Difference between revisions of "Stereo Vision Exam Questions"
m |
|||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <div id="BIO"> | ||
| + | <div class="b1"> | ||
| + | Stereo Vision Exam Questions | ||
| + | </div> | ||
| + | |||
| + | | ||
| + | | ||
| + | |||
__NOTOC__ | __NOTOC__ | ||
| | ||
| Line 5: | Line 13: | ||
;Molecules are three-dimensional entities and stereo-vision of images on paper and on the screen is one of the most powerful, intuitive ways to appreciate that. We have practiced stero-vision in this course; here are a number of situations that require spatial awareness. | ;Molecules are three-dimensional entities and stereo-vision of images on paper and on the screen is one of the most powerful, intuitive ways to appreciate that. We have practiced stero-vision in this course; here are a number of situations that require spatial awareness. | ||
</div> | </div> | ||
| − | | + | <br> |
| − | | + | <br> |
| + | |||
==2002== | ==2002== | ||
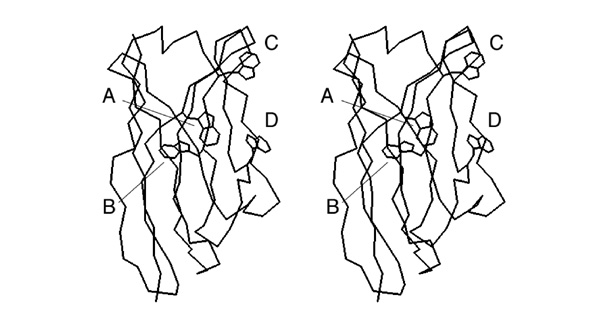
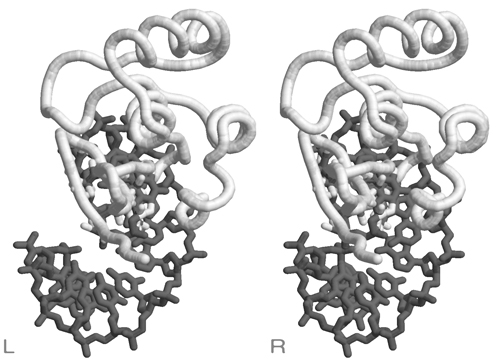
[[Image:Stereo_VH-domain.jpg|frame|none|This divergent stereo-view shows a trace of connected C<sup>α</sup> atoms of a protein domain (the VH domain of the anti-Fluorescein antibody 4-4-20, 4FAB.PDB) and a wireframe representation of all its tryptophan sidechains. ]] | [[Image:Stereo_VH-domain.jpg|frame|none|This divergent stereo-view shows a trace of connected C<sup>α</sup> atoms of a protein domain (the VH domain of the anti-Fluorescein antibody 4-4-20, 4FAB.PDB) and a wireframe representation of all its tryptophan sidechains. ]] | ||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
;Write down the label of the tryptophan that is a conserved element of the hydrophobic core of this domain. | ;Write down the label of the tryptophan that is a conserved element of the hydrophobic core of this domain. | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
==2002== | ==2002== | ||
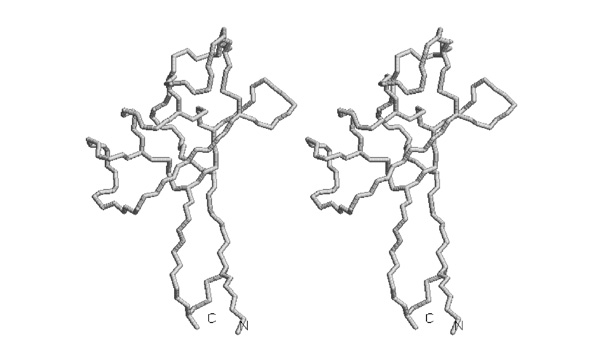
| − | [[Image: | + | [[Image:Stereo_Pleckstrin-domain.jpg|frame|none|This divergent stereo-view shows a trace of connected C<sup>α</sup> atoms of a Pleckstrin PH domain, 1PLS.PDB ]] |
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
;Trace the backbone from the N-terminus to amino acid 52 with pencil or pen in one of the images. | ;Trace the backbone from the N-terminus to amino acid 52 with pencil or pen in one of the images. | ||
| + | </div> | ||
| + | |||
| + | <small>''I wouldn't ask this type of question any longer - the drawing task seems a bit convoluted for the actual skill it is supposed to test.</small> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2003== | ||
| + | [[Image:Stereo_ATP-binding_site.jpg|frame|none|This divergent stereo-view shows selected helices from 1GTR.pdb (backbone only, light grey) and the substrate ATP (dark grey). ]] | ||
| + | |||
| + | As you know, a strong dipole moment is generated from the synergistic interactions of carbonyl groups in alpha helices. The carbonyls point towards the negative potential. | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | ;Which three alpha helices are oriented best, so that their helix-dipole moment is aligned for favourable interactions with the ATP phosphate groups? | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | ==2003== | ||
| + | [[Image:Stereo_DsbA_helices.jpg|frame|none|This divergent stereo-view shows selected helices from 1A2J.pdb (DsbA). ]] | ||
| + | |||
| + | The figure was generated with the following RasMol commands: | ||
| + | |||
| + | set background white | ||
| + | set stereo -5 | ||
| + | select all | ||
| + | color white | ||
| + | restrict helix and backbone | ||
| + | wireframe 90 | ||
| + | select glu,asp | ||
| + | color [80,80,80] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | ;Mark on this sheet the position of those Asp or Glu residues that are positioned to interact favourably with the helix dipole. If there are several plausible residues in a helix, mark the one closest to the correct terminus. | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2003== | ||
| + | [[Image:Stereo_Defensin_1.jpg|frame|none|This divergent stereo-view shows a trace of connected backbone atoms – N, C<sup>α</sup>, C and O – as well as the cysteine sidechains of the four disulfide bridges, of the pea defensin 1JKZ.pdb. ]] | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | ;Trace the disulfide bonded cysteine sidechains in one of the stereoviews of this picture. | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2004== | ||
| + | [[Image:Stereo_Defensin_2.jpg|frame|none|This divergent stereo-view shows a trace of connected backbone atoms – N, C<sup>α</sup> and C – as well as the cysteine sidechains of the four disulfide bridges, of the pea defensin 1JKZ.pdb. ]] | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | ;Number the cysteines from 1 to 8, from N- to C- terminus and determine the disulfide bonding topology of this protein. Write the disulfide bonded residue pairs into your exam booklet. | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | ==2004== | ||
| + | [[Image:Stereo_GlnRS-SaltBridges.jpg|frame|none|This divergent stereo-view shows a subdomain of GluRS from ''Thermus thermophilus'' (1N78.PDB) that is rich in charged amino acids. Sidechains are shown for aspartate, glutamate, lysine and arginine residues. The backbone is shaded in grey from light (N-terminus) to dark (C-terminus). Sequence numbers are given next to the sidechains.]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''List all pairs of residues that form salt-bridges according to this figure. Write the one-letter code for the negatively charged amino acid and its sequence number, then the same for the positively charged amino acid.''' | ||
| + | </div> | ||
| + | |||
| + | <small>Comment</small> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2004== | ||
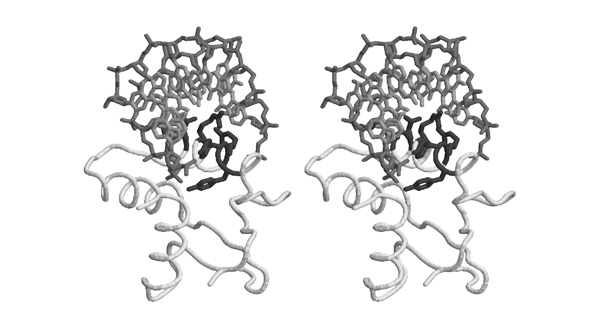
| + | [[Image:Stereo-Gln-RS_BindingSite.jpg|frame|none|This divergent stereo-view shows part of the substrate binding site of a subdomain of GluRS from ''Thermus thermophilus'' (1N78.PDB), including the substrate analogon <u>glutamol-AMP</u>. The "<code>HIGH</code>" region is included here, its sequence is <code>HVGT</code> in this case. Carbons are shown in light grey, nitrogen atoms are darker, oxygen atoms are darkest grey and sulphur and phosphorous are black.]] | ||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Based on this figure, argue briefly why "G" is a conserved residue in the "<code>HIGH</code>" motif. ''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2005== | ||
| + | |||
| + | Base specific recognition of canonically paired nucleotides can be mediated through specific recognition sites that contribute H-bond donors/acceptors or methyl groups. One example of such recognition sites for a particular basepair is sketched here: | ||
| + | |||
| + | [[Image:DNA_major-minor_groove.jpg|frame|none|In this sketch the major-groove recognition sites for a basepair are labelled A,B, C and D. ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | The sketch corresponds to the situation in the stereo figure below. It was generated with the following RasMol commands to demonstrate the fold of the domain and several important sidechains on the recognition helix that interact with various parts of the DNA. | ||
| + | |||
| + | set background white | ||
| + | select all | ||
| + | wireframe off | ||
| + | restrict *c | ||
| + | color white | ||
| + | wireframe off | ||
| + | trace 90 | ||
| + | select (58,59,62,65-67) and *c | ||
| + | select (*.ca or sidechain) and selected | ||
| + | color [80,80,80] | ||
| + | wireframe 100 | ||
| + | select (5-11 and *a) or (3-9 and *b) | ||
| + | color [190,190,190] | ||
| + | wireframe 80 | ||
| + | select (*a or *b) and backbone | ||
| + | color [140,140,140] | ||
| + | set stereo -5 | ||
| + | |||
| + | <br> | ||
| + | |||
| + | [[Image:DNA_Elk-1_RecognitionHelix.jpg|frame|none|This divergent stereo-view shows the recognition helix of the Elk-1 transcription factor (pdb|1DUX) inserted into the major groove of a cognate oligonucleotide. ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Identify the "C" and "D" recognition interactions depicted in the sketch above. Note the one-letter code(s) for the amino acid type(s) that is/are involved in these interactions. ''' | ||
| + | </div> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==2006== | ||
| + | <br> | ||
| + | |||
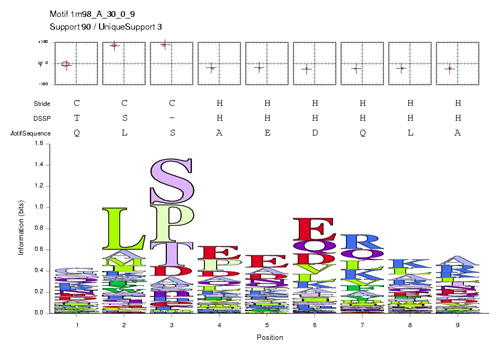
| + | In order to study structural and functional conservation patterns in the Mbp1 recognition helix, you obtain a structural motif analysis from a collaborator. The segment of residues 44 to 53 (numbering follows the 1MB1 structure) corresponds to a well defined helix N-cap motif. This motif is a structural pattern that recurrs 90 times in the Nh3D database of non-homologous proteins. The amino acid propensities of this pattern are summarized in the sequence-logo below: | ||
| + | |||
| + | <br> | ||
| + | |||
| + | [[Image:MotifSequenceLogoSmall.jpg|frame|none|Sequence logo for a nine-amino acid helix N-cap motif (1m98_A_30_0_9). ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | After loading the PDB coordinate file 1MB1, a stereo view was generated with the following RasMol commands: | ||
| + | |||
| + | set background white | ||
| + | set stereo -5 | ||
| + | restrict 44-53 | ||
| + | color white | ||
| + | wireframe 80 | ||
| + | select backbone | ||
| + | color [170,170,170] | ||
| + | select 46 | ||
| + | color [80,80,80] | ||
| + | select 53-60 and backbone | ||
| + | wireframe 80 | ||
| + | select 6 and HOH | ||
| + | cpk 100 | ||
| + | |||
| + | <br> | ||
| + | [[Image:Helix_N-cap.jpg|frame|none|Divergent stereo view of residues 44 to 53 of 1MB1 as well as the backbone atoms of residues 54 to 60. The view is from the solvent-accessible surface towards the core of the protein, it includes the putative recognition helix of Mbp1, the 3 residues leading up to it, as well as a water molecule from the solvent shell. This structured water molecule is observed in approximately the location that a sidechain hydroxyl in position 46 would occupy. To facilitate orientation, Ala46 has been colored dark; it corresponds to residue 3 of the N-cap motif sequence logo given above. ]] | ||
| + | |||
| + | <br> | ||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Briefly explain the high propensity for S/T in position 3 of the N-cap motif. This corresponds to Ala46 of 1MB1, which is not highly conserved (cf. question 1).''' | ||
| + | *'''Briefly explain the high propensity for R in position 7 of the N-cap motif . This corresponds to Arg50 of 1MB1, which is highly conserved (cf. question 1). Is this an example of structural or functional conservation?''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | ==2006== | ||
| + | The helix-turn-helix DNA binding domain of the Rfx protein was surprisingly found to bind its cognate DNA with a beta−strand inserted into the major groove, not the expected recognition helix. In order to evaluate this non-canonical binding mode as a model for APSES domain – DNA interactions, a researcher has downloaded the coordinate file 1DP7 from the PDB and openend it in a text editor. Here is an excerpt from the header of the file. | ||
| + | |||
| + | HEADER TRANSCRIPTION/DNA 23-DEC-99 1DP7 | ||
| + | TITLE COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE | ||
| + | TITLE 2 X-BOX BINDING SITE | ||
| + | COMPND MOL_ID: 1; | ||
| + | COMPND 2 MOLECULE: MHC CLASS II TRANSCRIPTION FACTOR HRFX1; | ||
| + | COMPND 3 CHAIN: P; | ||
| + | COMPND 8 MOL_ID: 2; | ||
| + | COMPND 9 MOLECULE: DNA (5'-D(*CP*GP*(BRO)UP*TP*AP*CP*CP*AP*(BRO) | ||
| + | COMPND 10 UP*GP*GP*TP*AP*AP*CP*G)-3'); | ||
| + | COMPND 11 CHAIN: D; | ||
| + | |||
| + | The researcher now copies the complete set of coordinate records for '''Chain D''' from this file and pastes it right before the "END" record of the 1MB1 coordinate set. The result is shown below. | ||
| + | |||
| + | Considering the scenario and the figure below, you should immediately recognize two fundamental errors in this procedure. | ||
| − | <small>'' | + | [[Image:Stereo_nonsense_complex.jpg|frame|none|Divergent stereo view of the result of an attempt to model a protein/DNA complex based on the 1MB1 coordinates (light) and the DNA chain from 1DP7 (dark). ]] |
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Briefly describe the problems that you can see in the figure. Explain the two steps that need to be done differently to model a protein DNA complex, based on structural similarity between the Mbp1 APSES domain and the Rfx /DNA complex structure (the 1DP7 coordinate set).''' | ||
| + | </div> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==2007== | ||
| + | DNA-helicases unwind DNA and separate the strands as a prerquisite for replication. A structure of an archeal helicase published this year by Büttner et al. ( Nat Struct Mol Biol. 2007 14:647) has provided fascinating insight into the mechanism of this crucial process. The mechanical analogy to the operation of a zipper is startling: ATP hydrolysis moves a helix that pulls on a DNA strand in a ratchet-like mechanism. And a small structural element takes on the role of the wedge that separates the zipper's teeth. | ||
| + | |||
| + | [[Image:2007exam_stereo_a.jpg|frame|none|Divergent stereo view of (2P6R) Archeaoglobus Fulgidus Superfamily 2 Helicase Hel308, in complex with partially unwound, separated duplex DNA. The protein is displayed in a cartoon representation, the DNA is shown in a liquorice representation. The crystal structure is of reasonable quality, 3.0Å resolution; it contains coordinates for 686 of the protein's 702 residues. ]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Describe in one sentence the structural elements that form the "wedge" which appears to cause the DNA-strand separation.''' | ||
| + | </div> | ||
| + | |||
| + | |||
| + | [[Image:2007exam_stereo_b.jpg|frame|none|Divergent stereo view of (2P6R) Archeaoglobus Fulgidus Superfamily 2 Helicase Hel308, in complex with partially unwound, separated duplex DNA. Close-up showing parts of the protein-DNA interaction interface, "liquorice" representation. The DNA strands are shown, as well as the backbone atoms of two secondary-structure elements of the protein (residues 240 to 265), and the sidechains of residue 252 and 253. ]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Briefly describe the interaction of the sidechain of residue 252 with the DNA strand, state whether this interaction stabilizes or destabilizes the complex, and why.''' | ||
| + | *'''Briefly describe the interaction of the DNA strand with the secondary structure element that extends from residue 251 to 265, state whether this interaction helps bind DNA or is more likely to aid in strand release and explain why.''' | ||
| + | :In your answers, refer to specific interactions by residue number (if given), residue name and by naming the type of interaction. <small>Example: "a Hydrogen-bond between the hydroxyl of Thr 123 and a DNA ribose oxygen").</small> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <small>The key here is to clearly identify and distinguish the protein backbone, amino acid sidechains and DNA strand. Once the nature of the sidechains is clear to you, and you remember where the phosphate groups in a DNA backbone are located, a the role of the two sidechains should suggest itself. Also, you should realize that you have seen the context of this scene in the first image in a different orientation and that the function of this element was described in the introduction. </small> | ||
| + | |||
| + | |||
| + | |||
| + | ==2008== | ||
| + | Zinc finger domains are ubiquitous DNA binding modules in a large number of transcription factors. Often such proteins contain a large number (ten or more) such domains. Here is the structure of the zinc-finger domain for which the sequence was given in the [[Amino_Acid_Exam_Questions#2008|2008 Amino Acid question]]. | ||
| + | |||
| + | [[Image:2008exam_stereo_a.jpg|frame|none|Divergent stereo view of one domain from the crystal structure of the human YY1 zinc finger domain in complex with a viral initator element (1UBD). Only the polypeptide is shown, displayed in a licorice representation. The Zn-atom is shown as a van der Waals radius sized sphere (gray). Carbon is colored black and N, O and S are colored white. ]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Describe in one sentence how the Zn-atom is coordinated i.e. which atoms from which types of residues participate.''' | ||
| + | :(You do not need to mention the PDB atom name - e.g. if a threonine were involved, you would not need to identify the "OG1" atom but only write e.g. "an oxygen from a threonine sidechain."). | ||
| + | *'''Briefly describe in general terms what role the Zn-atom appears to play for the structure or function of this domain. ''' | ||
| + | </div> | ||
| + | |||
| + | |||
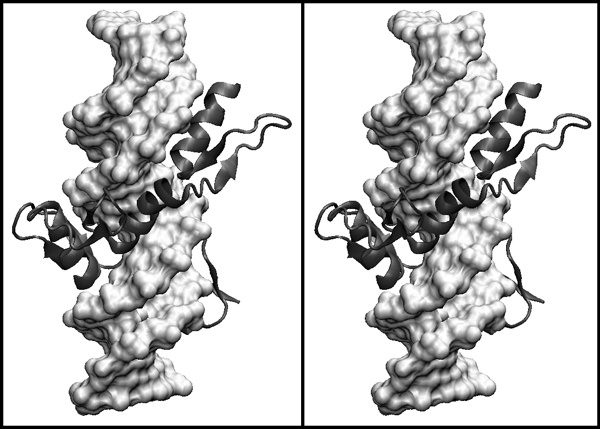
| + | [[Image:2008exam_stereo_b.jpg|frame|none|Divergent stereo view of the human YY1 zinc finger domain in complex with a viral initator element (1UBD). The polypeptide chain is shown in a cartoon presentation (gray), The DNA is shown in a surface representation. Three of the protein's four zinc finger domains are clearly visible, the fourth one is mostly hidden by DNA. The domain shown in the previous stereo image is the most C-terminal of the four domains. ]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Describe in one sentence the topology of a C2H2 zinc finger domain.''' | ||
| + | *'''Describe in one sentence how a single zinc finger domain contacts DNA in principle.''' | ||
| + | </div> | ||
| + | |||
| + | ==2009== | ||
| + | |||
| + | AT-hook motifs have been described in a large number of DNA-binding proteins. While they themselves don't appear to have strong sequence specificity, they are often ancillary to more specific DNA binding domains and increase the overall affinity of the protein to DNA. Here is the structure of the AT-hook motif shown in [[Amino_Acid_Exam_Questions#2009|the 2009 amino acid question]], in complex with a B-DNA double helix. | ||
| + | |||
| + | |||
| + | [[Image:2009exam-AT-HookStereoImage.jpg|frame|none|Stereo view of the AT-hook motif in the 2EZD NMR-structure, in complex with B-DNA. The polypeptide is shown in a liqorice representation. Carbon is coloured black and N, O and S are coloured white. Nucleic acids are shown in a solvent-accessible surface representation.]] | ||
| + | |||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *'''Explain the conservation of residue #2 of the AT-hook motif.''' | ||
| + | :<small>(Residue numbers refer to the [[Amino_Acid_Exam_Questions#209|sequence logo of the motif]].)</small> | ||
| + | *'''Identify the proline residue that is highly conserved in the AT-hook motif.''' | ||
| + | *'''Explain the conservation of residue #5 of the AT-hook motif.''' | ||
| + | </div> | ||
| + | |||
| + | <small>Refer to the [[Amino_Acid_Exam_Questions#209|sequence logo of the motif]] and the interpretation of the sequence in this structure and the canonical sequence of the domain to orient yourself. Once you know which residue is which, it should be obvious from the scene why particular residues cannot be mutated without significant loss of function. This provides the explanation that is asked for.</small> | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | ==2002== | ||
| + | [[Image:Stereo_000000.jpg|frame|none|Caption. ]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | Explanation ... | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <div style="padding: 5px; background: #DDDDDD; border:solid 1px #000000;"> | ||
| + | *''' ''' | ||
| + | </div> | ||
| + | |||
| + | <small>Comment</small> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | --> | ||
| + | |||
| + | [[Category: Bioinformatics]] | ||
| + | </div> | ||
Latest revision as of 01:57, 11 December 2012
Stereo Vision Exam Questions
- Molecules are three-dimensional entities and stereo-vision of images on paper and on the screen is one of the most powerful, intuitive ways to appreciate that. We have practiced stero-vision in this course; here are a number of situations that require spatial awareness.
2002
- Write down the label of the tryptophan that is a conserved element of the hydrophobic core of this domain.
2002
- Trace the backbone from the N-terminus to amino acid 52 with pencil or pen in one of the images.
I wouldn't ask this type of question any longer - the drawing task seems a bit convoluted for the actual skill it is supposed to test.
2003
As you know, a strong dipole moment is generated from the synergistic interactions of carbonyl groups in alpha helices. The carbonyls point towards the negative potential.
- Which three alpha helices are oriented best, so that their helix-dipole moment is aligned for favourable interactions with the ATP phosphate groups?
2003
The figure was generated with the following RasMol commands:
set background white set stereo -5 select all color white restrict helix and backbone wireframe 90 select glu,asp color [80,80,80]
- Mark on this sheet the position of those Asp or Glu residues that are positioned to interact favourably with the helix dipole. If there are several plausible residues in a helix, mark the one closest to the correct terminus.
2003
- Trace the disulfide bonded cysteine sidechains in one of the stereoviews of this picture.
2004
- Number the cysteines from 1 to 8, from N- to C- terminus and determine the disulfide bonding topology of this protein. Write the disulfide bonded residue pairs into your exam booklet.
2004
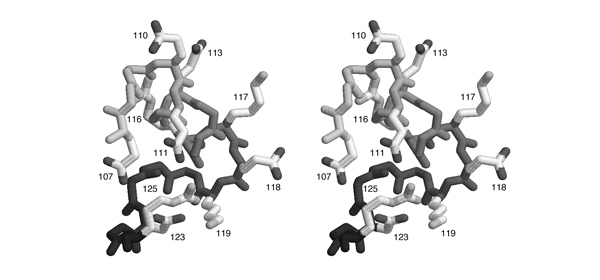
- List all pairs of residues that form salt-bridges according to this figure. Write the one-letter code for the negatively charged amino acid and its sequence number, then the same for the positively charged amino acid.
Comment
2004

HIGH" region is included here, its sequence is HVGT in this case. Carbons are shown in light grey, nitrogen atoms are darker, oxygen atoms are darkest grey and sulphur and phosphorous are black.
- Based on this figure, argue briefly why "G" is a conserved residue in the "
HIGH" motif.
2005
Base specific recognition of canonically paired nucleotides can be mediated through specific recognition sites that contribute H-bond donors/acceptors or methyl groups. One example of such recognition sites for a particular basepair is sketched here:
The sketch corresponds to the situation in the stereo figure below. It was generated with the following RasMol commands to demonstrate the fold of the domain and several important sidechains on the recognition helix that interact with various parts of the DNA.
set background white select all wireframe off restrict *c color white wireframe off trace 90 select (58,59,62,65-67) and *c select (*.ca or sidechain) and selected color [80,80,80] wireframe 100 select (5-11 and *a) or (3-9 and *b) color [190,190,190] wireframe 80 select (*a or *b) and backbone color [140,140,140] set stereo -5
- Identify the "C" and "D" recognition interactions depicted in the sketch above. Note the one-letter code(s) for the amino acid type(s) that is/are involved in these interactions.
2006
In order to study structural and functional conservation patterns in the Mbp1 recognition helix, you obtain a structural motif analysis from a collaborator. The segment of residues 44 to 53 (numbering follows the 1MB1 structure) corresponds to a well defined helix N-cap motif. This motif is a structural pattern that recurrs 90 times in the Nh3D database of non-homologous proteins. The amino acid propensities of this pattern are summarized in the sequence-logo below:
After loading the PDB coordinate file 1MB1, a stereo view was generated with the following RasMol commands:
set background white set stereo -5 restrict 44-53 color white wireframe 80 select backbone color [170,170,170] select 46 color [80,80,80] select 53-60 and backbone wireframe 80 select 6 and HOH cpk 100
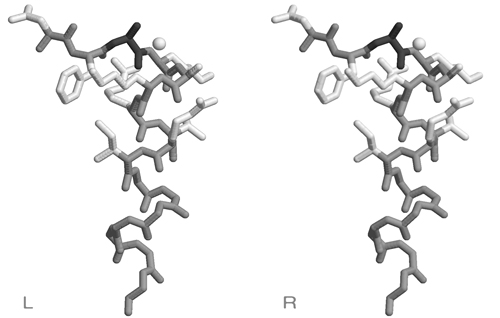
- Briefly explain the high propensity for S/T in position 3 of the N-cap motif. This corresponds to Ala46 of 1MB1, which is not highly conserved (cf. question 1).
- Briefly explain the high propensity for R in position 7 of the N-cap motif . This corresponds to Arg50 of 1MB1, which is highly conserved (cf. question 1). Is this an example of structural or functional conservation?
2006
The helix-turn-helix DNA binding domain of the Rfx protein was surprisingly found to bind its cognate DNA with a beta−strand inserted into the major groove, not the expected recognition helix. In order to evaluate this non-canonical binding mode as a model for APSES domain – DNA interactions, a researcher has downloaded the coordinate file 1DP7 from the PDB and openend it in a text editor. Here is an excerpt from the header of the file.
HEADER TRANSCRIPTION/DNA 23-DEC-99 1DP7 TITLE COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE TITLE 2 X-BOX BINDING SITE COMPND MOL_ID: 1; COMPND 2 MOLECULE: MHC CLASS II TRANSCRIPTION FACTOR HRFX1; COMPND 3 CHAIN: P; COMPND 8 MOL_ID: 2; COMPND 9 MOLECULE: DNA (5'-D(*CP*GP*(BRO)UP*TP*AP*CP*CP*AP*(BRO) COMPND 10 UP*GP*GP*TP*AP*AP*CP*G)-3'); COMPND 11 CHAIN: D;
The researcher now copies the complete set of coordinate records for Chain D from this file and pastes it right before the "END" record of the 1MB1 coordinate set. The result is shown below.
Considering the scenario and the figure below, you should immediately recognize two fundamental errors in this procedure.
- Briefly describe the problems that you can see in the figure. Explain the two steps that need to be done differently to model a protein DNA complex, based on structural similarity between the Mbp1 APSES domain and the Rfx /DNA complex structure (the 1DP7 coordinate set).
2007
DNA-helicases unwind DNA and separate the strands as a prerquisite for replication. A structure of an archeal helicase published this year by Büttner et al. ( Nat Struct Mol Biol. 2007 14:647) has provided fascinating insight into the mechanism of this crucial process. The mechanical analogy to the operation of a zipper is startling: ATP hydrolysis moves a helix that pulls on a DNA strand in a ratchet-like mechanism. And a small structural element takes on the role of the wedge that separates the zipper's teeth.
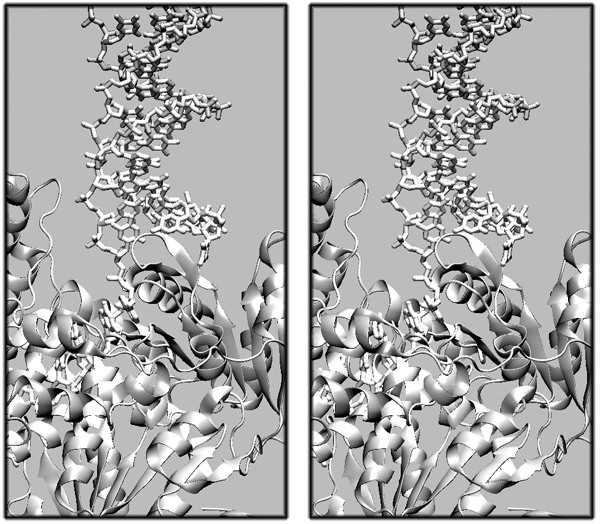
- Describe in one sentence the structural elements that form the "wedge" which appears to cause the DNA-strand separation.

- Briefly describe the interaction of the sidechain of residue 252 with the DNA strand, state whether this interaction stabilizes or destabilizes the complex, and why.
- Briefly describe the interaction of the DNA strand with the secondary structure element that extends from residue 251 to 265, state whether this interaction helps bind DNA or is more likely to aid in strand release and explain why.
- In your answers, refer to specific interactions by residue number (if given), residue name and by naming the type of interaction. Example: "a Hydrogen-bond between the hydroxyl of Thr 123 and a DNA ribose oxygen").
The key here is to clearly identify and distinguish the protein backbone, amino acid sidechains and DNA strand. Once the nature of the sidechains is clear to you, and you remember where the phosphate groups in a DNA backbone are located, a the role of the two sidechains should suggest itself. Also, you should realize that you have seen the context of this scene in the first image in a different orientation and that the function of this element was described in the introduction.
2008
Zinc finger domains are ubiquitous DNA binding modules in a large number of transcription factors. Often such proteins contain a large number (ten or more) such domains. Here is the structure of the zinc-finger domain for which the sequence was given in the 2008 Amino Acid question.

- Describe in one sentence how the Zn-atom is coordinated i.e. which atoms from which types of residues participate.
- (You do not need to mention the PDB atom name - e.g. if a threonine were involved, you would not need to identify the "OG1" atom but only write e.g. "an oxygen from a threonine sidechain.").
- Briefly describe in general terms what role the Zn-atom appears to play for the structure or function of this domain.
- Describe in one sentence the topology of a C2H2 zinc finger domain.
- Describe in one sentence how a single zinc finger domain contacts DNA in principle.
2009
AT-hook motifs have been described in a large number of DNA-binding proteins. While they themselves don't appear to have strong sequence specificity, they are often ancillary to more specific DNA binding domains and increase the overall affinity of the protein to DNA. Here is the structure of the AT-hook motif shown in the 2009 amino acid question, in complex with a B-DNA double helix.
- Explain the conservation of residue #2 of the AT-hook motif.
- (Residue numbers refer to the sequence logo of the motif.)
- Identify the proline residue that is highly conserved in the AT-hook motif.
- Explain the conservation of residue #5 of the AT-hook motif.
Refer to the sequence logo of the motif and the interpretation of the sequence in this structure and the canonical sequence of the domain to orient yourself. Once you know which residue is which, it should be obvious from the scene why particular residues cannot be mutated without significant loss of function. This provides the explanation that is asked for.