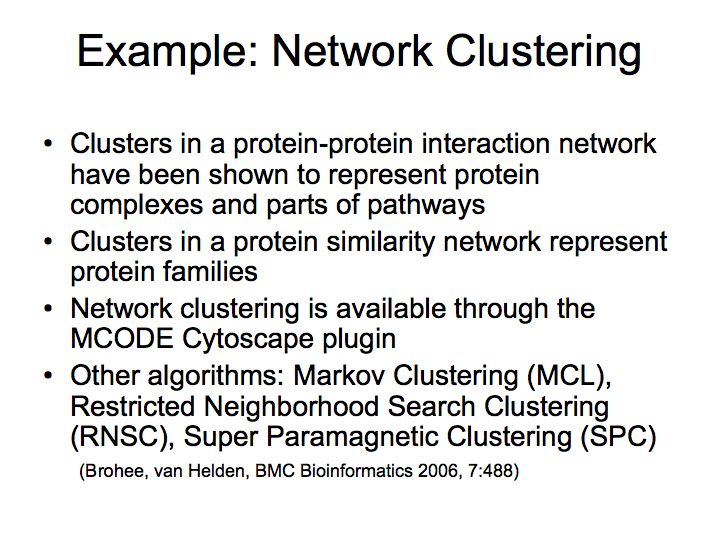
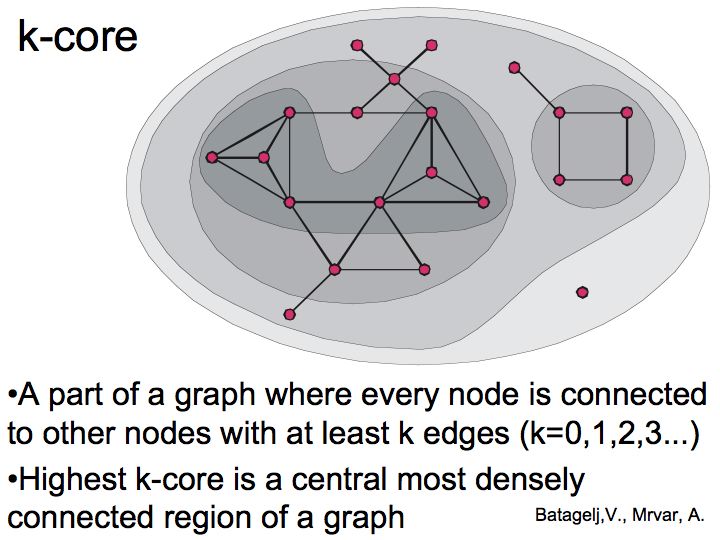
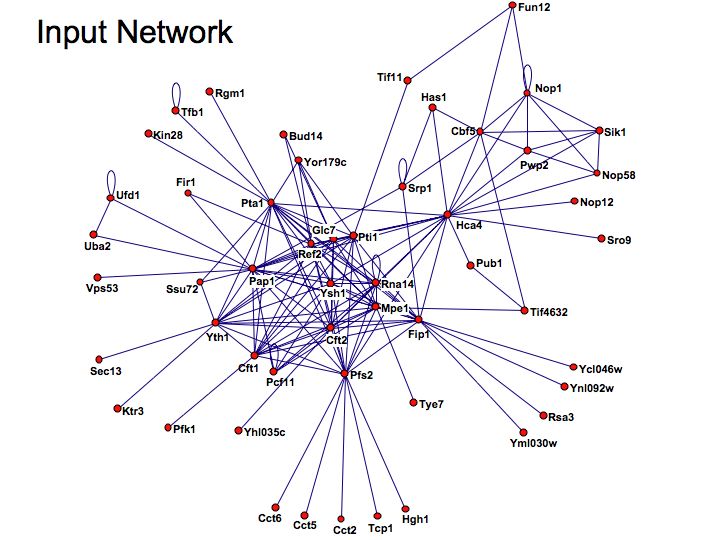
Difference between revisions of "Lecture 23"
Jump to navigation
Jump to search


| Line 1: | Line 1: | ||
| − | <div style="padding: 5px; background: #FF4560; border:solid 2px #000000;"> | + | |
| + | |||
| + | |||
| + | <!-- div style="padding: 5px; background: #FF4560; border:solid 2px #000000;"> | ||
'''Update Warning!''' | '''Update Warning!''' | ||
| − | This page has not been revised yet for the | + | This page has not been revised yet for the 2008 Fall term. |
| − | </div> | + | Some of the slides will probably be reused, but please consider the page as a whole out of date |
| + | as long as this warning appears here. Also, the lectures may be taught in a different sequence. | ||
| + | </div --> | ||
| | ||
| + | | ||
| + | __NOTOC__ | ||
| + | <small>[[Lecture_22|(Previous lecture)]] ... [[Lecture_24|(Next lecture)]]</small> | ||
| + | |||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 2px; background: #879BFA; border:solid 1px #AAAAAA;"> | ||
| + | ==Computational analysis of pathways and interaction networks== | ||
| + | </div><br> | ||
| | ||
| + | BCH441 Guest lecture: | ||
| + | |||
| + | ;[http://baderlab.org Gary Bader] | ||
| + | :Terrence Donnelly Center for Cellular and Biomolecular Research (DCCBR)<br> | ||
| + | :BBDMR, MedGen, DCS, SLRI<br> | ||
| + | :University of Toronto<br> | ||
| + | |||
| + | |||
| + | Book chapter:<br> | ||
| + | :Intermolecular Interactions and Biological Pathways<br> | ||
| + | :Gary D. Bader, Anton J. Enright<br> | ||
| + | :in Bioinformatics 3E Eds. Andreas D. Baxevanis and B.F.Francis Ouellette<br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ====Links summary==== | ||
| + | </div><br> | ||
| + | *[http://baderlab.org/Publications Bader Lab - publications]<br> | ||
| + | *[http://discover.nci.nih.gov/kohnk/interaction_maps.html Kurt Kohn's signalling maps at the NIH]<br> | ||
| + | *[http://sugp.caltech.edu/endomes Sea urchin development gene regulatory pathways]<br> | ||
| + | *[http://pathguide.org '''Pathguide''': the pahway resource list]<br> | ||
| + | *[http://cancer.cellmap.org The Cancer Cell Map]<br> | ||
| + | *[http://www.pathwaycommons.org Pathway Commons]<br> | ||
| + | *[http://cytoscape.org Cytoscape - biological graph visualization and analysis]<br> | ||
| + | *[http://baderlab.org/Software/MCODE MCODE - a cytoscape plugin]<br> | ||
| + | *[http://baderlab.org/Home?action=AttachFile&do=get&target=InnerCellLife.mov The Inner Cell Life movie - download link (large!)]<br> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #879BFA; border:solid 1px #AAAAAA;"> | ||
| + | ==Lecture slides== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ===What are pathways and networks?=== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | ======Slide 004====== | ||
| + | [[Image:23_slide004.jpg|frame|none|Lecture 23, Slide 004<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 005====== | ||
| + | [[Image:23_slide005.jpg|frame|none|Lecture 23, Slide 005<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 006====== | ||
| + | [[Image:23_slide006.jpg|frame|none|Lecture 23, Slide 006<br> | ||
| + | Ref: Pathway information for systems biology, Cary MP, Bader GD, Sander C, ''FEBS Lett.'' 2005 Mar 21; '''579'''(8):1815-20 (Available from [http://baderlab.org/Publications http://baderlab.org/Publications]) | ||
| + | ]] | ||
| + | ======Slide 007====== | ||
| + | [[Image:23_slide007.jpg|frame|none|Lecture 23, Slide 007<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 008====== | ||
| + | [[Image:23_slide008.jpg|frame|none|Lecture 23, Slide 008<br> | ||
| + | [http://discover.nci.nih.gov/kohnk/interaction_maps.html http://discover.nci.nih.gov/kohnk/interaction_maps.html] | ||
| + | ]] | ||
| + | ======Slide 009====== | ||
| + | [[Image:23_slide009.jpg|frame|none|Lecture 23, Slide 009<br> | ||
| + | [http://sugp.caltech.edu/endomes http://sugp.caltech.edu/endomes] | ||
| + | ]] | ||
| + | ======Slide 010====== | ||
| + | [[Image:23_slide010.jpg|frame|none|Lecture 23, Slide 010<br> | ||
| + | Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. ''Nature'' 2002 Jan 10; '''415'''(6868): 180-183 (Available from [http://baderlab.org/Publications http://baderlab.org/Publications]) | ||
| + | ]] | ||
| + | ======Slide 011====== | ||
| + | [[Image:23_slide011.jpg|frame|none|Lecture 23, Slide 011<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 012====== | ||
| + | [[Image:23_slide012.jpg|frame|none|Lecture 23, Slide 012<br> | ||
| + | A "pathway" is as arbitrary as our assignment of protein function, since proteins can certainly have more than one function, and be responsible for more than one phenotype | ||
| + | ]] | ||
| + | ======Slide 013====== | ||
| + | [[Image:23_slide013.jpg|frame|none|Lecture 23, Slide 013<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 014====== | ||
| + | [[Image:23_slide014.jpg|frame|none|Lecture 23, Slide 014<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 015====== | ||
| + | [[Image:23_slide015.jpg|frame|none|Lecture 23, Slide 015<br> | ||
| + | |||
| + | ]] | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ===Where is pathway information found?=== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | ======Slide 017====== | ||
| + | [[Image:23_slide017.jpg|frame|none|Lecture 23, Slide 017<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 018====== | ||
| + | [[Image:23_slide018.jpg|frame|none|Lecture 23, Slide 018<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 019====== | ||
| + | [[Image:23_slide019.jpg|frame|none|Lecture 23, Slide 019<br> | ||
| + | [http://pathguide.org '''Pathguide''': the pahway resource list] | ||
| + | ]] | ||
| + | ======Slide 020====== | ||
| + | [[Image:23_slide020.jpg|frame|none|Lecture 23, Slide 020<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 021====== | ||
| + | [[Image:23_slide021.jpg|frame|none|Lecture 23, Slide 021<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 022====== | ||
| + | [[Image:23_slide022.jpg|frame|none|Lecture 23, Slide 022<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 023====== | ||
| + | [[Image:23_slide023.jpg|frame|none|Lecture 23, Slide 023<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 024====== | ||
| + | [[Image:23_slide024.jpg|frame|none|Lecture 23, Slide 024<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 025====== | ||
| + | [[Image:23_slide025.jpg|frame|none|Lecture 23, Slide 025<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 026====== | ||
| + | [[Image:23_slide026.jpg|frame|none|Lecture 23, Slide 026<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 027====== | ||
| + | [[Image:23_slide027.jpg|frame|none|Lecture 23, Slide 027<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 028====== | ||
| + | [[Image:23_slide028.jpg|frame|none|Lecture 23, Slide 028<br> | ||
| + | [http://cancer.cellmap.org The Cancer Cell Map] | ||
| + | ]] | ||
| + | ======Slide 029====== | ||
| + | [[Image:23_slide029.jpg|frame|none|Lecture 23, Slide 029<br> | ||
| + | [http://cancer.cellmap.org The Cancer Cell Map] | ||
| + | ]] | ||
| + | ======Slide 030====== | ||
| + | [[Image:23_slide030.jpg|frame|none|Lecture 23, Slide 030<br> | ||
| + | [http://cancer.cellmap.org The Cancer Cell Map] | ||
| + | ]] | ||
| + | ======Slide 031====== | ||
| + | [[Image:23_slide031.jpg|frame|none|Lecture 23, Slide 031<br> | ||
| + | [http://cancer.cellmap.org The Cancer Cell Map] | ||
| + | ]] | ||
| + | ======Slide 032====== | ||
| + | [[Image:23_slide032.jpg|frame|none|Lecture 23, Slide 032<br> | ||
| + | [http://www.pathwaycommons.org Pathway Commons] | ||
| + | ]] | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ===How can we use pathway information?=== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | ======Slide 034====== | ||
| + | [[Image:23_slide034.jpg|frame|none|Lecture 23, Slide 034<br> | ||
| + | Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne | ||
| + | S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, | ||
| + | Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366-82. PMID: 17947979 | ||
| + | <br> | ||
| + | <br> | ||
| + | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, | ||
| + | Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498-504. PMID: 14597658 | ||
| + | ]] | ||
| + | ======Slide 035====== | ||
| + | [[Image:23_slide035.jpg|frame|none|Lecture 23, Slide 035<br> | ||
| − | + | ]] | |
| − | < | + | ======Slide 036====== |
| + | [[Image:23_slide036.jpg|frame|none|Lecture 23, Slide 036<br> | ||
| + | [http://cytoscape.org Cytoscape - biological graph visualization and analysis] | ||
| + | ]] | ||
| + | ======Slide 037====== | ||
| + | [[Image:23_slide037.jpg|frame|none|Lecture 23, Slide 037<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 038====== | ||
| + | [[Image:23_slide038.jpg|frame|none|Lecture 23, Slide 038<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 039====== | ||
| + | [[Image:23_slide039.jpg|frame|none|Lecture 23, Slide 039<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 040====== | ||
| + | [[Image:23_slide040.jpg|frame|none|Lecture 23, Slide 040<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 041====== | ||
| + | [[Image:23_slide041.jpg|frame|none|Lecture 23, Slide 041<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 042====== | ||
| + | [[Image:23_slide042.jpg|frame|none|Lecture 23, Slide 042<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 043====== | ||
| + | [[Image:23_slide043.jpg|frame|none|Lecture 23, Slide 043<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 044====== | ||
| + | [[Image:23_slide044.jpg|frame|none|Lecture 23, Slide 044<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 045====== | ||
| + | [[Image:23_slide045.jpg|frame|none|Lecture 23, Slide 045<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 046====== | ||
| + | [[Image:23_slide046.jpg|frame|none|Lecture 23, Slide 046<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 047====== | ||
| + | [[Image:23_slide047.jpg|frame|none|Lecture 23, Slide 047<br> | ||
| + | |||
| + | ]] | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div style="padding: 10 px; background: #B0B8D7; border:solid 1px #AAAAAA;"> | ||
| + | ===Detailed example: detecting protein complexes=== | ||
| + | </div><br> | ||
| + | <br> | ||
| + | |||
| + | ======Slide 049====== | ||
| + | [[Image:23_slide049.jpg|frame|none|Lecture 23, Slide 049<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 050====== | ||
| + | [[Image:23_slide050.jpg|frame|none|Lecture 23, Slide 050<br> | ||
| + | [http://baderlab.org/Software/MCODE MCODE - a cytoscape plugin] | ||
| + | ]] | ||
| + | ======Slide 051====== | ||
| + | [[Image:23_slide051.jpg|frame|none|Lecture 23, Slide 051<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 052====== | ||
| + | [[Image:23_slide052.jpg|frame|none|Lecture 23, Slide 052<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 053====== | ||
| + | [[Image:23_slide053.jpg|frame|none|Lecture 23, Slide 053<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 054====== | ||
| + | [[Image:23_slide054.jpg|frame|none|Lecture 23, Slide 054<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 055====== | ||
| + | [[Image:23_slide055.jpg|frame|none|Lecture 23, Slide 055<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 056====== | ||
| + | [[Image:23_slide056.jpg|frame|none|Lecture 23, Slide 056<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 057====== | ||
| + | [[Image:23_slide057.jpg|frame|none|Lecture 23, Slide 057<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 058====== | ||
| + | [[Image:23_slide058.jpg|frame|none|Lecture 23, Slide 058<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 059====== | ||
| + | [[Image:23_slide059.jpg|frame|none|Lecture 23, Slide 059<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 060====== | ||
| + | [[Image:23_slide060.jpg|frame|none|Lecture 23, Slide 060<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 061====== | ||
| + | [[Image:23_slide061.jpg|frame|none|Lecture 23, Slide 061<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 062====== | ||
| + | [[Image:23_slide062.jpg|frame|none|Lecture 23, Slide 062<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 063====== | ||
| + | [[Image:23_slide063.jpg|frame|none|Lecture 23, Slide 063<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 064====== | ||
| + | [[Image:23_slide064.jpg|frame|none|Lecture 23, Slide 064<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 065====== | ||
| + | [[Image:23_slide065.jpg|frame|none|Lecture 23, Slide 065<br> | ||
| + | |||
| + | ]] | ||
| + | ======Slide 066====== | ||
| + | [[Image:23_slide066.jpg|frame|none|Lecture 23, Slide 066<br> | ||
| + | [http://baderlab.org/Home?action=AttachFile&do=get&target=InnerCellLife.mov The Inner Cell Life movie - download link (large!)] | ||
| + | ]] | ||
| − | + | <br> | |
| − | |||
| − | < | + | <br> |
| − | + | <br> | |
| − | + | ---- | |
| − | + | <small>[[Lecture_22|(Previous lecture)]] ... [[Lecture_24|(Next lecture)]]</small> | |
| − | |||
| − | --> | ||
Revision as of 18:03, 9 December 2007
(Previous lecture) ... (Next lecture)
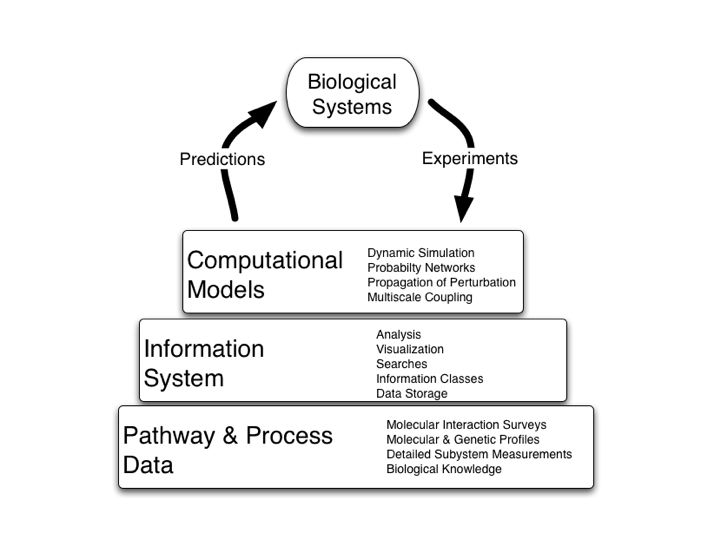
Computational analysis of pathways and interaction networks
BCH441 Guest lecture:
- Gary Bader
- Terrence Donnelly Center for Cellular and Biomolecular Research (DCCBR)
- BBDMR, MedGen, DCS, SLRI
- University of Toronto
Book chapter:
- Intermolecular Interactions and Biological Pathways
- Gary D. Bader, Anton J. Enright
- in Bioinformatics 3E Eds. Andreas D. Baxevanis and B.F.Francis Ouellette
Links summary
- Bader Lab - publications
- Kurt Kohn's signalling maps at the NIH
- Sea urchin development gene regulatory pathways
- Pathguide: the pahway resource list
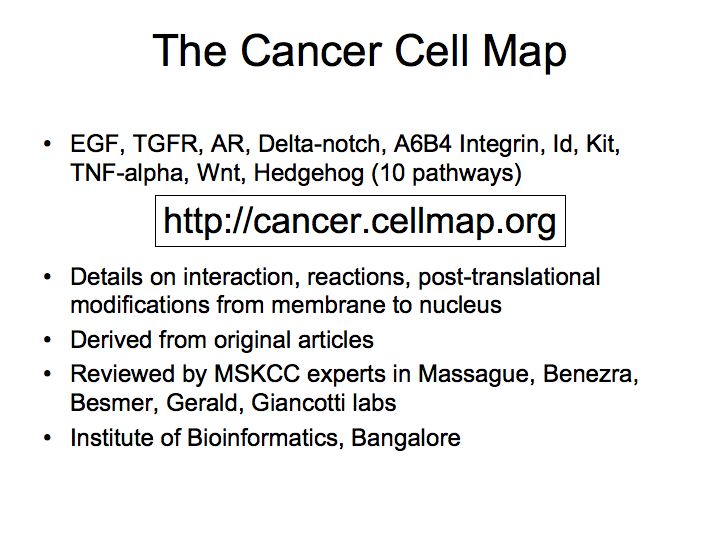
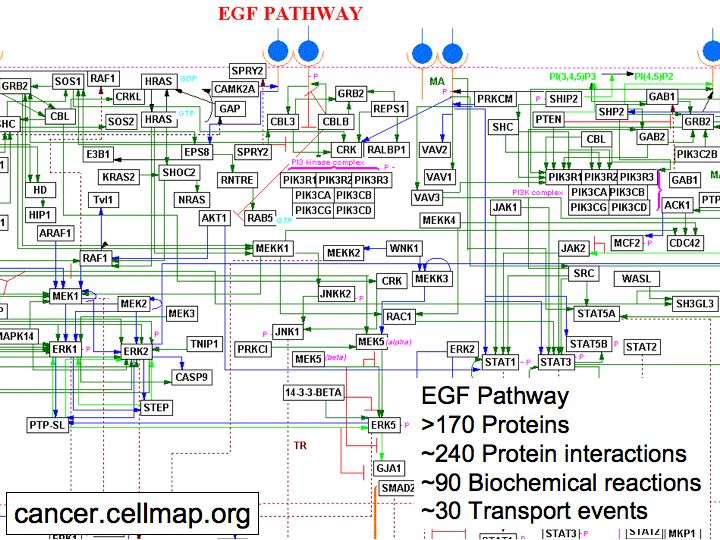
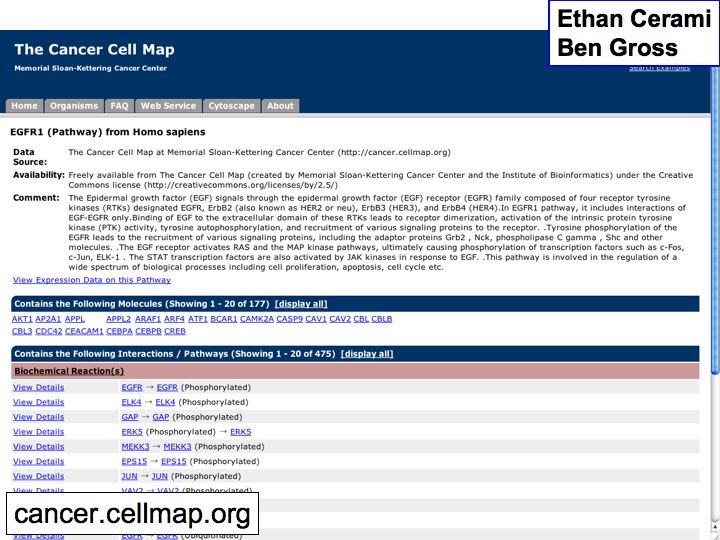
- The Cancer Cell Map
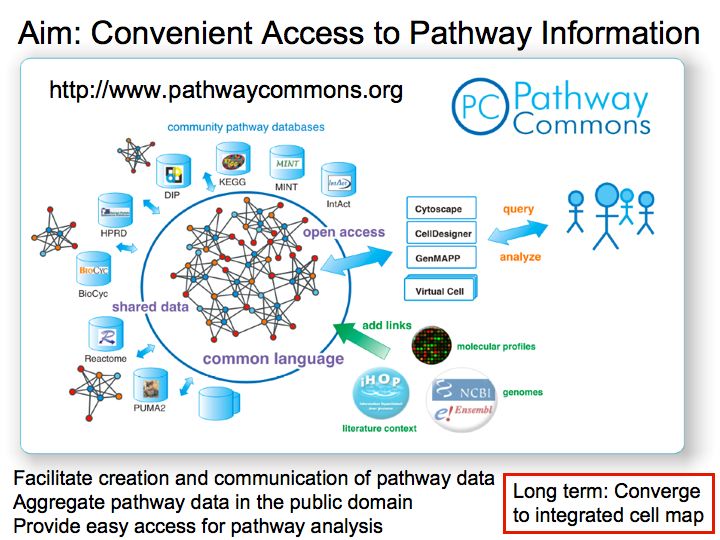
- Pathway Commons
- Cytoscape - biological graph visualization and analysis
- MCODE - a cytoscape plugin
- The Inner Cell Life movie - download link (large!)
Lecture slides
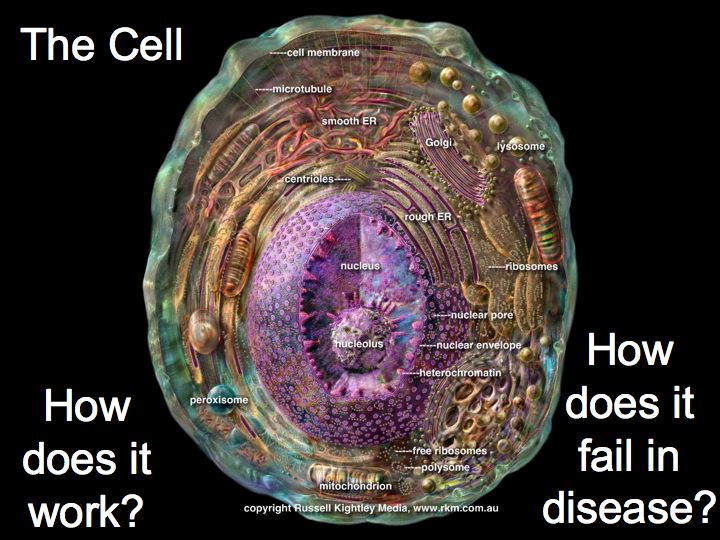
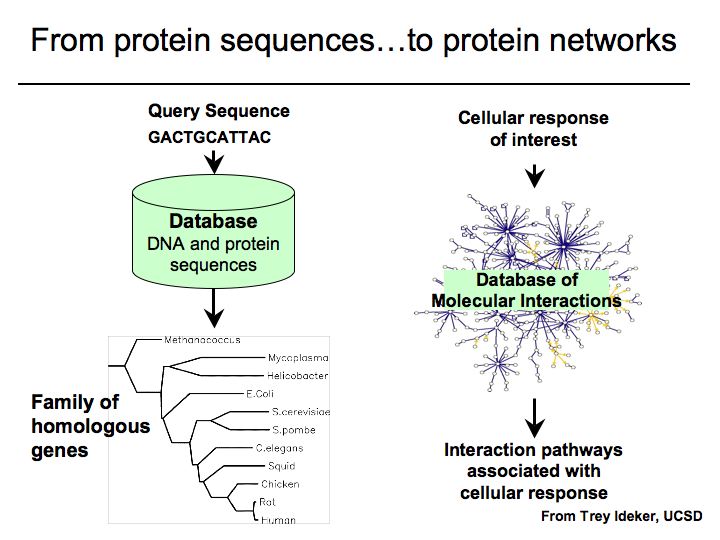
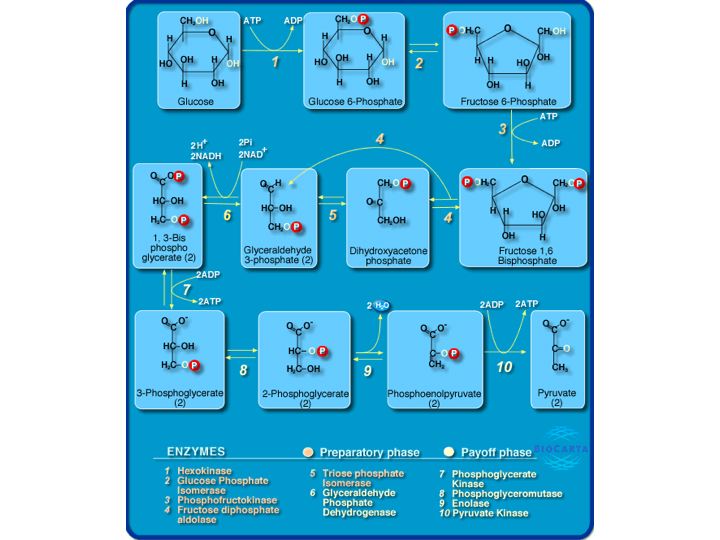
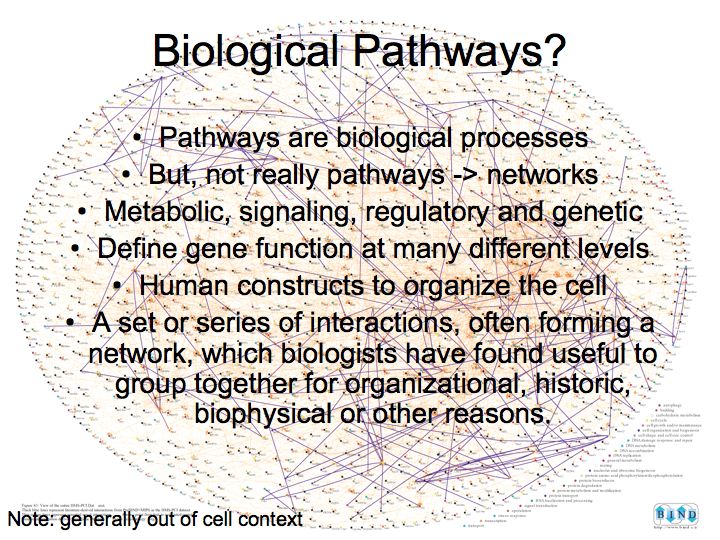
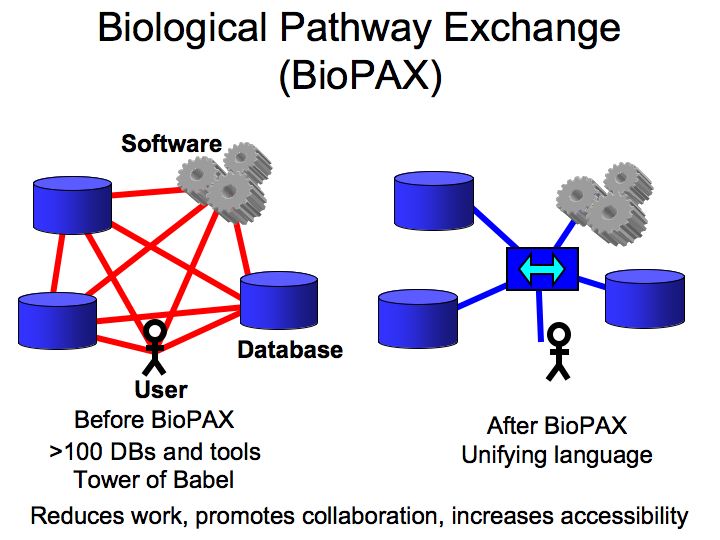
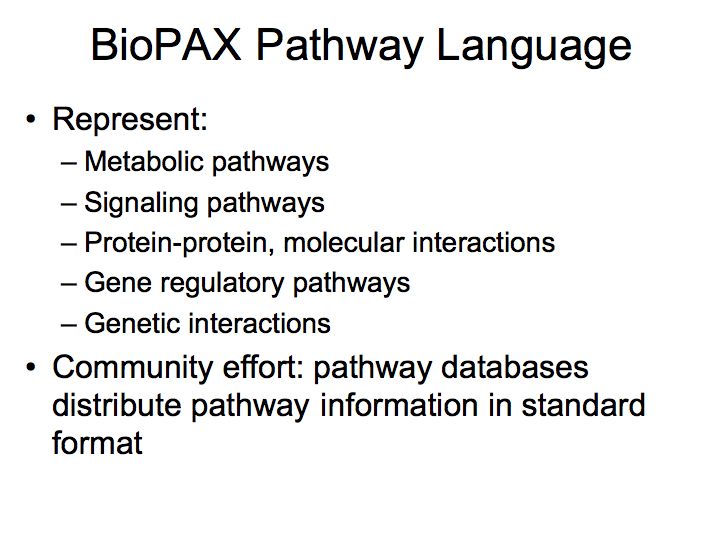
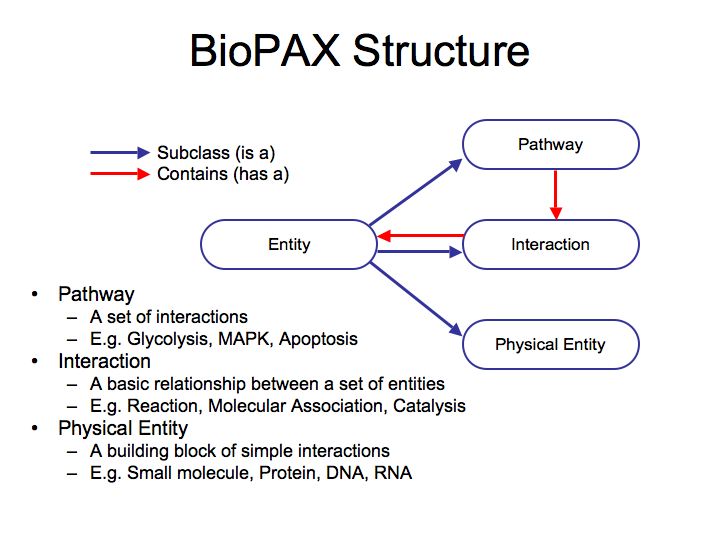
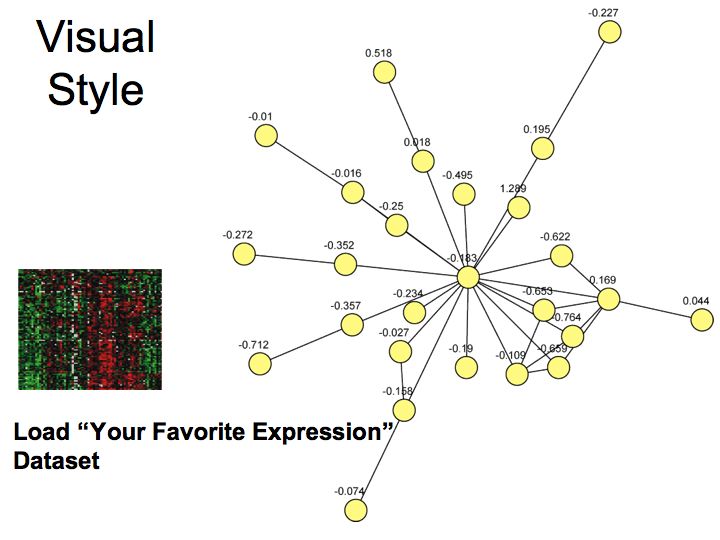
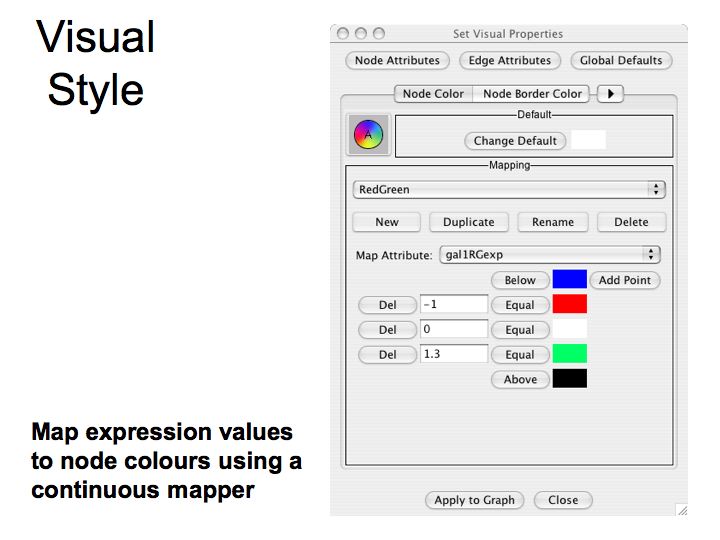
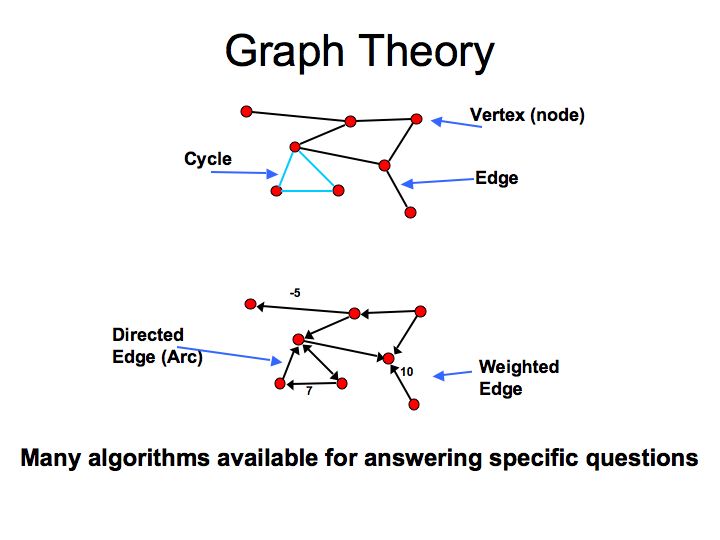
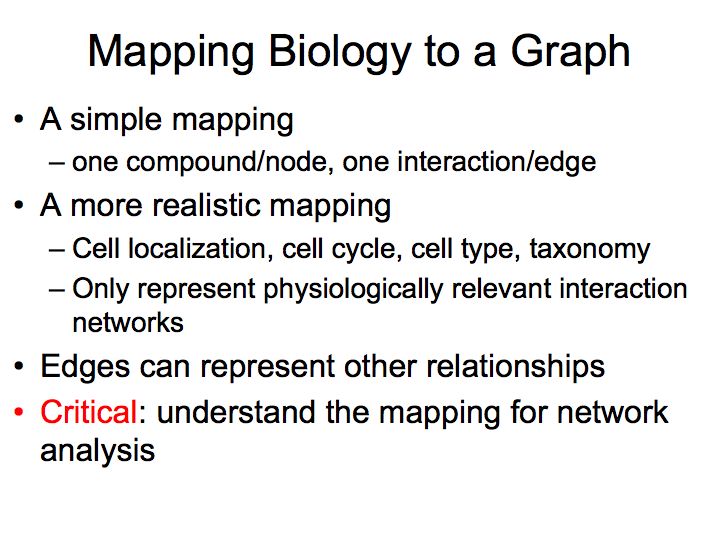
What are pathways and networks?
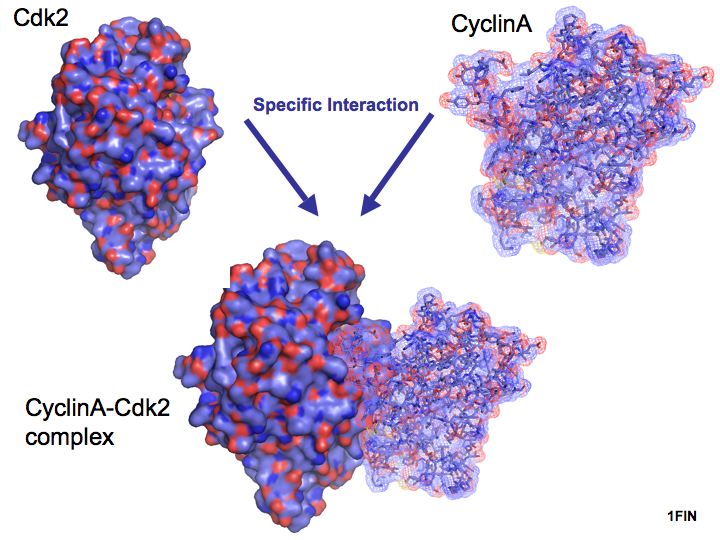
Slide 004
Slide 005
Slide 006

Lecture 23, Slide 006
Ref: Pathway information for systems biology, Cary MP, Bader GD, Sander C, FEBS Lett. 2005 Mar 21; 579(8):1815-20 (Available from http://baderlab.org/Publications)
Ref: Pathway information for systems biology, Cary MP, Bader GD, Sander C, FEBS Lett. 2005 Mar 21; 579(8):1815-20 (Available from http://baderlab.org/Publications)
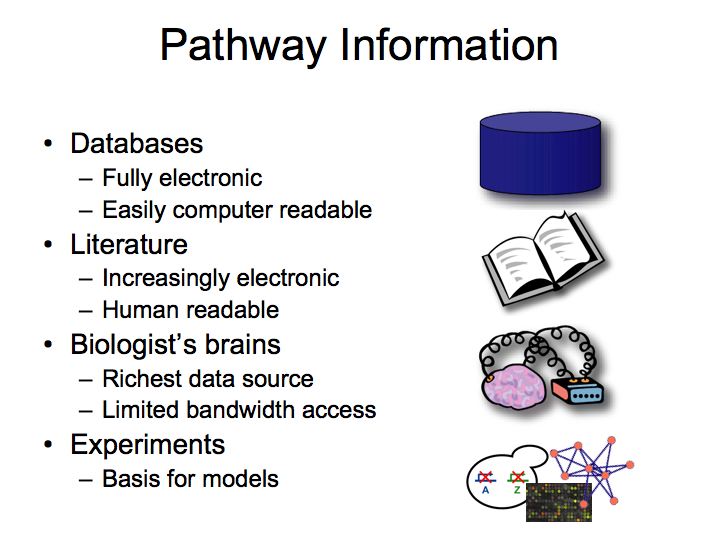
Slide 007
Slide 008

Lecture 23, Slide 008
http://discover.nci.nih.gov/kohnk/interaction_maps.html
http://discover.nci.nih.gov/kohnk/interaction_maps.html
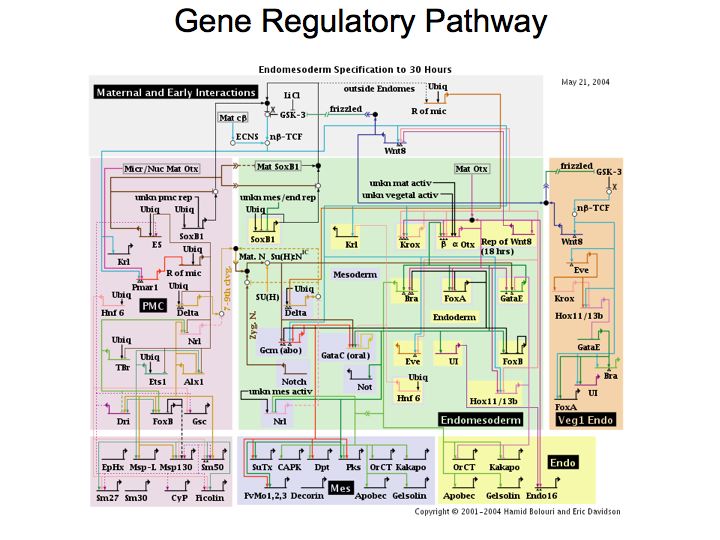
Slide 009

Lecture 23, Slide 009
http://sugp.caltech.edu/endomes
http://sugp.caltech.edu/endomes
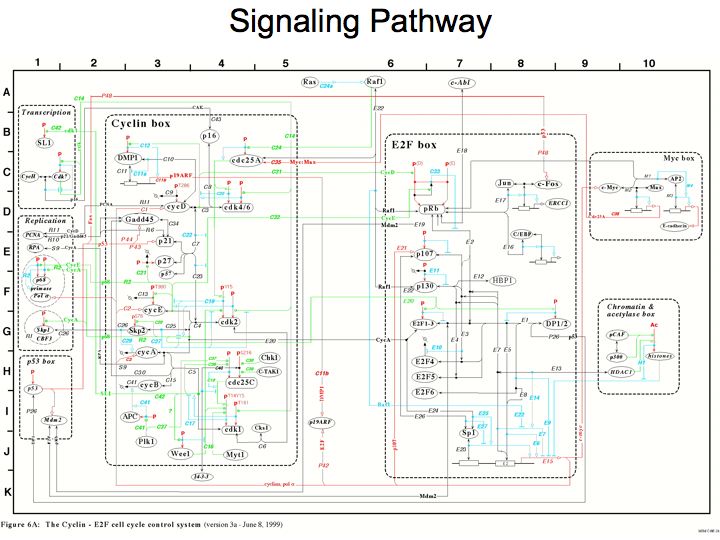
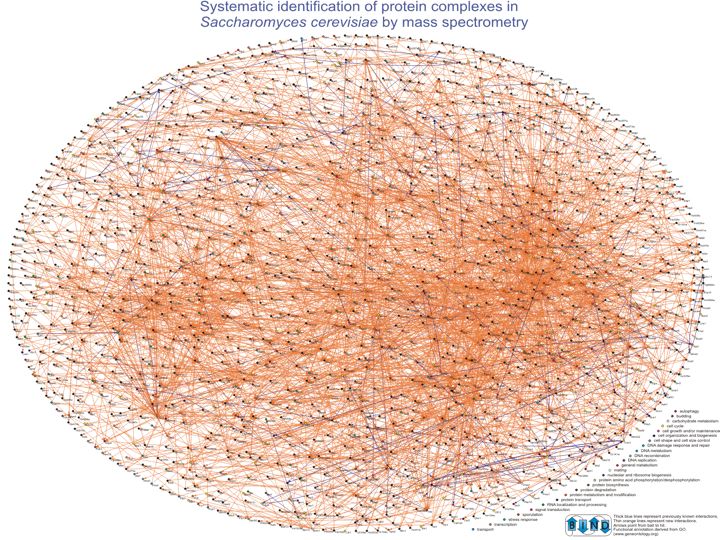
Slide 010

Lecture 23, Slide 010
Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Nature 2002 Jan 10; 415(6868): 180-183 (Available from http://baderlab.org/Publications)
Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Nature 2002 Jan 10; 415(6868): 180-183 (Available from http://baderlab.org/Publications)
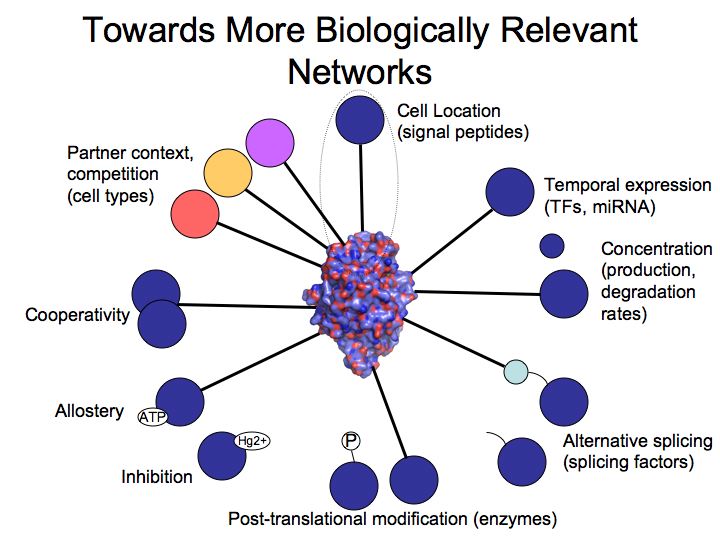
Slide 011
Slide 012
Slide 013
Slide 014
Slide 015
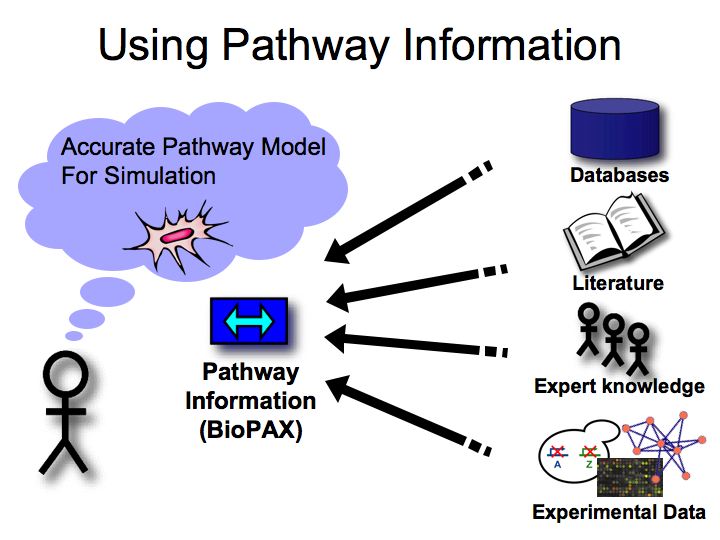
Where is pathway information found?
Slide 017
Slide 018
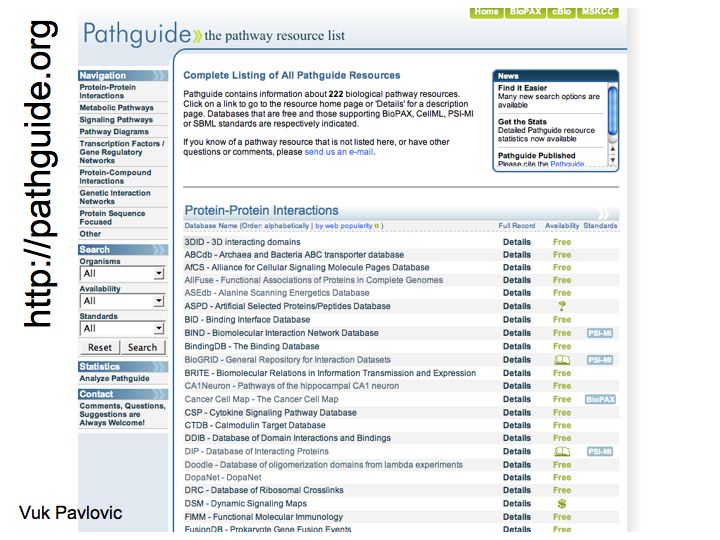
Slide 019

Lecture 23, Slide 019
Pathguide: the pahway resource list
Pathguide: the pahway resource list
Slide 020
Slide 021
Slide 022
Slide 023
Slide 024
Slide 025
Slide 026
Slide 027
Slide 028

Lecture 23, Slide 028
The Cancer Cell Map
The Cancer Cell Map
Slide 029

Lecture 23, Slide 029
The Cancer Cell Map
The Cancer Cell Map
Slide 030

Lecture 23, Slide 030
The Cancer Cell Map
The Cancer Cell Map
Slide 031

Lecture 23, Slide 031
The Cancer Cell Map
The Cancer Cell Map
Slide 032

Lecture 23, Slide 032
Pathway Commons
Pathway Commons
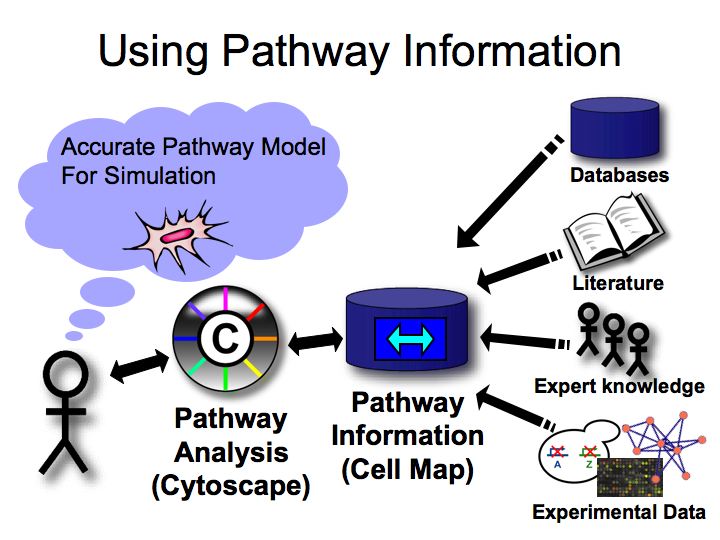
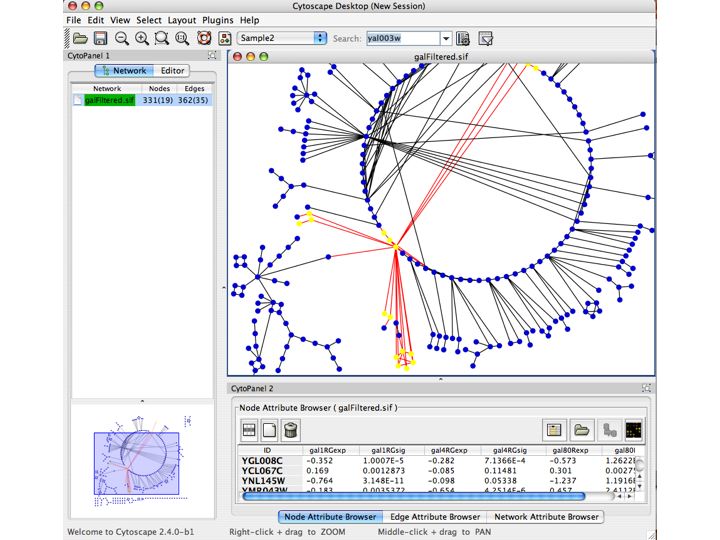
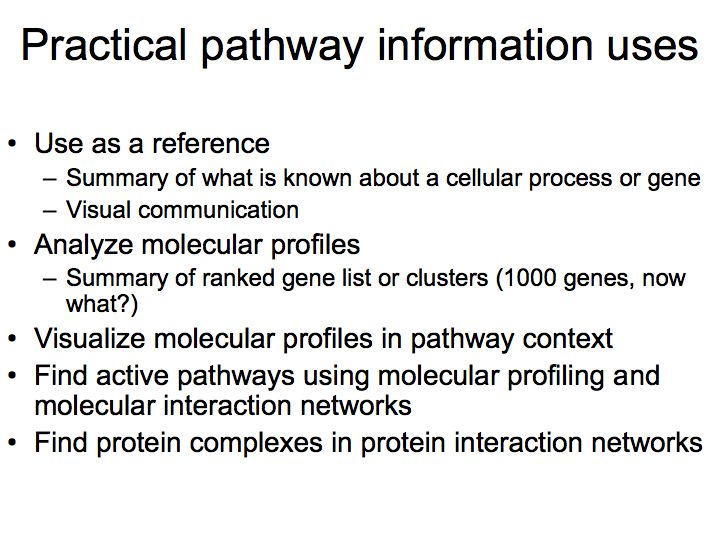
How can we use pathway information?
Slide 034

Lecture 23, Slide 034
Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366-82. PMID: 17947979
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498-504. PMID: 14597658
Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366-82. PMID: 17947979
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498-504. PMID: 14597658
Slide 035
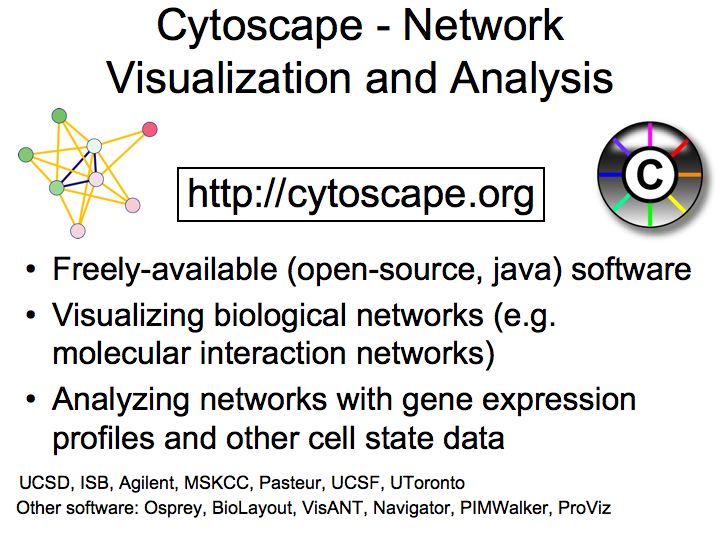
Slide 036

Lecture 23, Slide 036
Cytoscape - biological graph visualization and analysis
Cytoscape - biological graph visualization and analysis
Slide 037
Slide 038
Slide 039
Slide 040
Slide 041
Slide 042
Slide 043
Slide 044
Slide 045
Slide 046
Slide 047
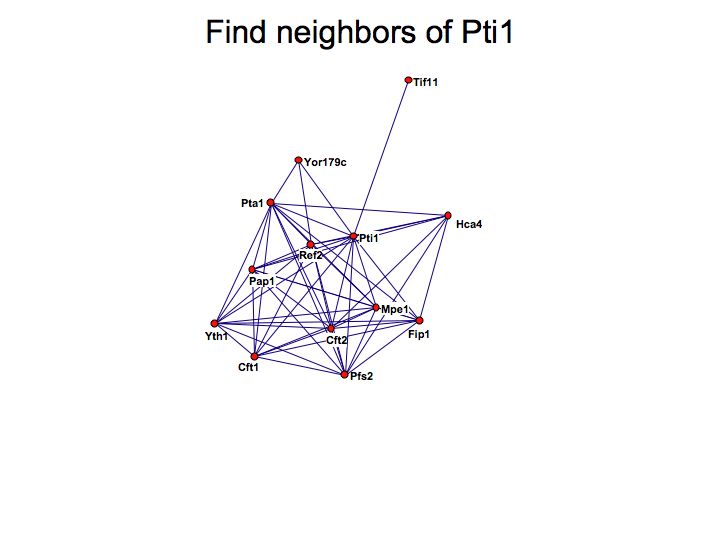
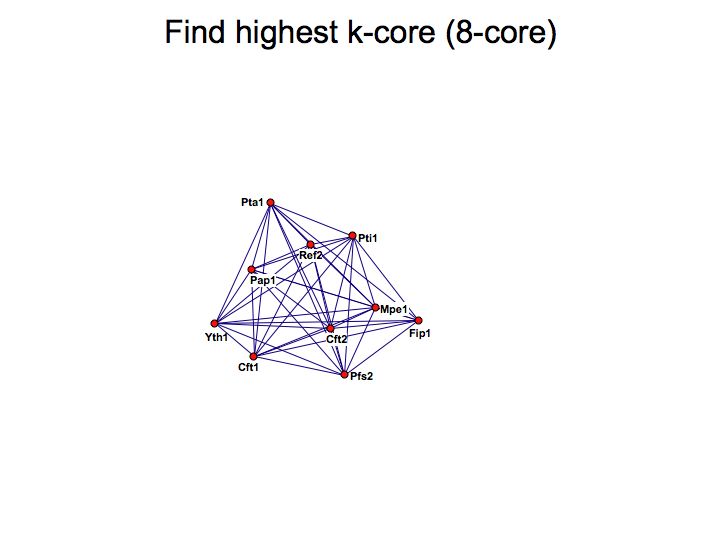
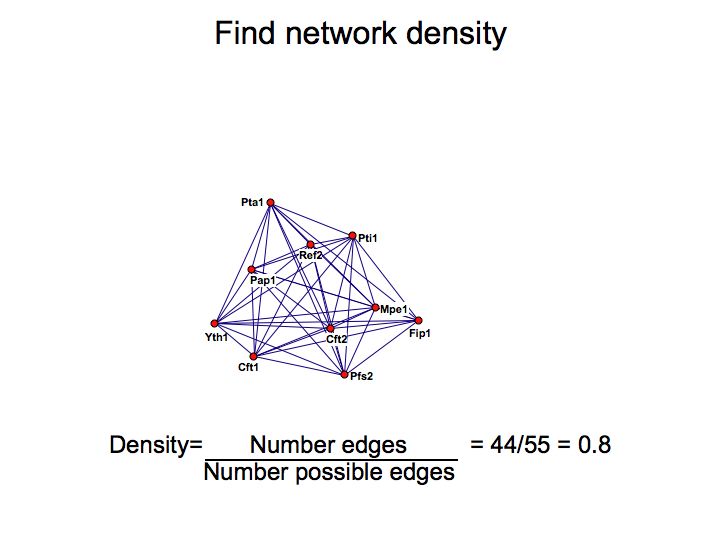
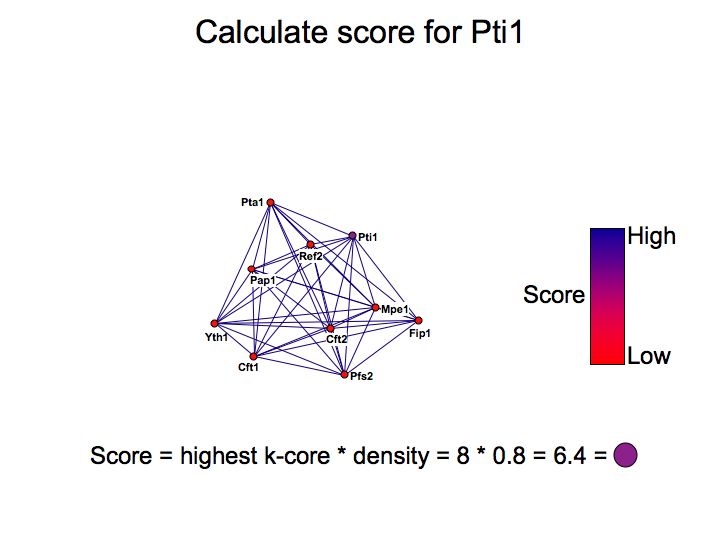
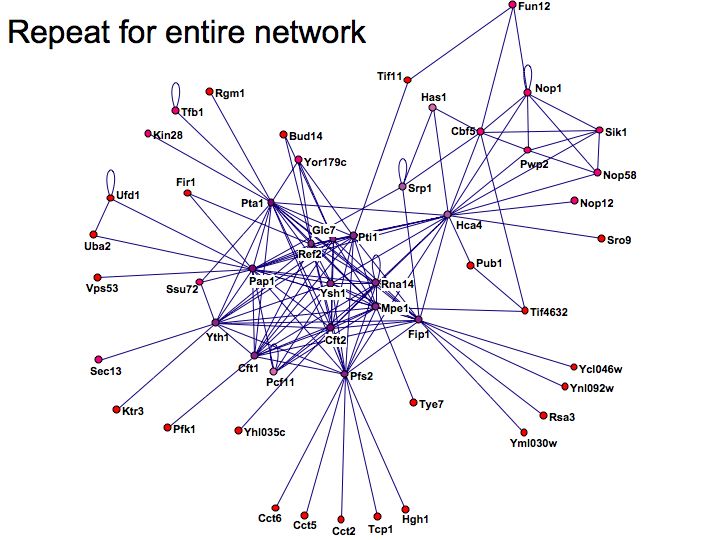
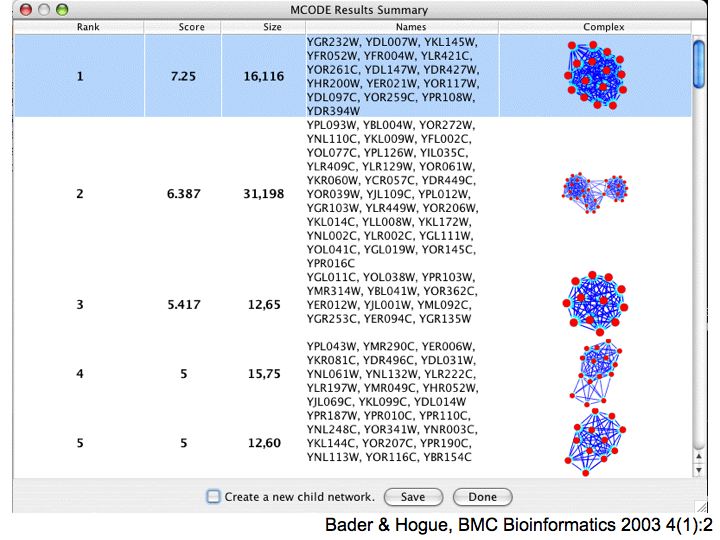
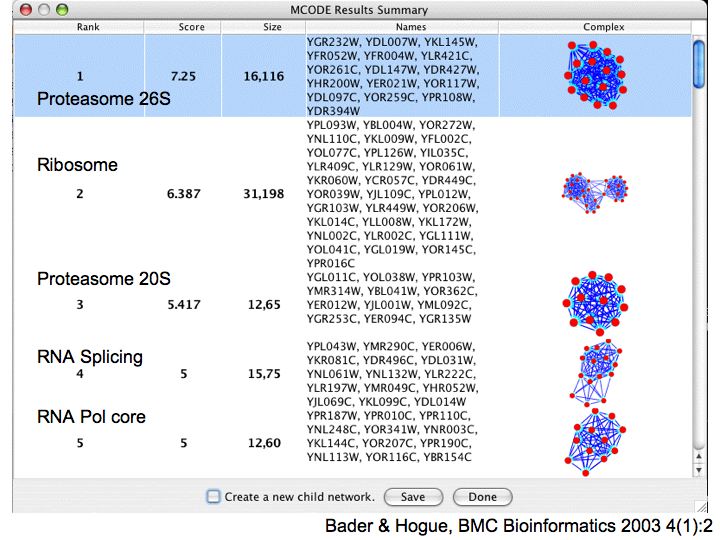
Detailed example: detecting protein complexes
Slide 049
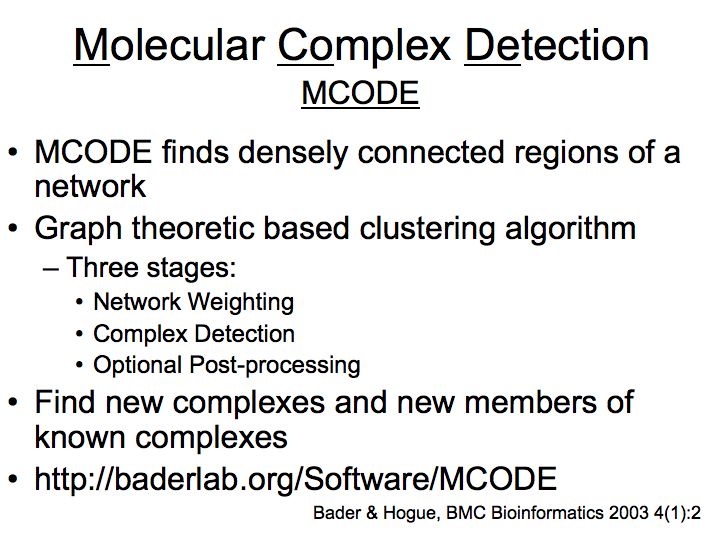
Slide 050

Lecture 23, Slide 050
MCODE - a cytoscape plugin
MCODE - a cytoscape plugin
Slide 051
Slide 052
Slide 053
Slide 054
Slide 055
Slide 056
Slide 057
Slide 058
Slide 059
Slide 060
Slide 061
Slide 062
Slide 063
Slide 064
Slide 065
Slide 066

Lecture 23, Slide 066
The Inner Cell Life movie - download link (large!)
The Inner Cell Life movie - download link (large!)