Difference between revisions of "Amino Acid Exam Questions"
(→2004) |
m |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <div id="BIO"> | ||
| + | <div class="b1"> | ||
| + | BCH441 / BCH1441 Exam Principles | ||
| + | </div> | ||
| + | |||
| + | | ||
| + | | ||
| + | |||
| + | |||
__NOTOC__ | __NOTOC__ | ||
| | ||
| Line 66: | Line 75: | ||
<br> | <br> | ||
<br> | <br> | ||
| + | |||
| + | ==2005== | ||
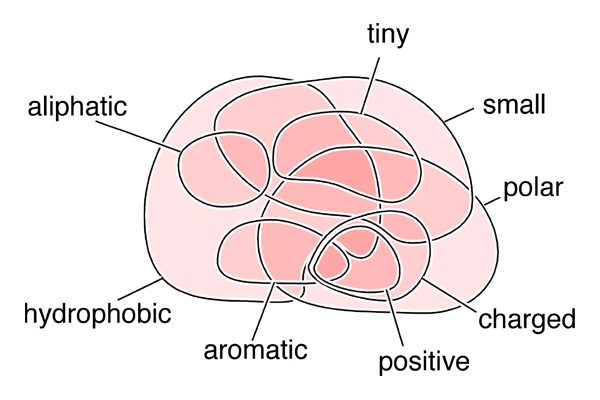
| + | [[Image:20_AA_Venn.jpg|frame|none|Venn-diagram of biophysical properties of amino acids. ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Please complete the Venn diagram by entering the one-letter code for the standard proteinogenic amino acids.''' | ||
| + | </div> | ||
| + | |||
| + | <small>Note that some of the properties in this diagram are quantitative, not qualitative, thus there may be some perceived ambiguity in assigning amino acids to these categories and more than one solution may be considered correct.</small> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2005== | ||
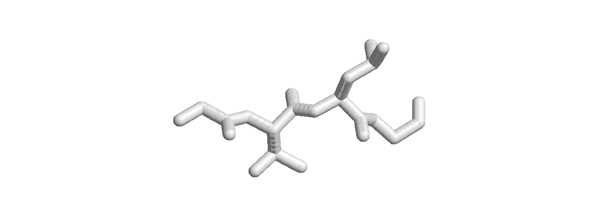
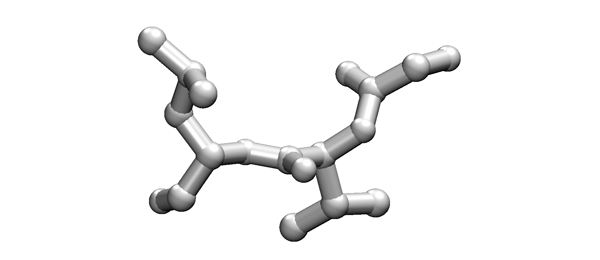
| + | [[Image:Tetrapeptide.jpg|frame|none|This sketch shows the backbone atoms of four amino acids, as well as two sidechains (no hydrogen atoms). ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''What could the sequence(s) of the central dipeptide be?''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2006== | ||
| + | [[Image:MSA.jpg|frame|none|Excerpt from the T-Coffee multiple sequence alignment of Mbp1 orthologues. ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | Consider the following qualitative scales of some amino acid properties: | ||
| + | *volume: small - intermediate - bulky | ||
| + | *hydrophobicity: hydrophobic - polar - charged (+)/charged (-) | ||
| + | *H-bond donor/H-bond acceptor | ||
| + | *propensity for: β-strand, α-helix, turn | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''For each of the columns marked with numbers at the bottom, please note the property (or properties) that appears to be favoured in that position. Write "none" if no property matches well.''' | ||
| + | </div> | ||
| + | |||
| + | <small>Bringing the abstract knowledge about amino acids to bear on the biological context is the way to go for future questions on this topic.</small> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | ==2007== | ||
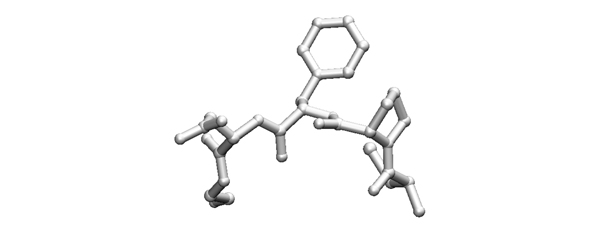
| + | [[Image:2007exam_tripeptide.jpg|frame|none|Model of an amino acid fragment, taken from a protein structure file. The structure has been determined by X-ray crystallography. Note, that this is not a free peptide: this fragment has only one carbonyl oxygen at its C-terminus since the structure continues with a peptide bond. ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Write the possible sequences (from N to C, one-letter code) that the fragment shown above could have. If amino acids are ambiguous, write all possibilities in brackets.''' | ||
| + | </div> | ||
| + | |||
| + | <small>The key to identify particular residues or local sequence while studying protein structure, is to first orient yourself regarding chain direction by identifying CA(CB)-C(O) units. This is most easily done by starting from something other than alanine or glycine.</small> | ||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''What is the most important amino acid property that distinguishes between residues that are part of a protein's surface and residues that are part of its core?''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | ==2008== | ||
| + | |||
| + | <small>This question integrates a multiple sequence alignment with knowledge about amino acids.</small> | ||
| + | |||
| + | [[Image:2008exam_peptide.jpg|frame|none|Model of an amino acid fragment, taken from the PDB file 1UBD. The structure has been determined by X-ray crystallography. Note, that this is not a free peptide: this fragment has only one carbonyl oxygen at its C-terminus since the structure continues with a peptide bond. ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Identify the sequence of this fragment in the alignment below and draw a box around it.''' | ||
| + | </div> | ||
| + | The alignment below is the CDD profile for pfam domain 00096: the C2H2 zinc finger domain. The following pattern describes the zinc finger. | ||
| + | #-X-C-X(1-5)-C-X3-#-X5-#-X2-H-X(3-6)-[H/C] | ||
| + | Where <code>X</code> can be any amino acid, numbers (and ranges of numbers in brackets) indicate the number of residues, and "<code>#</code>" marks those positions that are '''important for the stable fold''' of the zinc finger domain. | ||
| + | |||
| + | 10 20 | ||
| + | ....'....|....'....|....'. | ||
| + | 1UBD_C 93 YVCPfdGCNKKFAQSTNLKSHILT-H 117 | ||
| + | gi 1706178 555 FVCS--KCGKTFTRRNTMARHADN-C 577 | ||
| + | gi 586050 221 HVCG--KCYKTFRRLMSLKKHLEF-C 243 | ||
| + | gi 1723485 132 YICE--SCLKYMNSDHVLQRHKMK-C 154 | ||
| + | gi 465512 301 FICE--FCLKYMTSRYTFYRHQLK-C 323 | ||
| + | gi 730713 101 FVCE--YCFKYTDDQTRFVGHVAS-C 123 | ||
| + | gi 141697 958 FECE--TCRNRYRKLENFENHKKFyC 981 | ||
| + | gi 138336 541 MVCE--ICKYECETLKELKAHNLS-C 563 | ||
| + | gi 138340 549 MVCE--VCQQECEMAKELESHKKS-C 571 | ||
| + | gi 401357 768 CVCP--FCEKQALDLVTLEEHVKE-C 790 | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Clearly mark the three columns that are "important for the stable fold" with an "#".''' | ||
| + | *'''For each of these three columns note what appear to be their two most important conserved properties.''' | ||
| + | :(Choose from the following list: Polarity, Charge, Size, Backbone torsional angles, H-bond donor or acceptor, Metal ion coordination. Note: the conserved properties are not necessarily different for each column.) | ||
| + | *'''Since the conserved H residues coordinate a Zn-ion, do you expect them to be protonated or not?''' | ||
| + | </div> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2009== | ||
| + | <small>This question integrates a sequence logo with knowledge about amino acids.</small> | ||
| + | [[Image:2009exam-Peptide.jpg|frame|none|Model of a nonapeptide fragment, taken from the PDB file 2EZD. This is part of an AT-hook DNA binding motif. Such motifs can be found as an accessory domain to other DNA binding domains. In our course two AT-hook motifs were annotated by SMART for the ''Candida glabrata'' Mbp1 orthologue, for residues 128 to 140 and residues 173 to 185. | ||
| + | The structure from which the fragment above was taken has been determined by NMR spectroscopy. It was visualized in VMD in a CPK representation, carbons coloured black and non-carbons grey, with the following selection: '''protein and resid 7 to 15 and not element H''' | ||
| + | ]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Identify the N-terminal N atom, the C-terminal C atom, and trace the peptide backbone.''' <small>This is actualy an instruction to help you structure your analysis.</small> | ||
| + | *'''Write down the sequence of the fragment in the direction from N to C in one-letter code.''' | ||
| + | </div> | ||
| + | |||
| + | |||
| + | [[Image:2009exam-AT-HookLogo.jpg|frame|none|Sequence logo of 51 AT-hook sequences taken from the SMART database, plus two instances of the sequence motif found in the Candida Glabrata Mbp1 orthologue. | ||
| + | ]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Match the motif represented by the logo to the sequence you have identified above.''' | ||
| + | *'''For position 2 of the motif, what is the conserved amino acid property (if any)? For position 5?''' | ||
| + | :(Choose from the following list: Polarity, Charge, Size, Backbone torsional angles, H-bond donor or acceptor, Metal ion coordination.) | ||
| + | *'''Are these properties conserved in the 2EZD AT-hook or not?'''</div> | ||
| + | <br> <br> | ||
| + | |||
| + | |||
| + | Analysis of this peptide and its function as a binding motif is continued in the [[Stereo_Vision_Exam_Questions#2009|'''2009 stereo-vision exam questions''']]. | ||
| + | |||
| + | |||
<!-- | <!-- | ||
| − | == | + | ==2006== |
[[Image:Stereo_000000.jpg|frame|none|Caption. ]] | [[Image:Stereo_000000.jpg|frame|none|Caption. ]] | ||
| Line 86: | Line 225: | ||
--> | --> | ||
| + | |||
| + | [[Category: Bioinformatics]] | ||
| + | </div> | ||
Latest revision as of 01:57, 11 December 2012
BCH441 / BCH1441 Exam Principles
- Of all data abstractions in bioinformatics, the one-letter amino acid code is the most important one. Whether we are evaluating multiple sequence alignments, database searches or mutational studies, this all requires a confident understanding of the physicochemical nature of the residues we are considering and of course knowing which letters correspond to which amino acids.
2002
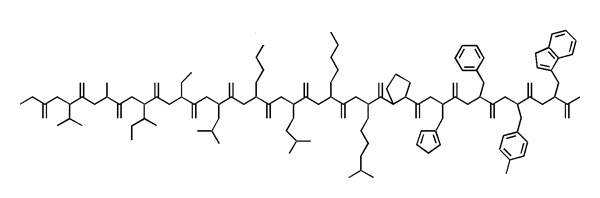
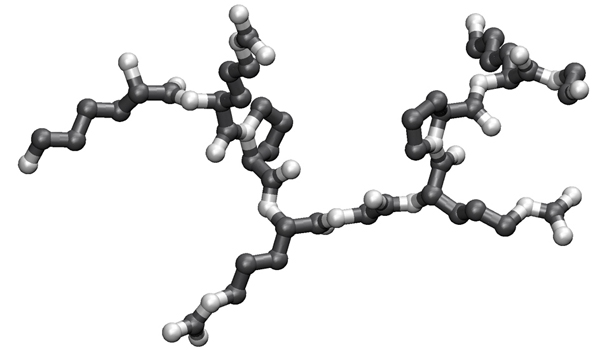
- Write the sequence of this polypeptide into your exam booklet in one-letter code. Where the sidechains are ambiguous, write all possible one letter codes for the residue in square brackets. Annotate residues that are > 80 % charged at physiological pH with a "+" or "-".
- Example:
AB+CD[EFG]HIJ[KL]M-N-OPQ
2003
- List the three letter and one letter code for the two sulfur containing amino acids.
- List the three letter and one letter code for the two amino acids that you would expect more frequently near the N-terminus of an alpha helix than its C-terminus.
- Which one of the above is larger ?
- List the three letter and one letter code for all the amino acids with aromatic side chains.
- List the three letter code for the amino acids K and M. What do these two have in common?
- List the one letter codes for glutamine, glutamic acid and aspartic acid.
- List the one letter code for the residue that cannot donate a hydrogen bond from its backbone.
- List the three letter code and one letter code for the three β-branched amino acids.
- You find the following residues in a position of a multiple alignment: V, I, L. What secondary structure could this position be a part of ? (Write "none" if these are not typical for any secondary structure.)
- In the amino acid substitution H→T, which is the most important property that is conserved? (Write "none" if this is a non-conservative substitution.)
2004
- List the three letter and one letter code for the two negatively charged amino acids.
- List the three letter and one letter code for the three amino acids that you would expect to form salt bridges with a DNA phosphate backbone.
- Which one of the above is the largest ?
- List the three letter and one letter code for all the amino acids that have side chains which cannot participate in hydrogen bonds.
(Ignore Glycine.)
- List the three letter code for the amino acids K, E, D. What do these three have in common?
- List the one letter codes for tryptophan, arginine and leucine.
- List the three letter and one letter code for the residue that cannot donate a hydrogen bond from its backbone.
- List the three letter code and one letter code for the three β-branched amino acids.
- In the amino acid substitution T→S, which is the most important property that is conserved? (Write "none" if this is a non-conservative substitution.)
- In the amino acid substitution E→D, which is the most important property that is conserved? (Write "none" if this is a non-conservative substitution.)
2004
- List the three letter and one-letter code for the smaller of the two amino acids that have a side-chain carboxylate group.
- List the three-letter and one-letter code for the three amino acids that you would expect to form salt bridges with the phosphate group of a DNA strand.
- In the amino acid substitution F→N, which is the most important property that is conserved? (Write "none" if this is a non-conservative substitution.)
- List the three-letter and one-letter code for all the amino acids with aromatic side chains and note those that cannot accept a hydrogen bond with their sidechain. (Write "all" if all these sidechains can function as hydrogen bond acceptors.)
- List the three-letter and one-letter code for the one residue that cannot participate in the regular i→i+4 hydrogen bonding pattern of an α-helix backbone and note why it can't.
- List the three-letter code and one-letter code for the β-branched amino acids and explain with a simple sketch what "β-branched" means.
2005
- Please complete the Venn diagram by entering the one-letter code for the standard proteinogenic amino acids.
Note that some of the properties in this diagram are quantitative, not qualitative, thus there may be some perceived ambiguity in assigning amino acids to these categories and more than one solution may be considered correct.
2005
- What could the sequence(s) of the central dipeptide be?
2006
Consider the following qualitative scales of some amino acid properties:
- volume: small - intermediate - bulky
- hydrophobicity: hydrophobic - polar - charged (+)/charged (-)
- H-bond donor/H-bond acceptor
- propensity for: β-strand, α-helix, turn
- For each of the columns marked with numbers at the bottom, please note the property (or properties) that appears to be favoured in that position. Write "none" if no property matches well.
Bringing the abstract knowledge about amino acids to bear on the biological context is the way to go for future questions on this topic.
2007
- Write the possible sequences (from N to C, one-letter code) that the fragment shown above could have. If amino acids are ambiguous, write all possibilities in brackets.
The key to identify particular residues or local sequence while studying protein structure, is to first orient yourself regarding chain direction by identifying CA(CB)-C(O) units. This is most easily done by starting from something other than alanine or glycine.
- What is the most important amino acid property that distinguishes between residues that are part of a protein's surface and residues that are part of its core?
2008
This question integrates a multiple sequence alignment with knowledge about amino acids.
- Identify the sequence of this fragment in the alignment below and draw a box around it.
The alignment below is the CDD profile for pfam domain 00096: the C2H2 zinc finger domain. The following pattern describes the zinc finger.
#-X-C-X(1-5)-C-X3-#-X5-#-X2-H-X(3-6)-[H/C]
Where X can be any amino acid, numbers (and ranges of numbers in brackets) indicate the number of residues, and "#" marks those positions that are important for the stable fold of the zinc finger domain.
10 20
....'....|....'....|....'.
1UBD_C 93 YVCPfdGCNKKFAQSTNLKSHILT-H 117
gi 1706178 555 FVCS--KCGKTFTRRNTMARHADN-C 577
gi 586050 221 HVCG--KCYKTFRRLMSLKKHLEF-C 243
gi 1723485 132 YICE--SCLKYMNSDHVLQRHKMK-C 154
gi 465512 301 FICE--FCLKYMTSRYTFYRHQLK-C 323
gi 730713 101 FVCE--YCFKYTDDQTRFVGHVAS-C 123
gi 141697 958 FECE--TCRNRYRKLENFENHKKFyC 981
gi 138336 541 MVCE--ICKYECETLKELKAHNLS-C 563
gi 138340 549 MVCE--VCQQECEMAKELESHKKS-C 571
gi 401357 768 CVCP--FCEKQALDLVTLEEHVKE-C 790
- Clearly mark the three columns that are "important for the stable fold" with an "#".
- For each of these three columns note what appear to be their two most important conserved properties.
- (Choose from the following list: Polarity, Charge, Size, Backbone torsional angles, H-bond donor or acceptor, Metal ion coordination. Note: the conserved properties are not necessarily different for each column.)
- Since the conserved H residues coordinate a Zn-ion, do you expect them to be protonated or not?
2009
This question integrates a sequence logo with knowledge about amino acids.
- Identify the N-terminal N atom, the C-terminal C atom, and trace the peptide backbone. This is actualy an instruction to help you structure your analysis.
- Write down the sequence of the fragment in the direction from N to C in one-letter code.
- Match the motif represented by the logo to the sequence you have identified above.
- For position 2 of the motif, what is the conserved amino acid property (if any)? For position 5?
- (Choose from the following list: Polarity, Charge, Size, Backbone torsional angles, H-bond donor or acceptor, Metal ion coordination.)
- Are these properties conserved in the 2EZD AT-hook or not?
Analysis of this peptide and its function as a binding motif is continued in the 2009 stereo-vision exam questions.