Difference between revisions of "WWW UniProt"
(Created page with "<div style="display: none"> <section begin=title />UniProt<section end=title /> <section begin=file />ImageFile...<section end=file /> <section begin=url /> http://www.unip...") |
m |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<section begin=title />UniProt<section end=title /> | <section begin=title />UniProt<section end=title /> | ||
| − | <section begin=file /> | + | <section begin=file />Uniprot.org.jpg<section end=file /> |
| − | <section begin=url /> | + | <section begin=url />http://www.uniprot.org/uniprot/P39678<section end=url /> |
| − | http://www.uniprot.org/uniprot/P39678 | ||
| − | <section end=url /> | ||
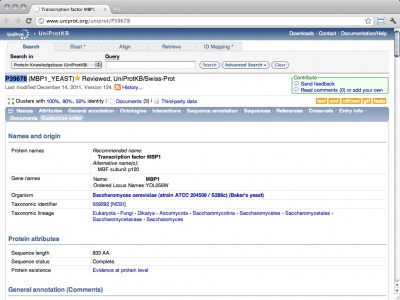
| − | <section begin=abstract /> | + | <section begin=abstract />UniProt is the protein sequence database of the European Bioinformatics Institute. It is an extraordinarily well constructed, curated, and integrated resource. As a public resource, its results are freely accessible world-wide. The "Knowledge Base" (UniProtKB), which is the database proper, contains two subsections: '''SwissProt''', the manually curated and heavily annotated protein sequence repository; it is approximately equivalent to the NCBI Refseq protein database, albeit with usually higher annotation levels. '''TrEMBL''' is much larger and contains sequences that have been computationally translated from the EMBL nucleotide sequence collection. It is approximately equivalent to the NCBI's Entrez protein database. The URL links to the entry for the ''Saccharomyces cerevisiae'' cell-cycle regulation transcription factor Mbp1.<section end=abstract /> |
| − | UniProt is the protein sequence database of the European Bioinformatics Institute. It is an extraordinarily well constructed, curated, and integrated resource. As a public resource, its results are freely accessible world-wide. The "Knowledge Base" (UniProtKB), which is the database proper, contains two subsections: '''SwissProt''', the manually curated and heavily annotated protein sequence repository; it is approximately equivalent to the NCBI Refseq protein database, albeit with usually higher annotation levels. '''TrEMBL''' is much larger and contains sequences that have been computationally translated from the EMBL nucleotide sequence collection. It is approximately equivalent to the NCBI's Entrez protein database. The URL links to the entry for the ''Saccharomyces cerevisiae'' cell-cycle regulation transcription factor Mbp1. | ||
| − | <section end=abstract /> | ||
| − | <section begin=reference /> | + | <section begin=reference />{{WWW_resource_reference_section|22102590 }}<section end=reference /> |
| − | {{WWW_resource_reference_section| | ||
| − | <section end=reference /> | ||
Latest revision as of 17:02, 28 January 2012
UniProt |
URL
http://www.uniprot.org/uniprot/P39678
Abstract
UniProt is the protein sequence database of the European Bioinformatics Institute. It is an extraordinarily well constructed, curated, and integrated resource. As a public resource, its results are freely accessible world-wide. The "Knowledge Base" (UniProtKB), which is the database proper, contains two subsections: SwissProt, the manually curated and heavily annotated protein sequence repository; it is approximately equivalent to the NCBI Refseq protein database, albeit with usually higher annotation levels. TrEMBL is much larger and contains sequences that have been computationally translated from the EMBL nucleotide sequence collection. It is approximately equivalent to the NCBI's Entrez protein database. The URL links to the entry for the Saccharomyces cerevisiae cell-cycle regulation transcription factor Mbp1.
Reference
| UniProt Consortium (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40:D71-5. (pmid: 22102590) |