Difference between revisions of "Tools Exam Questions"
(→2002) |
m |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <div id="BIO"> | ||
| + | <div class="b1"> | ||
| + | Tools Exam Questions | ||
| + | </div> | ||
| + | |||
| + | | ||
| + | | ||
| + | |||
__NOTOC__ | __NOTOC__ | ||
| | ||
| Line 14: | Line 22: | ||
;Briefly discuss the key parameters that such a program needs and how they influence the result. | ;Briefly discuss the key parameters that such a program needs and how they influence the result. | ||
</div> | </div> | ||
| − | + | <br> | |
| + | <br> | ||
==2003== | ==2003== | ||
| Line 70: | Line 79: | ||
*'''Would the same search using PSI-BLAST rather than BLAST have helped for your task?''' | *'''Would the same search using PSI-BLAST rather than BLAST have helped for your task?''' | ||
</div> | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2003 - Clustal W== | ||
| + | |||
| + | In order to run a multiple alignment from a Web interface to the ClustalW program, you are requested to specify a number of parameters. | ||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Briefly discuss gap and weight-matrix parameters, their relationship and sensible choices.''' | ||
| + | *'''Briefly list the key steps of the ClustalW algorithm.''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
==2003 - PSI-Blast== | ==2003 - PSI-Blast== | ||
| Line 112: | Line 133: | ||
*'''Are the two genes that "Query" and "Sbjct" refer to homologous ? Explain. ''' | *'''Are the two genes that "Query" and "Sbjct" refer to homologous ? Explain. ''' | ||
*'''Should you include this protease inhibitor in your next iteration of PSI-BLAST ? Why or why not? ''' | *'''Should you include this protease inhibitor in your next iteration of PSI-BLAST ? Why or why not? ''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2004 - Sequence alignments== | ||
| + | <br> | ||
| + | |||
| + | Typically, sequence alignments are used to measure similarity between sequences, in order to infer homology. In this course, we have used many different methods for sequence alignment. I hope, by now you are quite confident what method to use under which circumstances. | ||
| + | |||
| + | Please be brief in your answers and restrict yourself to the one or two most important inferences. However, you must be specific, eg. in case you argue that you could infer a property such as homology from an alignment, you must state what you would consider sufficient evidence for that conclusion. | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Briefly state what input data and other data resources and/or parameters are needed to perform a Needleman-Wunsch or Smith-Waterman sequence alignment and what you can infer from the results.''' | ||
| + | *'''Briefly state when you would use a BLAST search rather than one of the algorithms stated above and what you can infer from the results.''' | ||
| + | *'''Briefly state (i) when you would use a multiple sequence alignment program rather than any of the above algorithms and (ii) how a pairwise alignment taken from a multiple sequence alignment differs from one produced by a Needleman-Wunsch or Smith-Waterman sequence alignment.''' | ||
| + | *'''Briefly state what criteria you could use to improve a multiple sequence alignment "by hand" and how the sequence of a known protein structure could contribute useful information.''' | ||
| + | </div> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | ==2005== | ||
| + | A BLAST search was performed with the full-length (833aa) yeast Mbp1 protein (refseq database, default parameters, results restricted to Fungi with an Entrez filter). The highest scoring hit from ''Cryptococcus neoformans'' is shown here: | ||
| + | |||
| + | >gi|58266778|ref|XP_570545.1| transcription factor [Cryptococcus neoformans] | ||
| + | Length=925 | ||
| + | |||
| + | Score = 174 bits (440), Expect = 2e-42, Method: Composition-based stats. | ||
| + | Identities = 173/602 (28%), Positives = 263/602 (43%), Gaps = 76/602 (12%) | ||
| + | |||
| + | Query 1 MSNQ--IYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEV 58 | ||
| + | MS Q +Y++ YSGV V+E + S+M+R D WVNAT ILK A K+ RT+ILEKEV | ||
| + | Sbjct 108 MSTQPKVYASVYSGVPVFEAMIRGISVMRRASDSWVNATQILKVAGVHKSARTKILEKEV 167 | ||
| + | |||
| + | Query 59 LKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDFTQTDGSASPPPAPKHH 118 | ||
| + | L HEK+QGG+GKYQGTWVPL+ + LAE++ V L +FDF + | ||
| + | Sbjct 168 LNGIHEKIQGGYGKYQGTWVPLDRGRDLAEQYGVGSYLSSVFDFVPS------------- 214 | ||
| + | |||
| + | Query 119 HASKVDRKKAIRSASTSAIMETKRNNKKAEENQFQSSKILGNPTAAPRKRGRPVGSTRGS 178 | ||
| + | AS + IR+ + + + NQ S + P P +G+ | ||
| + | Sbjct 215 -ASVIAALPVIRTGTPDRSGQQTPSGLPGHPNQRVISPFANHGQTTPHMP-PPQFIHQGN 272 | ||
| + | |||
| + | Query 179 RRKLGVNLQRSQSDMGFPRPAIPNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQ 238 | ||
| + | + + NL S + +P P S+ ++ T+GPQ +ERH+ | ||
| + | Sbjct 273 EQMM--NLPPHPSSLAYPTQPKPYFSM------PLQHTVGPQY-------DERHEGMTMT 317 | ||
| + | |||
| + | Query 239 PQQNNSAQFKEIDLED-GL---SSDVEPSQQLQ-------QVFNQNTGFVPQQQSSLIQT 287 | ||
| + | P + D+ G SD+ Q Q + + +G ++Q S + | ||
| + | Sbjct 318 PTMSMDGLAPPADIARMGFPYNPSDIYIDQYGQPHATYQASPYGKESGHPSKRQRSDAEG 377 | ||
| + | |||
| + | Query 288 QQTESMATSVSSSPSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDIN 347 | ||
| + | ES A + + + + P ++P P+ RP+ + N | ||
| + | Sbjct 378 SYIESGAAVQQHVEQDEEADDGLDNDSTASDDARDPPPLPSSMLLPHKPI--RPKATPAN 435 | ||
| + | |||
| + | Query 348 DKVNKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPY-IDAPIDPELHTAFHWACSMG 406 | ||
| + | ++ S+LV F ++ +L V P + ID ID + H+A HWAC++ | ||
| + | Sbjct 436 GRIK---SRLVQIF---NVEGQVNLRSVFGLAPDQLPNFDIDMVIDDQGHSALHWACALA 489 | ||
| + | |||
| + | Query 407 NLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRIFQLLHETVFDIDSQSQ 466 | ||
| + | L I + L E G I N G+TPL+R+ L N +F + LL ++ +D + | ||
| + | Sbjct 490 RLSIVQQLIELGADIHRGNYAGETPLIRAVLTSNHAEAGSFTDLLHLLSPSIRTLDHAYR 549 | ||
| + | |||
| + | Query 467 TVIHHI---VKRKSTTPSAVYYLDVVL-------------SKIKDFSPQYRIEL------ 504 | ||
| + | TV+HHI K P+A Y+ VL S +P R EL | ||
| + | Sbjct 550 TVLHHIALVAGVKGRVPAARTYMASVLEWVAREQQANNTHSITNPPNPADRNELAPINLR 609 | ||
| + | |||
| + | Query 505 -LLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTANEIMNQQYEQMMIQ 563 | ||
| + | L++ QD +GDTAL++A++ G+ L+ GA T +NK GL E + E + I | ||
| + | Sbjct 610 TLVDVQDVHGDTALNVAARVGNKGLVGLLLDAGADKTRANKLGLRP-ENFGLEIEALKIS 668 | ||
| + | |||
| + | Query 564 NG 565 | ||
| + | NG | ||
| + | Sbjct 669 NG 670 | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | |||
| + | *'''Is this probably a homologue? Why or why not?''' | ||
| + | *'''Could this be a full-length homologue or has the BLAST alignment excluded this possibility?''' | ||
| + | *'''Describe how to further analyze whether the two sequences are homologous over their full length.''' | ||
| + | *'''Could this be an orthologue? Describe the steps that you would need to perform to test this.''' | ||
| + | *'''What further information could an RPS-BLAST or SMART analysis of the two proteins contribute?''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==2009== | ||
| + | You know that most fungal species have several proteins with APSES domains that are homologous to Mbp1. But do they also share the AT-hook domain? In order to find that out, you perform a BLAST search with the Refseq identifier of the ''Candida glabrata'' Mbp1 orthologue: XP_445458. | ||
| + | |||
| + | |||
| + | On the BLAST entry page, you find links to the following programs: nucleotide BLAST, protein BLAST, BLASTX, TBLASN, and TBLASTX. | ||
| + | |||
| + | |||
| + | On the "Choose Search Set" section of the search form, you are asked to select from the database options: "Non-redundant protein sequences (nr), "Reference proteins (refseq_protein), "Swissprot protein sequences(swissprot), "Patented protein sequences(pat), "Protein Data Bank proteins(pdb)," and "Environmental samples(env_nr)". | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Which BLAST program should you choose?''' | ||
| + | *'''Which database option should you select? ''' | ||
| + | :<small>Write your answer in one word each and one sentence to justify your choice.</small> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | From the hits that were returned, you are interested in the hit to an ''Eremothecium gossypii'' protein, (it is NP_986147). You would like to get an optimal sequence alignment between that part of the query sequence that contains the APSES domain and the annotated AT-hook motifs, and the homologous sequence from XP_444966. | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Describe briefly how this can be achieved: how and where can you retrieve the sequences, where and how can you obtain the alignment.''' | ||
| + | </div> | ||
| + | |||
| + | |||
| + | Here is the relevant part of the optimal sequence alignment result: it includes the APSES domain of the of the ''Candida glabrata'' Mbp1 orthologue (<code>QIYSAKY...LKPLF</code>) and the first (<code>AKKAGRSVSSPAM</code>) and second (<code>TRRRGRPPNSTLT</code>) annotated AT-hook motif. | ||
| + | |||
| + | |||
| + | # Program: needle | ||
| + | # Aligned_sequences: 2 | ||
| + | # 1: XP_445458 | ||
| + | # 2: NP_986147 | ||
| + | # Matrix: EBLOSUM62 | ||
| + | # Gap_penalty: 10.0 | ||
| + | # Extend_penalty: 0.5 | ||
| + | # | ||
| + | # Length: 884 | ||
| + | # Identity: 358/884 (40.5%) | ||
| + | # Similarity: 506/884 (57.2%) | ||
| + | # Gaps: 151/884 (17.1%) | ||
| + | #======================================= | ||
| + | XP_445458.1 1 -------MSNQIYSAKYSGVDVYEFIHPTGSIMKRKNDGWVNATHILKAA 43 | ||
| + | .:.||||||||||:||||:||||||||||.|.||||||||||| | ||
| + | NP_986147.2 1 MSAGSAVSATQIYSAKYSGVEVYEFLHPTGSIMKRKADDWVNATHILKAA 50 | ||
| + | |||
| + | XP_445458.1 44 NFAKAKRTRILEKEVLKEMHEKVQGGFGKYQGTWVPLNIAINLAEKFDVY 93 | ||
| + | .||||||||||||||:|:.||||||||||||||||||:||..||:||:|. | ||
| + | NP_986147.2 51 KFAKAKRTRILEKEVIKDTHEKVQGGFGKYQGTWVPLDIARRLAQKFEVL 100 | ||
| + | |||
| + | XP_445458.1 94 QDLKPLFDFSEENGDAAPPPAPKHHHASKASSAKAKKAGRSVSSPAMNDS 143 | ||
| + | ::|:|||||:..:|..:||.||||||||:|.||:. |:..||.:... | ||
| + | NP_986147.2 101 EELRPLFDFTRRDGSESPPQAPKHHHASRADSARK----RTTKSPPLPHG 146 | ||
| + | |||
| + | XP_445458.1 144 KTRASTRKANTPSSNDITSDSGAVVNPVVTRRRGRPPNSTLTNKRKLG-- 191 | ||
| + | :..| :.:||||||.: |||. | ||
| + | NP_986147.2 147 QLDA------------------------LPKRRGRPPRA-----RKLSDV 167 | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Based on the alignment results, do the two sequences appear homologous over most of the aligned length, or only in the APSES domain?''' | ||
| + | *'''Are either or both AT-hook domains conserved and/or are other AT-hook motifs present in NP_986147 (but not aligned to XP_445458)?''' | ||
| + | *'''Are there other features present in the sequences that could indicate similar function in the region between the APSES domain and the second AT-hook motif?''' | ||
</div> | </div> | ||
| + | |||
| + | <!-- | ||
==2002== | ==2002== | ||
[[Image:Stereo_000000.jpg|frame|none|Caption. ]] | [[Image:Stereo_000000.jpg|frame|none|Caption. ]] | ||
| + | <br> | ||
| + | |||
| + | Explanation ... | ||
| + | |||
| + | <br> | ||
<div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| − | + | *''' ''' | |
</div> | </div> | ||
<small>Comment</small> | <small>Comment</small> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | --> | ||
| + | |||
| + | [[Category: Bioinformatics]] | ||
| + | </div> | ||
Latest revision as of 01:56, 11 December 2012
Tools Exam Questions
One aspect of bioinformatics concerns algorithms: computational tools that allow us to analyse the data and support our inferences.
2003
WWW servers for the multiple alignment program T-Coffee require only a set of sequences as input for their task. Obviously, the important parameters the program uses are hidden - they have been set to a reasonable default.
- Briefly discuss the key parameters that such a program needs and how they influence the result.
2003
- " Magnaporthe grisea, the causal agent of rice blast disease, is one of the most devastating threats to food security worldwide. Conservatively, each year enough rice is destroyed by rice blast disease to feed 60 million people [...]. Indeed, the Centers for Disease Control and Prevention has recently recognized and listed rice blast as a significant biological weapon. No part of the world is now safe from this disease. It was long thought of as being confined to developing nations, but over the past decade it has emerged as a serious problem in the United States. [...] Widespread devastation of golf courses, particularly in the Midwest, where it has been attacking cool season grasses, is of particular concern. "
[... excerpt from the Web pages of the US Magnaporthe grisea genome project of the Center for Genome Research]
In an effort to annotate the M. grisea genome, you have done a BLAST search of the E. coli Glutaminyl tRNA synthetase gene against the predicted M. grisea open reading frames: your goal is to find the orthologue of this gene. You have chosen the "nr" database and have limited the search output to magnaporthe grisea"[organism] in the appropriate advanced-options field of the Web form. Here are excerpts from the output you receive:
Sequences producing significant alignments: (bits) Value gi|38104873|gb|EAA51376.1| hypothetical protein MG09393.4 [... 391 e-109 gi|38106536|gb|EAA52828.1| hypothetical protein MG05956.4 [... 268 3e-72 gi|38106250|gb|EAA52583.1| hypothetical protein MG05275.4 [... 59 2e-09 gi|38101579|gb|EAA48524.1| hypothetical protein MG00182.4 [... 30 1.7
- Comment briefly on each of the portions of the above excerpt from the BLAST output, that is formatted in bold and red.
>gi|38106250|gb|EAA52583.1| hypothetical protein MG05275.4 [Magnaporthe grisea ]
Length = 594
Score = 59.3 bits (142), Expect = 2e-09
Identities = 61/243 (25%), Positives = 102/243 (41%), Gaps = 34/243 (13%)
Query: 30 TRFPPEPNGYLHIGHAKSICLNFGIAQDYKGQCNLRFDDTNPVKEDIEYVESIKNDVEWL 89
TRF P P G+LH+G ++ N+ +A+ GQ LR +DT+ + + + D+ W
Sbjct: 61 TRFAPSPTGFLHLGSLRTALFNYLLAKATGGQFLLRLEDTDRTRIVPDAEARLYQDLRWA 120
Query: 90 GFHW---------SGNVRYSSDYFDQLHAYAIELINKGLAYVDELTPEQIREYR-GTLTQ 139
G W SG R S+ YA +L++ G AY T E++ + G+
Sbjct: 121 GLVWDEGPDVGGPSGPYR-QSERLGHYSKYAQQLLDSGRAYRCFCTREELAASQLGSQAD 179
Query: 140 PGKNSPYRDRSVEENLALFEKMRAGGFEEGKACLRAKIDMASPFIVMRDPVLYRIKFAEH 199
G Y + + E+ A G +R + + +PF V P L +F +
Sbjct: 180 SGAGGRYPGTCLAVSADESEERAARG---DAHVIRFRSN-TTPFTV---PDLVYRRFRKK 232
Query: 200 HQTGN----KWCIYPMYDFTHCISDALEGITHSLCTLEFQDNRRLYDWVLDNITIPVHPR 255
H + K +P Y F + + D L +TH + R +W+ I+ P+H
Sbjct: 233 HMEDDFIIMKSDGFPTYHFANVVDDHLMDVTHVI---------RGAEWL---ISTPMHCD 280
Query: 256 QYE 258
Y+
Sbjct: 281 LYD 283
- Briefly discuss what you can conclude about MG05275.4 from the above excerpt of the BLAST report.
- Describe at least two approaches for functional annotation that are not based on homology that you can use to annotate MG05275.4 ?
- Would the same search using PSI-BLAST rather than BLAST have helped for your task?
2003 - Clustal W
In order to run a multiple alignment from a Web interface to the ClustalW program, you are requested to specify a number of parameters.
- Briefly discuss gap and weight-matrix parameters, their relationship and sensible choices.
- Briefly list the key steps of the ClustalW algorithm.
2003 - PSI-Blast
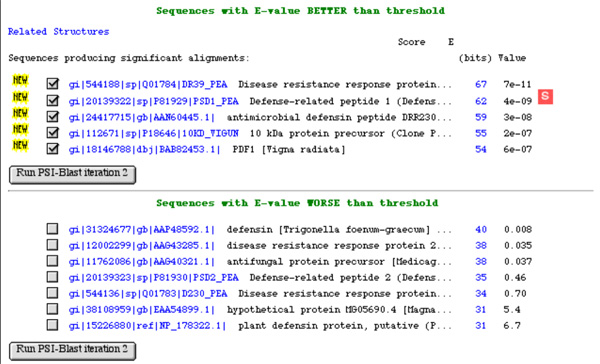
Defensins are small proteins of about 50 amino acids with a characteristic fold and disulfide bonding pattern. Plants have large families of defensins in their genome conferring resistance against fungal and bacterial pathogens. While resistance against fungi appears to involve specific binding to membrane targets, antibacterial effects seem to involve non-specific membrane permeabilization. In order to establish the relative importance of specific binding to target proteins and non-specific, physicochemical mode of action, you reason that specific binding should be compromised when you change defensin sequences towards the consensus sequence, while the non-specific effects should be enhanced. You thus decide to perform a sensitive PSI-BLAST search with the sequence of pea defensin I, as a basis for the multiple alignment of defensin sequences, in order to obtain a consensus sequence of defensin orthologs.
As you know, PSI-BLAST (Position Specific Iterated ...) scans a sequence database with a BLAST search, then builds a profile from the similar sequences it retrieves and repeats the search, then repeats this procedure, refinining the profile at every step, until no more sequences can be added.
- What key steps has the program gone through at this stage ?
- What is the "E-value" that is referred to here ?
- What will the program do in iteration 2 ?
- What input can you give the program before running iteration 2 and why is it necessary to manually adjust the input (i.e. what happens if a false positive is selected )?
- Which of these sequences are probably homologs to your query ? Explain.
Here is an excerpt from the alignments this PSI-BLAST search has produced in its first round:
gi|15226880|ref|NP_178322.1| plant defensin protein, putative (PDF2.6)
gi|11387216|sp|Q9ZUL8|THG4_ARATH Gamma-thionin homolog At2g02140 precursor
gi|25330850|pir||D84433 proteinase inhibitor II [imported] - Arabidopsis thaliana
gi|4038038|gb|AAC97220.1| protease inhibitor II [Arabidopsis thaliana]
gi|21592674|gb|AAM64623.1| protease inhibitor II [Arabidopsis thaliana]
Length = 73
Score = 30.8 bits (68), Expect = 6.7
Identities = 14/46 (30%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query: 1 KTCEHLADTYRGVCFTNASCDDHCKNKAHLISGTCHNWKCFCTQNC 46
+TCE ++ ++GVC + SC C ++ G C + +C+C++ C
Sbjct: 29 RTCESPSNKFQGVCLNSQSCAKACPSEG-FSGGRCSSLRCYCSKAC 73
gi|15226880|ref|NP_178322.1|is a piece of hypertext with a link. What does the link lead to?- What is a "gi" and what is a "ref" ?
- Why are there five records in front of one alignement here that begin with "gi|..." ?
- What does "Expect = 6.7" mean ?
- Are the two genes that "Query" and "Sbjct" refer to homologous ? Explain.
- Should you include this protease inhibitor in your next iteration of PSI-BLAST ? Why or why not?
2004 - Sequence alignments
Typically, sequence alignments are used to measure similarity between sequences, in order to infer homology. In this course, we have used many different methods for sequence alignment. I hope, by now you are quite confident what method to use under which circumstances.
Please be brief in your answers and restrict yourself to the one or two most important inferences. However, you must be specific, eg. in case you argue that you could infer a property such as homology from an alignment, you must state what you would consider sufficient evidence for that conclusion.
- Briefly state what input data and other data resources and/or parameters are needed to perform a Needleman-Wunsch or Smith-Waterman sequence alignment and what you can infer from the results.
- Briefly state when you would use a BLAST search rather than one of the algorithms stated above and what you can infer from the results.
- Briefly state (i) when you would use a multiple sequence alignment program rather than any of the above algorithms and (ii) how a pairwise alignment taken from a multiple sequence alignment differs from one produced by a Needleman-Wunsch or Smith-Waterman sequence alignment.
- Briefly state what criteria you could use to improve a multiple sequence alignment "by hand" and how the sequence of a known protein structure could contribute useful information.
2005
A BLAST search was performed with the full-length (833aa) yeast Mbp1 protein (refseq database, default parameters, results restricted to Fungi with an Entrez filter). The highest scoring hit from Cryptococcus neoformans is shown here:
>gi|58266778|ref|XP_570545.1| transcription factor [Cryptococcus neoformans]
Length=925
Score = 174 bits (440), Expect = 2e-42, Method: Composition-based stats.
Identities = 173/602 (28%), Positives = 263/602 (43%), Gaps = 76/602 (12%)
Query 1 MSNQ--IYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEV 58
MS Q +Y++ YSGV V+E + S+M+R D WVNAT ILK A K+ RT+ILEKEV
Sbjct 108 MSTQPKVYASVYSGVPVFEAMIRGISVMRRASDSWVNATQILKVAGVHKSARTKILEKEV 167
Query 59 LKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDFTQTDGSASPPPAPKHH 118
L HEK+QGG+GKYQGTWVPL+ + LAE++ V L +FDF +
Sbjct 168 LNGIHEKIQGGYGKYQGTWVPLDRGRDLAEQYGVGSYLSSVFDFVPS------------- 214
Query 119 HASKVDRKKAIRSASTSAIMETKRNNKKAEENQFQSSKILGNPTAAPRKRGRPVGSTRGS 178
AS + IR+ + + + NQ S + P P +G+
Sbjct 215 -ASVIAALPVIRTGTPDRSGQQTPSGLPGHPNQRVISPFANHGQTTPHMP-PPQFIHQGN 272
Query 179 RRKLGVNLQRSQSDMGFPRPAIPNSSISTTQLPSIRSTMGPQSPTLGILEEERHDSRQQQ 238
+ + NL S + +P P S+ ++ T+GPQ +ERH+
Sbjct 273 EQMM--NLPPHPSSLAYPTQPKPYFSM------PLQHTVGPQY-------DERHEGMTMT 317
Query 239 PQQNNSAQFKEIDLED-GL---SSDVEPSQQLQ-------QVFNQNTGFVPQQQSSLIQT 287
P + D+ G SD+ Q Q + + +G ++Q S +
Sbjct 318 PTMSMDGLAPPADIARMGFPYNPSDIYIDQYGQPHATYQASPYGKESGHPSKRQRSDAEG 377
Query 288 QQTESMATSVSSSPSLPTSPGDFADSNPFEERFPGGGTSPIISMIPRYPVTSRPQTSDIN 347
ES A + + + + P ++P P+ RP+ + N
Sbjct 378 SYIESGAAVQQHVEQDEEADDGLDNDSTASDDARDPPPLPSSMLLPHKPI--RPKATPAN 435
Query 348 DKVNKYLSKLVDYFISNEMKSNKSLPQVLLHPPPHSAPY-IDAPIDPELHTAFHWACSMG 406
++ S+LV F ++ +L V P + ID ID + H+A HWAC++
Sbjct 436 GRIK---SRLVQIF---NVEGQVNLRSVFGLAPDQLPNFDIDMVIDDQGHSALHWACALA 489
Query 407 NLPIAEALYEAGTSIRSTNSQGQTPLMRSSLFHNSYTRRTFPRIFQLLHETVFDIDSQSQ 466
L I + L E G I N G+TPL+R+ L N +F + LL ++ +D +
Sbjct 490 RLSIVQQLIELGADIHRGNYAGETPLIRAVLTSNHAEAGSFTDLLHLLSPSIRTLDHAYR 549
Query 467 TVIHHI---VKRKSTTPSAVYYLDVVL-------------SKIKDFSPQYRIEL------ 504
TV+HHI K P+A Y+ VL S +P R EL
Sbjct 550 TVLHHIALVAGVKGRVPAARTYMASVLEWVAREQQANNTHSITNPPNPADRNELAPINLR 609
Query 505 -LLNTQDKNGDTALHIASKNGDVVFFNTLVKMGALTTISNKEGLTANEIMNQQYEQMMIQ 563
L++ QD +GDTAL++A++ G+ L+ GA T +NK GL E + E + I
Sbjct 610 TLVDVQDVHGDTALNVAARVGNKGLVGLLLDAGADKTRANKLGLRP-ENFGLEIEALKIS 668
Query 564 NG 565
NG
Sbjct 669 NG 670
- Is this probably a homologue? Why or why not?
- Could this be a full-length homologue or has the BLAST alignment excluded this possibility?
- Describe how to further analyze whether the two sequences are homologous over their full length.
- Could this be an orthologue? Describe the steps that you would need to perform to test this.
- What further information could an RPS-BLAST or SMART analysis of the two proteins contribute?
2009
You know that most fungal species have several proteins with APSES domains that are homologous to Mbp1. But do they also share the AT-hook domain? In order to find that out, you perform a BLAST search with the Refseq identifier of the Candida glabrata Mbp1 orthologue: XP_445458.
On the BLAST entry page, you find links to the following programs: nucleotide BLAST, protein BLAST, BLASTX, TBLASN, and TBLASTX.
On the "Choose Search Set" section of the search form, you are asked to select from the database options: "Non-redundant protein sequences (nr), "Reference proteins (refseq_protein), "Swissprot protein sequences(swissprot), "Patented protein sequences(pat), "Protein Data Bank proteins(pdb)," and "Environmental samples(env_nr)".
- Which BLAST program should you choose?
- Which database option should you select?
- Write your answer in one word each and one sentence to justify your choice.
From the hits that were returned, you are interested in the hit to an Eremothecium gossypii protein, (it is NP_986147). You would like to get an optimal sequence alignment between that part of the query sequence that contains the APSES domain and the annotated AT-hook motifs, and the homologous sequence from XP_444966.
- Describe briefly how this can be achieved: how and where can you retrieve the sequences, where and how can you obtain the alignment.
Here is the relevant part of the optimal sequence alignment result: it includes the APSES domain of the of the Candida glabrata Mbp1 orthologue (QIYSAKY...LKPLF) and the first (AKKAGRSVSSPAM) and second (TRRRGRPPNSTLT) annotated AT-hook motif.
# Program: needle
# Aligned_sequences: 2
# 1: XP_445458
# 2: NP_986147
# Matrix: EBLOSUM62
# Gap_penalty: 10.0
# Extend_penalty: 0.5
#
# Length: 884
# Identity: 358/884 (40.5%)
# Similarity: 506/884 (57.2%)
# Gaps: 151/884 (17.1%)
#=======================================
XP_445458.1 1 -------MSNQIYSAKYSGVDVYEFIHPTGSIMKRKNDGWVNATHILKAA 43
.:.||||||||||:||||:||||||||||.|.|||||||||||
NP_986147.2 1 MSAGSAVSATQIYSAKYSGVEVYEFLHPTGSIMKRKADDWVNATHILKAA 50
XP_445458.1 44 NFAKAKRTRILEKEVLKEMHEKVQGGFGKYQGTWVPLNIAINLAEKFDVY 93
.||||||||||||||:|:.||||||||||||||||||:||..||:||:|.
NP_986147.2 51 KFAKAKRTRILEKEVIKDTHEKVQGGFGKYQGTWVPLDIARRLAQKFEVL 100
XP_445458.1 94 QDLKPLFDFSEENGDAAPPPAPKHHHASKASSAKAKKAGRSVSSPAMNDS 143
::|:|||||:..:|..:||.||||||||:|.||:. |:..||.:...
NP_986147.2 101 EELRPLFDFTRRDGSESPPQAPKHHHASRADSARK----RTTKSPPLPHG 146
XP_445458.1 144 KTRASTRKANTPSSNDITSDSGAVVNPVVTRRRGRPPNSTLTNKRKLG-- 191
:..| :.:||||||.: |||.
NP_986147.2 147 QLDA------------------------LPKRRGRPPRA-----RKLSDV 167
- Based on the alignment results, do the two sequences appear homologous over most of the aligned length, or only in the APSES domain?
- Are either or both AT-hook domains conserved and/or are other AT-hook motifs present in NP_986147 (but not aligned to XP_445458)?
- Are there other features present in the sequences that could indicate similar function in the region between the APSES domain and the second AT-hook motif?