Difference between revisions of "BIO Assignment Week 4"
m |
m |
||
| (24 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
</tr></table> | </tr></table> | ||
| − | {{Template: | + | {{Template:Inactive}} |
Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz. | Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz. | ||
| Line 19: | Line 19: | ||
| | ||
| − | <div | + | <div class="quote-box"> |
| − | |||
| − | |||
<br> | <br> | ||
| Line 29: | Line 27: | ||
| − | + | {{Vspace}} | |
==Introduction== | ==Introduction== | ||
| + | {{Vspace}} | ||
<div class="colmask doublepage"> | <div class="colmask doublepage"> | ||
| Line 40: | Line 39: | ||
Sequence alignment is a '''very''' large, and important topic. | Sequence alignment is a '''very''' large, and important topic. | ||
| − | One of the foundations of bioinformatics is the empirical observation that related sequences conserve structure, and often function. Much of what we know about a protein's physiological function is based on the '''conservation''' of that function as the species evolves. | + | One of the foundations of bioinformatics is the empirical observation that related sequences conserve structure, and often function. Much of what we know about a protein's physiological function is based on the '''conservation''' of that function as the species evolves. Indeed, conservation is a defining aspect of what can rightly be said to be a protein's "function" in the first place. Conservation - or its opposite: ''variation'' - is a consequence of '''selection under constraints''': protein sequences change as a consequence of DNA mutations, this changes the protein's structure, this in turn changes functions and that has multiple effects on a species' reproductive fitness. Detrimental variants may be removed. Variation that is tolerated is largely neutral and therefore found only in positions that are neither structurally nor functionally critical. Conservation patterns can thus provide evidence for many different questions: structural conservation among proteins with similar 3D-structures, functional conservation among homologues with comparable roles, or amino acid propensities as predictors for protein engineering and design tasks. |
| − | This is the basis on which we can make inferences from well-studied model organisms | + | We assess conservation by comparing sequences between related proteins. This is the basis on which we can make inferences from well-studied model organisms for species that have not been studied as deeply. The foundation is to measure protein sequence similarity. If two sequences are much more similar than we could expect from chance, we hypothesize that their similarity comes from shared ancestry plus conservation. The measurement of sequence similarity however requires sequence alignment<ref>This is not strictly true in all cases: some algorithms measure similarity through an alignment-free approach, for example by comparing structural features, or domain annotations. These methods are less sensitive, but important when sequences are so highly diverged that no meaningful sequence alignment can be produced.</ref>. |
<!-- Column 1 end --> | <!-- Column 1 end --> | ||
| Line 49: | Line 48: | ||
<!-- Column 2 start --> | <!-- Column 2 start --> | ||
| − | + | A carefully done sequence alignment is a cornerstone for the annotation of the essential properties a gene or protein. It can already tell us a lot about which proteins we expect to have similar functions in different species. | |
| − | Multiple sequence alignments ('''MSAs''') are further useful to resolve ambiguities in the precise placement of indels and to ensure that columns in alignments actually contain amino acids that evolve in a similar context. MSAs serve as input for | + | Multiple sequence alignments ('''MSAs''') are further useful to resolve ambiguities in the precise placement of "indels"<ref>"indel": '''in'''sertion / '''del'''etion – a difference in sequence length between two aligned sequences that is accommodated by gaps in the alignment. Since we can't tell from the comparison of two sequences whether such a change was introduced by ''insertion into'' or ''deletion from'' the ancestral sequence, we join both into a {{WP|Portmanteau|''portmanteau''}}.</ref> and to ensure that columns in alignments actually contain amino acids that evolve in a similar context. MSAs serve as input for |
* functional annotation; | * functional annotation; | ||
* protein homology modelling; | * protein homology modelling; | ||
| Line 60: | Line 59: | ||
<div class="emphasis-box"> | <div class="emphasis-box"> | ||
| − | * optimal global and local alignment; | + | * optimal global and local pairwise alignment; |
| − | * BLAST searches | + | * Fast BLAST searches to determine best matches in large databases, and reciprocal best matches; |
| − | * PSI BLAST searches for exhaustive matches; and | + | * PSI BLAST searches for exhaustive matches; |
| + | * Domain annotation by sequence alignment to statistical models; and | ||
* Multiple sequence alignments. | * Multiple sequence alignments. | ||
</div> | </div> | ||
| Line 69: | Line 69: | ||
As usual, the focus will be on practical, hands on approaches. | As usual, the focus will be on practical, hands on approaches. | ||
| − | This is the scenario: you have previously identified a best match for a Mbp1 relative in YFO. Is this the most closely related protein? Is its DNA binding domain conserved? How can we identify '''all''' related genes in YFO? And, what can we learn from | + | This is the scenario: you have previously identified a best match for a Mbp1 relative in YFO. Is this the most closely related protein? Is its DNA binding domain conserved? How can we identify '''all''' related genes in YFO? And, what can we learn from that collection of sequences? |
<!-- Column 2 end --> | <!-- Column 2 end --> | ||
</div> | </div> | ||
| Line 75: | Line 75: | ||
</div> | </div> | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Preparation: Updated Database Functions === | === Preparation: Updated Database Functions === | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | The database contents and tables will change over time in this course. This means we need a mechanism to update the database, without throwing away previous work. | |
| − | + | {{task|1 = | |
| − | |||
| − | |||
| − | |||
| − | + | * Open the BCH441 project scripts in RStudio by selecting '''File''' → '''Recent Projects''' → '''BCH441_216''' | |
| + | * Load the newest versions of scripts and data by pulling from the master file on GitHub. | ||
| + | * Study the code in the <code>Database maintenance</code> section of the <code>BCH441_A04.R</code> script | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| + | {{Vspace}} | ||
| − | + | === DotPlots and the Mutation Data Matrix === | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Before we start calculating alignments, we should get a better sense of the underlying sequence similarity. A Dotplot is a perfect tool for that, because it displays alignment-free similarity information. Let's make a dotplot that uses the BLOSUM62 Mutation Data Matrix to measure pairwise amino acid similarity. The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the [ftp://ftp.ncbi.nih.gov/blast/matrices/BLOSUM62 '''BLOSUM62 matrix''']<ref>That directory also contains sourcecode to generate the PAM matrices. This may be of interest if you ever want to produce scoring matrices from your own datasets.</ref>. | |
| − | + | {{Vspace}} | |
The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the [ftp://ftp.ncbi.nih.gov/blast/matrices/BLOSUM62 '''BLOSUM62 matrix''']<ref>That directory also contains sourcecode to generate the PAM matrices. This may be of interest if you ever want to produce scoring matrices from your own datasets.</ref>. | The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the [ftp://ftp.ncbi.nih.gov/blast/matrices/BLOSUM62 '''BLOSUM62 matrix''']<ref>That directory also contains sourcecode to generate the PAM matrices. This may be of interest if you ever want to produce scoring matrices from your own datasets.</ref>. | ||
| Line 534: | Line 103: | ||
Scoring matrices are also available in the Bioconductor Biostrings package. | Scoring matrices are also available in the Bioconductor Biostrings package. | ||
| − | <source lang=" | + | <source lang="text"> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
BLOSUM62 | BLOSUM62 | ||
| Line 573: | Line 133: | ||
X -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -4 | X -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -4 | ||
* -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 1 | * -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 1 | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</source> | </source> | ||
| Line 584: | Line 138: | ||
{{task| | {{task| | ||
| − | * Study this and make sure you understand what this table is, how it can be used, and what a reasonable range of values for identities and pairscores for non-identical, similar and dissimilar residues is. Ask on the mailing list in case you have questions. | + | * Study this and make sure you understand what this table is, how it can be used, and what a reasonable range of values for identities and pairscores for non-identical, similar and dissimilar residues is. Ask on the mailing list in case you have questions. '''This piece of data is the foundation of any sequence alignment. without it, no sensible alignment could be produced!''' |
| + | * Figure out the following values: | ||
| + | ** Compare an identical match of histidine with an identical match of serine. What does this mean? | ||
| + | ** How similar are lysine and leucine, as compared to leucine and isoleucine? Is this what you expect? | ||
| + | ** PAM matrices are sensitive to an interesting artefact. Since W and R can be interchanged with a single point mutation, the probability of observing W→R and R→W exchanges in '''closely related sequences''' is much higher than one would expect from the two amino acid's biophysical properties. (Why?) PAM matrices were compiled from hypothetical point exchanges and then extrapolated. Therefore these matrices assign a relatively high degree of similarity to (W, R), that is not warranted considering what actually happens in nature. Do you see this problem in the BLOSUM matrix? If BLOSUM does not have this issue, why not? | ||
}} | }} | ||
| + | {{Vspace}} | ||
| + | |||
| + | Next, let's apply the scoring matrix for actual comparison: | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | {{task|1 = | ||
| + | * Return to your RStudio session. | ||
| + | * If you've been away from it for a while, it's probably a good idea to update to the newest versions of scripts and data by pulling from the master file on GitHub. | ||
| + | * Study and work through the code in the <code>Dotplot and MDM</code> section of the <code>BCH441_A04.R</code> script | ||
| − | + | }} | |
| − | + | {{Vspace}} | |
| − | + | == Pairwise Alignments: Optimal == | |
| + | {{Vspace}} | ||
| − | < | + | Optimal pairwise sequence alignment is the mainstay of sequence comparison. To consider such alignments in practice, we'll align the same sequences that we have just mapped in the dotplot exercise: Mbp1 and its YFO relative. For simplicity, I will call the two proteins <code>MBP1_SACCE</code> and <code>MBP1_YFO</code> through the remainder of the assignment. Your dotplots should have shown you two regions of similarity: a highly similar region focussed somewhere around the N-terminal 100 amino acids, and a more extended, but somewhat less similar region in the middle of the sequences. You can think of the sequence alignment algorithm as building the similarity matrix, and then discovering the best path along high-scoring diagonals. |
| − | + | {{Vspace}} | |
| − | |||
| − | + | === Optimal Sequence Alignment: EMBOSS online tools=== | |
| − | |||
| + | {{Vspace}} | ||
| − | + | Online programs for optimal sequence alignment are part of the EMBOSS tools. The programs take FASTA files or raw text files as input. | |
| − | + | '''Local''' optimal sequence alignment using "water" | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{task|1= | |
| + | # Fetch the sequences for <code>MBP1_SACCE</code> and <code>MBP1_YFO</code> from your database. You can simply select them by name (if you have given your sequence the suggested name when you eneterd it into your database): paste the following into the console: | ||
| − | + | * to print the <code>MBP1_SACCE</code> protein sequence to the console | |
| − | + | <source lang="R"> | |
| − | + | myDB$protein$sequence[myDB$protein$name == "MBP1_SACCE"] | |
| − | + | </source> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * to print the <code>MBP1_YFO</code> protein sequence to the console: | |
| + | <source lang="R"> | ||
| + | YFOseq <- paste("MBP1_", biCode(YFO), sep="") | ||
| + | myDB$protein$sequence[myDB$protein$name == YFOseq] | ||
</source> | </source> | ||
| + | (If this didn't work, fix it. Did you give your sequence the right '''name'''?) | ||
| + | |||
| + | # Access the [http://emboss.bioinformatics.nl/ EMBOSS Explorer site] (if you haven't done so yet, you might want to bookmark it.) | ||
| + | # Look for '''ALIGNMENT LOCAL''', click on '''water''', paste your sequences and run the program with default parameters. | ||
| + | # Study the results. You will probably find that the alignment extends over most of the protein, but does not include the termini. | ||
| + | # Considering the sequence identity cutoff we discussed in class (25% over the length of a domain), do you believe that the N-terminal domains (the APSES domains) are homologous? | ||
| + | # Change the '''Gap opening''' and '''Gap extension''' parameters to high values (e.g. 30 and 5). Then run the alignment again. | ||
| + | # Note what is different. | ||
| + | }} | ||
| + | |||
| + | |||
| + | '''Global''' optimal sequence alignment using "needle" | ||
{{task|1= | {{task|1= | ||
| − | + | # Look for '''ALIGNMENT GLOBAL''', click on '''needle''', paste the <code>MBP1_SACCE</code> and <code>MBP1_YFO</code> sequences again and run the program with default parameters. | |
| + | # Study the results. You will find that the alignment extends over the entire protein, likely with long ''indels'' at the termini. | ||
}} | }} | ||
| − | |||
| − | |||
| + | {{Vspace}} | ||
| + | |||
| + | |||
| + | === Optimal Sequence Alignment with '''R''': Biostrings === | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | Biostrings has extensive functions for sequence alignments. They are generally well written and tightly integrated with the rest of Bioconductor's functions. There are a few quirks however: for example alignments won't work with lower-case sequences<ref>While this seems like an unnecessary limitation, given that we could easily write such code to transform to-upper when looking up values in the MDM, perhaps it is meant as an additional sanity check that we haven't inadvertently included text in the sequence that does not belong there, such as the FASTA header line perhaps.</ref>. | ||
| + | |||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | {{task|1 = | ||
| + | |||
| + | * Return to your RStudio session. | ||
| + | * Once again, if you've been away from it for a while, it's always a good idea to update to pull updtaes from the master file on GitHub. | ||
| + | * Study and work through the code in the <code>Biostrings Pairwise Alignment</code> section of the <code>BCH441_A04.R</code> script | ||
| + | |||
| + | }} | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ==Heuristic pairwise alignments: BLAST== | ||
| + | {{Vspace}} | ||
| Line 644: | Line 241: | ||
<div class="col1"> | <div class="col1"> | ||
<!-- Column 1 start --> | <!-- Column 1 start --> | ||
| − | [http://www.ncbi.nlm.nih.gov/blast '''BLAST'''] is by a margin the most important computational tool of molecular biology. It is so important, that we have already used BLAST in [[ | + | [http://www.ncbi.nlm.nih.gov/blast '''BLAST'''] is by a margin the most important computational tool of molecular biology. It is so important, that we have already used BLAST in [[BIO_Assignment_Week_3#Selecting_the_YFO_.22Mbp1.22|Assignment 3]] even before properly introducing the algorithm and the principles, to find the most similar sequence to <code>MBP1_SACCE</code> in YFO. |
In this part of the assignment we will use BLAST to perform '''Reciprocal Best Matches'''. | In this part of the assignment we will use BLAST to perform '''Reciprocal Best Matches'''. | ||
| Line 650: | Line 247: | ||
One of the important questions of ''model-organism based inference'' is: which genes perform the same function in two different organisms. In the absence of other information, our best guess is that these are the two genes that are '''mutually''' most similar. The keyword here is '''mutually'''. If <code>MBP1_SACCE</code> from ''S. cerevisiae'' is the best match to <code>RES2_SCHPO</code> in ''S. pombe'', the two proteins are only mutually most similar if <code>RES2_SCHPO</code> is more similar to <code>MBP1_SACCE</code> than to any other ''S. cerevisiae'' protein. We call this a '''Reciprocal Best Match''', or "RBM"<ref>Note that RBMs are usually orthologues, but the definition of orthologue and RBM is not the same. Most importantly, many orthologues are not RBMs. We will explore this more when we discuss phylogenetic inference.</ref>. | One of the important questions of ''model-organism based inference'' is: which genes perform the same function in two different organisms. In the absence of other information, our best guess is that these are the two genes that are '''mutually''' most similar. The keyword here is '''mutually'''. If <code>MBP1_SACCE</code> from ''S. cerevisiae'' is the best match to <code>RES2_SCHPO</code> in ''S. pombe'', the two proteins are only mutually most similar if <code>RES2_SCHPO</code> is more similar to <code>MBP1_SACCE</code> than to any other ''S. cerevisiae'' protein. We call this a '''Reciprocal Best Match''', or "RBM"<ref>Note that RBMs are usually orthologues, but the definition of orthologue and RBM is not the same. Most importantly, many orthologues are not RBMs. We will explore this more when we discuss phylogenetic inference.</ref>. | ||
| − | The argument is summarized in the figure on the right: genes that evolve under continuos selective pressure on their function have relatively lower mutation rates and are thus more similar to each other, than genes that undergo neo- or sub- functionalization after duplication. | + | The argument is summarized in the figure on the right: genes that evolve under continuos selective pressure on their function have relatively lower mutation rates and are thus more similar to each other, than genes that undergo neo- or sub-functionalization after duplication. |
| − | + | However, there is a catch: proteins are often composed of multiple domains that implement distinct roles of their function. Under the assumptions above we could hypothesize: | |
* a gene in YFO that has the "same" function as the Mbp1 cell-cycle checkpoint switch in yeast should be an RBM to Mbp1; | * a gene in YFO that has the "same" function as the Mbp1 cell-cycle checkpoint switch in yeast should be an RBM to Mbp1; | ||
* a gene that binds to the same DNA sites as Mbp1 should have a DNA-binding domain that is an RBM to the DNA binding domain of Mbp1. | * a gene that binds to the same DNA sites as Mbp1 should have a DNA-binding domain that is an RBM to the DNA binding domain of Mbp1. | ||
| Line 671: | Line 268: | ||
| − | + | {{Vspace}} | |
| + | |||
| + | ===Full-length RBM=== | ||
| + | {{Vspace}} | ||
| − | |||
You have already performed the first half of the experiment: matching from ''S. cerevisiae'' to YFO. The backward match is simple. | You have already performed the first half of the experiment: matching from ''S. cerevisiae'' to YFO. The backward match is simple. | ||
{{task|1= | {{task|1= | ||
# Access [http://www.ncbi.nlm.nih.gov/blast '''BLAST'''] and follow the link to the '''protein blast''' program. | # Access [http://www.ncbi.nlm.nih.gov/blast '''BLAST'''] and follow the link to the '''protein blast''' program. | ||
| − | # Enter the | + | # Enter the RefSeq ID for <code>MBP1_YFO</code> in the '''Query sequence''' field. |
# Select <code>refseq_protein</code> as the '''database''' to search in, and enter <code>Saccharomyces cerevisiae (taxid:4932)</code> to restrict the '''organism''' for which hits are reported. | # Select <code>refseq_protein</code> as the '''database''' to search in, and enter <code>Saccharomyces cerevisiae (taxid:4932)</code> to restrict the '''organism''' for which hits are reported. | ||
# Run BLAST. Examine the results. | # Run BLAST. Examine the results. | ||
| − | If your top-hit is <code>NP_010227</code>, you have confirmed the RBM between <code>Mbp1_SACCE</code> and <code> | + | If your top-hit is <code>NP_010227</code>, you have confirmed the RBM between <code>Mbp1_SACCE</code> and <code>Mbp1_YFO</code>. If it is not, let me know. I expect this to be the same and would like to verify your results if it is not<ref>One such case we encountered involved a protein that has a corrupted annotation for the DNA binding domain. It appears to be the correct orthologue, but it only has the second highest BLAST score.</ref>. |
}} | }} | ||
| + | {{Vspace}} | ||
| − | + | ===RBM for the DNA binding domain=== | |
| − | + | {{Vspace}} | |
| − | The DNA-binding domain of <code>Mbp1_SACCE</code> is called an '''APSES''' domain. | + | The DNA-binding domain of <code>Mbp1_SACCE</code> is called an '''APSES''' domain. If the RBM between ''Saccharomyces cerevisiae'' Mbp1 and YFO is truly an orthologue, we expect all of the protein's respective domains to have the RBM property as well. But let's not simply assume what we can easily test. We'll define the sequence of the APSES domain in MBP1_SACCE and YFO and see how these definitions reflect in a BLAST search. |
| + | {{Vspace}} | ||
| − | + | ====Defining the range of the APSES domain annotation==== | |
| − | ====Defining the domain | ||
{{#lst:Reference annotation yeast Mbp1|CDD_APSES}} | {{#lst:Reference annotation yeast Mbp1|CDD_APSES}} | ||
| − | + | {{Vspace}} | |
====Executing the forward search==== | ====Executing the forward search==== | ||
| + | {{Vspace}} | ||
{{task|1= | {{task|1= | ||
# Access [http://www.ncbi.nlm.nih.gov/blast '''BLAST'''] and follow the link to the '''protein blast''' program. | # Access [http://www.ncbi.nlm.nih.gov/blast '''BLAST'''] and follow the link to the '''protein blast''' program. | ||
# '''Forward search:''' | # '''Forward search:''' | ||
| − | ## | + | ## Paste only the APSES domain sequence for <code>MBP1_SACCE</code> in the '''Query sequence''' field (copy the sequence from above). |
| − | ## Select <code>refseq_protein</code> as the '''database''' to search in, and enter the correct taxonomy ID for YFO. | + | ## Select <code>refseq_protein</code> as the '''database''' to search in, and enter the correct taxonomy ID for YFO in the '''Organism''' field. |
## Run BLAST. Examine the results. | ## Run BLAST. Examine the results. | ||
| − | ## If | + | ## If the top hit is the same protein you have already seen, oK. If it's not '''add it to your protein database in RStudio'''. |
}} | }} | ||
| + | With this we have confirmed the sequence with the most highly conserved APSES domain in YFO. Can we take the sequence for the reverse search from the alignment that BLAST returns? Actually, that is not a good idea. The BLAST alignment is not guaranteed to be optimal. We should do an optimal sequnece alignment instead. That is: we use two different tools here for two different purposes: we use BLAST to identify one protein as the most similar among many alternatives and we use optimal sequence alignment to determine the best alignment between two sequences. That best alignment is what we will annotate as the YFO APSES domain. | ||
| − | + | {{Vspace}} | |
| − | ====Alignment to define the | + | ====Alignment to define the YFO APSES domain for the reverse search==== |
| − | {{ | + | {{Vspace}} |
| − | |||
| − | |||
| − | + | {{task|1 = | |
| − | |||
| − | |||
| − | |||
| − | + | * Return to your RStudio session. | |
| − | + | * Study and work through the code in the <code>APSES Domain annotation by alignment</code> section of the <code>BCH441_A04.R</code> script | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| − | + | {{Vspace}} | |
| − | |||
====Executing the reverse search==== | ====Executing the reverse search==== | ||
| + | {{Vspace}} | ||
{{task|1= | {{task|1= | ||
| − | # | + | #Paste the the APSES domain sequence for the YFO best-match and enter it into '''Query sequence''' field of the BLAST form. |
## Select <code>refseq_protein</code> as the '''database''' to search in, and enter <code>Saccharomyces cerevisiae (taxid:4932)</code> to restrict the '''organism''' for which hits are reported. | ## Select <code>refseq_protein</code> as the '''database''' to search in, and enter <code>Saccharomyces cerevisiae (taxid:4932)</code> to restrict the '''organism''' for which hits are reported. | ||
## Run BLAST. Examine the results. | ## Run BLAST. Examine the results. | ||
| − | If your top-hit is again <code>NP_010227</code>, you have confirmed the RBM between the APSES domain of <code>Mbp1_SACCE</code> and <code> | + | If your top-hit is again <code>NP_010227</code>, you have confirmed the RBM between the APSES domain of <code>Mbp1_SACCE</code> and <code>Mbp1_<YFO></code>. If it is not, let me know. There may be some organisms for which the full-length and APSES RBMs are different and I would like to discuss these cases. |
}} | }} | ||
| − | + | {{Vspace}} | |
| + | |||
| + | ==Heuristic profile-based alignment: PSI BLAST== | ||
| − | + | {{Vspace}} | |
<div class="colmask doublepage"> | <div class="colmask doublepage"> | ||
| Line 826: | Line 400: | ||
| + | {{Vspace}} | ||
| − | + | ===Selecting species for a PSI-BLAST search=== | |
| − | + | {{Vspace}} | |
| Line 837: | Line 412: | ||
<!-- Column 1 start --> | <!-- Column 1 start --> | ||
| − | To | + | To select species, we will use an approach that is conceptually simple: select a set of species according to their shared taxonomic rank in the tree of life. {{WP|Biological classification|'''Biological classification'''}} provides a hierarchical system that describes evolutionary relatedness for all living entities. The levels of this hierarchy are so called {{WP|Taxonomic rank|'''taxonomic ranks'''}}. These ranks are defined in ''Codes of Nomenclature'' that are curated by the self-governed international associations of scientists working in the field. The number of ranks is not specified: there is a general consensus on seven principal ranks (see below, in bold) but many subcategories exist and may be newly introduced. It is desired–but not mandated–that ranks represent ''clades'' (a group of related species, or a "branch" of a phylogeny), and it is desired–but not madated–that the rank is sharply defined. The system is based on subjective dissimilarity. Needless to say that it is in flux. |
If we follow a link to an entry in the NCBI's Taxonomy database, eg. [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=559292 ''Saccharomyces cerevisiae S228c''], the strain from which the original "yeast genome" was sequenced in the late 1990s, we see the following specification of its taxonomic lineage: | If we follow a link to an entry in the NCBI's Taxonomy database, eg. [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=559292 ''Saccharomyces cerevisiae S228c''], the strain from which the original "yeast genome" was sequenced in the late 1990s, we see the following specification of its taxonomic lineage: | ||
| Line 970: | Line 545: | ||
subclass-, order-, or family rank (hover over the names to see their taxonomic rank.) | subclass-, order-, or family rank (hover over the names to see their taxonomic rank.) | ||
| − | + | I have chosen the 10 species below to define a well-distributed search-space for PSI-BLAST. Of course '''you must also include YFO in the selection''' (<small>if YFO is not in this list already</small>). | |
| − | To enter these | + | To enter these 10 species as an Entrez restriction, they need to be formatted as below. (<small>One could also enter species one by one, by pressing the '''(+)''' button after the organism list</small>) |
<source lang="text"> | <source lang="text"> | ||
| − | + | ||
| − | + | "Wallemia mellicola"[organism] OR | |
| − | + | "Puccinia Graminis"[organism] OR | |
| − | + | "Ustilago maydis"[organism] OR | |
| − | + | "Cryptococcus neoformans"[organism] OR | |
| − | + | "Coprinopsis cinerea"[organism] OR | |
| − | + | "Schizosaccharomyces pombe"[organism] OR | |
| − | OR | + | "Aspergillus nidulans"[organism] OR |
| − | + | "Neurospora crassa"[organism] OR | |
| − | OR | + | "Bipolaris oryzae"[organism] OR |
| − | + | "Saccharomyces cerevisiae"[organism] | |
| − | OR | + | |
| − | |||
| − | |||
| − | |||
| − | |||
</source> | </source> | ||
| Line 1,000: | Line 571: | ||
| − | + | {{Vspace}} | |
===Executing the PSI-BLAST search=== | ===Executing the PSI-BLAST search=== | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
We have a list of species. Good. Next up: how do we '''use''' it. | We have a list of species. Good. Next up: how do we '''use''' it. | ||
| Line 1,011: | Line 585: | ||
# Navigate to the [http://www.ncbi.nlm/nih.gov/blast/ BLAST homepage]. | # Navigate to the [http://www.ncbi.nlm/nih.gov/blast/ BLAST homepage]. | ||
# Select '''protein BLAST'''. | # Select '''protein BLAST'''. | ||
| − | # Paste the APSES domain sequence into the search field. | + | # Paste the MBP1_SACCE APSES domain sequence into the search field: |
| + | >APSES_MBP1 Residues 4-102 of S. cerevisiae Mbp1 | ||
| + | QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI | ||
| + | LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF | ||
# Select '''refseq''' as the database. | # Select '''refseq''' as the database. | ||
| − | # Copy the Entrez restrictions from above '''and | + | # Copy the Entrez restrictions from above '''and add the correct name for YFO''' to the list if it is not there already. (Obviously, you can't find sequences in YFO if YFO is not included among the genomes you are searching in.) Paste the list into the '''Entrez Query''' field. |
# In the '''Algorithm''' section, select PSI-BLAST. | # In the '''Algorithm''' section, select PSI-BLAST. | ||
#Click on '''BLAST'''. | #Click on '''BLAST'''. | ||
| Line 1,019: | Line 596: | ||
| − | Evaluate the results carefully. Since we did not change the algorithm parameters, the threshold for inclusion was set at an '''E-value''' of 0.005 by default, and that may be a bit too lenient, i.e. include sequences that are not homologous. If you look at the table of your hits– in the '''Sequences producing significant alignments...''' section– there may also be a few sequences that have a low query coverage of less than 80%. Let's exclude these from the profile initially: not to worry, if they are true positives, the will come back with improved E-values and greater coverage in subsequent iterations. But if they were false positives, their E-values will rise and they | + | Evaluate the results carefully. Since we did not change the algorithm parameters, the threshold for inclusion was set at an '''E-value''' of 0.005 by default, and that may be a bit too lenient, i.e. it might include sequences that are not homologous. If you look at the table of your hits– in the '''Sequences producing significant alignments...''' section– there may also be a few sequences that have a low query coverage of less than 80%. Let's exclude these from the profile initially: not to worry, if they are true positives, the will come back with improved E-values and greater coverage in subsequent iterations. But if they were false positives, their E-values will rise and they will drop out of the profile and not contaminate it. |
| Line 1,039: | Line 616: | ||
#Again, study the table of hits. Sequences highlighted in yellow have met the search criteria in the second iteration and are proposed for inclusion in the next iteration. Note that the coverage of (some) of the previously excluded sequences is now above 80%. These are the ones you need to check carefully: do you agree that they should be included? If there is any doubt, perhaps because of a really marginal E-value, poor coverage or a function annotation that is not compatible with your query, it is safer to exclude a sequence than to risk profile corruption. If the sequence is a true positive, it will return to the list in later iterations, usually with a better E-value as the profile improves. It's a good idea to note such sequences in your journal so you can keep track of how their E-values change. | #Again, study the table of hits. Sequences highlighted in yellow have met the search criteria in the second iteration and are proposed for inclusion in the next iteration. Note that the coverage of (some) of the previously excluded sequences is now above 80%. These are the ones you need to check carefully: do you agree that they should be included? If there is any doubt, perhaps because of a really marginal E-value, poor coverage or a function annotation that is not compatible with your query, it is safer to exclude a sequence than to risk profile corruption. If the sequence is a true positive, it will return to the list in later iterations, usually with a better E-value as the profile improves. It's a good idea to note such sequences in your journal so you can keep track of how their E-values change. | ||
# Let's exclude partial matches one more time. Again, deselect all sequences with less than 80% coverage. Then run the third iteration. | # Let's exclude partial matches one more time. Again, deselect all sequences with less than 80% coverage. Then run the third iteration. | ||
| − | # Iterate the search in this way until no more "New" sequences are added to the profile. The search has converged. | + | # Iterate the search in this way, successively relaxing the coverage threshold, until no more "New" sequences are added to the profile. The search has converged. Obviously the result depends on your data, but it would be unusual if the search had not converged after 6 iterations or so, and there is probably a mistake if there are more than 70 hits or so. |
# Now look at the list of excluded hits (if any), the hits that are reasonable but didn't quite make the cut. Are there any from YFO that seem like they should actually be included? Perhaps their E-value is only marginally above the threshold? If that's the case, try returning the E-value threshold to the default 0.005 and see what happens... | # Now look at the list of excluded hits (if any), the hits that are reasonable but didn't quite make the cut. Are there any from YFO that seem like they should actually be included? Perhaps their E-value is only marginally above the threshold? If that's the case, try returning the E-value threshold to the default 0.005 and see what happens... | ||
}} | }} | ||
| Line 1,048: | Line 625: | ||
{{task|1= | {{task|1= | ||
| − | # | + | # In the header section of the BLAST report, click on '''Taxonomy reports''' and find YFO in the '''Organism Report''' section. These are '''your APSES domain homologs'''. All of them. There is a link to the alignment, the BLAST score, the E-value, and a link to the entry in RefSeq. |
# From the report copy the sequence identifiers from YFO, with E-values above your defined threshold to your notebook. | # From the report copy the sequence identifiers from YFO, with E-values above your defined threshold to your notebook. | ||
}} | }} | ||
| − | For example, the list of ''Saccharomyces'' genes | + | For example, the list of ''Saccharomyces'' genes contains the following information: |
<code> | <code> | ||
| − | + | [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=559292 Saccharomyces cerevisiae S288c]</b> [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=4890 [ascomycetes]] taxid 559292<br \> | |
| − | [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6320147&dopt=GenPept ref|NP_010227.1|] Mbp1p [Saccharomyces cerevisiae S288c] | + | 4e-37 [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6320147&dopt=GenPept ref|NP_010227.1|] Mbp1p [Saccharomyces cerevisiae S288c]<br \> |
| − | [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6320957&dopt=GenPept ref|NP_011036.1|] Swi4p [Saccharomyces cerevisiae S288c] | + | 2e-30 [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6320957&dopt=GenPept ref|NP_011036.1|] Swi4p [Saccharomyces cerevisiae S288c]<br \> |
| − | [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6322808&dopt=GenPept ref|NP_012881.1|] Phd1p [Saccharomyces cerevisiae S288c] | + | 4e-27 [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6322808&dopt=GenPept ref|NP_012881.1|] Phd1p [Saccharomyces cerevisiae S288c]<br \> |
| − | [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6323658&dopt=GenPept ref|NP_013729.1|] Sok2p [Saccharomyces cerevisiae S288c] | + | 4e-27 [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6323658&dopt=GenPept ref|NP_013729.1|] Sok2p [Saccharomyces cerevisiae S288c]<br \> |
| − | [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6322090&dopt=GenPept ref|NP_012165.1|] Xbp1p [Saccharomyces cerevisiae S288c] | + | 7e-06 [http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=6322090&dopt=GenPept ref|NP_012165.1|] Xbp1p [Saccharomyces cerevisiae S288c]<br \> |
</code> | </code> | ||
| − | [[Saccharomyces cerevisiae Xbp1|Xbp1]] is a special case. It has only very low coverage, but that is because it has a long domain insertion and the N-terminal match often is not recognized by alignment because the gap scores for long indels are unrealistically large. For now, I keep that sequence with the others. | + | <small>[[Saccharomyces cerevisiae Xbp1|Xbp1]] is a special case. It has only very low coverage, but that is because it has a long domain insertion and the N-terminal match often is not recognized by alignment because the gap scores for long indels are unrealistically large. For now, I keep that sequence with the others.</small> |
{{task|1= | {{task|1= | ||
| − | # To add the sequences to your database, open each of the links for | + | # To add the sequences to your database, open each of the links to the RefSeq record for YFO organism into a separate tab. |
| − | # | + | # Find the UniProt IDs |
| − | + | # Go through the (short) section <code>add PSI BLAST results</code> in the Assignment 04 R-script. | |
| + | |||
| + | }} | ||
| + | |||
| + | <!-- | ||
| + | |||
| + | Add to this assignment: | ||
| + | - study the BLAST output format, links, tools, scores ... | ||
| + | - compare the improvement in E-values to standard BLAST | ||
| + | - examine this in terms of sensitivity and specificity | ||
| + | --> | ||
| − | + | So much for using PSI-BLAST. The last step seems a bit tedious, adding all this information by hand. There's got to be a better way, right? | |
| − | + | But for now, we'll have a look at what the sequences tell us. | |
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Model Based Alignments: PSSMs and HMMs== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <div class="colmask doublepage"> | |
| − | + | <div class="colleft"> | |
| − | + | <div class="col1"> | |
| − | + | <!-- Column 1 start --> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ;Position Specific Scoring Matrices (PSSMs) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | The sensitivity of PSI-BLAST is based on the alignment of profiles of related sequences. The profiles are represented as position specific scoring matrices compiled from the alignment of hits, first to the original sequence and then to the profile. Incidentally, this process can also be turned around, and a collection of pre-compiled PSSMs can be used to annotate protein sequence: this is the principle employed by RPS-BLAST, the tool that identifies conserved domains at the beginning of every BLAST search, and has been used to build the CDD database of conserved domains (for a very informative help-page on CDD [https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml '''see here''']. | |
| − | |||
| − | |||
| − | |||
| − | |||
<!-- | <!-- | ||
| + | === CDD domain annotation === | ||
| + | |||
| + | In the last assignment, you followed a link to '''CDD Search Results''' from the [http://www.ncbi.nlm.nih.gov/protein/NP_010227 RefSeq record for yeast Mbp1] and briefly looked at the information offered by the NCBI's Conserved Domain Database, a database of ''Position Specific Scoring Matrices'' that embody domain definitions. Rather than access precomputed results, you can also search CDD with sequences: assuming you have saved the YFO Mbp1 sequence in FASTA format, this is straightforward. If you did not save this sequence, return to [[BIO_Assignment_Week_3|Assignment 3]] and retrieve it again. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | {{task|1= | ||
| + | # Access the [http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml '''CDD database'''] at http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml | ||
| + | # Read the information. CDD is a superset of various other database domain annotations as well as NCBI-curated domain definitions. | ||
| + | # Copy the YFO Mbp1 FASTA sequence, paste it into the search form and click '''Submit'''. | ||
| + | ## On the result page, clik on '''View full result''' | ||
| + | ## Note that there are a number of partially overlapping ankyrin domain modules. We will study ankyrin domains in a later assignment. | ||
| + | ## Also note that there may be blocks of sequence colored cyan in the sequence bar. Hover your mouse over the blocks to see what these blocks signify. | ||
| + | ## Open the link to '''Search for similar domain architecture''' in a separate window and study it. This is the '''CDART''' database. Think about what these results may be useful for. | ||
| + | ## Click on one of the ANK superfamily graphics and see what the associated information looks like: there is a summary of structure and function, links to specific literature and a tree of the relationship of related sequences. | ||
| + | }} | ||
--> | --> | ||
| − | |||
| − | + | <!-- Column 1 end --> | |
| + | </div> | ||
| + | <div class="col2"> | ||
| + | <!-- Column 2 start --> | ||
| + | |||
| + | ; Hidden Markov Models (HMMs) | ||
| + | |||
| + | An approach to represent such profile information that is more general than PSSMs is a {{WP|Hidden Markov model|'''Hidden Markov model (HMM)'''}} and the standard tool to use HMMs in Bioinformatics is [http://hmmer.org/ '''HMMER'''], written by Sean Eddy. HMMER has allowed to represent the entirety of protein sequences as a collection of profiles, stored in databases such as [http://pfam.xfam.org/ '''Pfam'''], [https://www.ebi.ac.uk/interpro/ '''Interpro'''], and [http://smart.embl-heidelberg.de/ '''SMART''']. While the details are slightly different, all of these services allow to scan sequences for the presence of domains. Importantly thus, the alignment results are not collections of full-length protein families, but annotate to domain families, i.e. full length proteins are decomposed into their homologous domains. This is a very powerful approach towards the functional annotation of unknown sequences. | ||
| + | |||
| + | In this section, we will annotate the YFO sequence with the domains it contains, using the database of domain HMMs curated by SMART in Heidelberg and Pfam at the EMBL. We will then compare these annotations with those determined for the orthologues in the reference species. In this way we can enhance the information about one protein by determining how its features are conserved. | ||
| + | |||
| + | <!-- Column 2 end --> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | == SMART domain annotation == | ||
| + | |||
| + | |||
| + | The [http://smart.embl-heidelberg.de/ SMART database] at the EMBL in Heidelberg integrates a number of feature detection tools including Pfam domain annotation and its own, HMM based SMART domain database. You can search by sequence, or by accession number and retrieve domain annotations and more. | ||
| + | |||
| + | |||
| + | ===SMART search=== | ||
| + | |||
| + | {{task|1= | ||
| + | # Access the [http://smart.embl-heidelberg.de/ '''SMART database'''] at http://smart.embl-heidelberg.de/ | ||
| + | # Click the lick to access SMART in the '''normal''' mode. | ||
| + | # Paste the YFO Mbp1 UniProtKB Accession number into the '''Sequence ID or ACC''' field. If you were not able to find a UniProt ID, paste the sequence instead. | ||
| + | # Check all the boxes for: | ||
| + | ## '''outlier homologues''' (also including homologues in the PDB structure database) | ||
| + | ## '''PFAM domains''' (domains defined by sequence similarity in the PFAM database) | ||
| + | ## '''signal peptides''' (using the Gunnar von Heijne's SignalP 4.0 server at the Technical University in Lyngby, Denmark) | ||
| + | ## '''internal repeats''' (using the programs ''ariadne'' and ''prospero'' at the Wellcome Trust Centre for Human Genetics at Oxford University, England) | ||
| + | # Click on '''Sequence SMART''' to run the search and annotation. <small>(In case you get an error like: "Sorry, your entry seems to have no SMART domain ...", try again with the actual sequence instead of the accession number.)</small> | ||
| + | |||
| + | Study the results. | ||
| + | |||
| + | # Note down the following information so you can enter the annotation in the protein database for YFO: | ||
| + | ## From the section on "Confidently predicted domains ..." | ||
| + | ### The start and end coordinates of the '''KilA-N''' domain <small>(...according to SMART, not Pfam, in case the two differ)</small>. | ||
| + | ### All start and end coordinates of '''low complexity segments''' | ||
| + | ### All start and end coordinates of '''ANK''' (Ankyrin) domains | ||
| + | ### Start and end coordinates of '''coiled coil''' domain(s) <small>I expect only one.</small> | ||
| + | ### Start and end coordinates of '''AT hook''' domain(s) <small>I expect at most one - not all Mbp1 orthologues have one.</small> | ||
| + | ## From the section on "Features NOT shown ..." | ||
| + | ### All start and end coordinates of '''low complexity segments''' for which the ''Reason'' is "overlap". | ||
| + | ### Any start and end coordinates of overlapping '''coiled coil''' segments. | ||
| + | ### <small>I expect all other annotations - besides the overlapping KilA-N domain defined by Pfam - to arise from the succession of ankyrin domains that the proteins have, both '''Pfam_ANK..''' domains, as well as internal repeats. However, if there are other features I have not mentioned here, feel encouraged to let me know.</small> | ||
| + | ## From the section on "Outlier homologues ..." | ||
| + | ### Start and end coordinates of a '''PDB:1SW6{{!}}B''' annotation (if you have one): this is a region of sequence similarity to a protein for which the 3D structural coordinate are known. | ||
| + | ### <small>Of course there should also be annotations to the structure of 1BM8 / 1MB1 and/or 1L3G - all of which are structures of the Mbp1 APSES domain that we have already annotated as an"APSES fold" feature previously. And there will be BLAST annotations to Ankyrin domains. We will not annotate these separately either.</small> | ||
| + | # Follow the links to the database entries for the information so you know what these domains and features are. | ||
| + | }} | ||
| − | + | Next we'll enter the features into our database, so we can compare them with the annotations that I have prepared from SMART annotations of Mbp1 orthologues from the ten reference fungi. | |
| + | {{Vspace}} | ||
| − | + | === Visual comparison of domain annotations in '''R''' === | |
| − | + | The versatile plotting functions of '''R''' allow us to compare domain annotations. The distribution of segments that are annotated as "low-complexity, presumably disordered, is particularly interesting: these are functional features that are often not associated with sequence similarity but may have arisen from convergent evolution. Those would not be detectable through sequence alignment - which is after all based on amino acid pair scores and therefore context independent. | |
| − | In | + | In the following code tutorial, we create a plot similar to the CDD and SMART displays. It is based on the SMART domain annotations of the six-fungal reference species for the course. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{task|1 = | |
| − | + | * Return to your RStudio session. | |
| + | * Make sure you have saved <code>myDB</code> as instructed previously. Then quit the program, restart, and re-open the project via the '''File''' → '''Recent projects ...''' menu. This is to clear out-of-date assignments and functions from the workspace. | ||
| + | * Do not type <code>init()</code> yet, but '''pull''' the most recent version of files from github. Then type <code>init()</code>. | ||
| + | * Study and work through the code in the <code>SMART domain annotations</code> section of the <code>BCH441_A04.R</code> script. This includes entering your domain and other feature annotations into the database. | ||
| + | * At the end of the script, print out your plot of the domain annotations for MB1_YFO and the reference proteins. Bring this plot with you for the next quiz. | ||
| + | * Can this plot be improved? What would you do differently to maximize its utility from an information-design point of view? | ||
| + | }} | ||
| − | + | When you execute the code, your plot should look similar to this one: | |
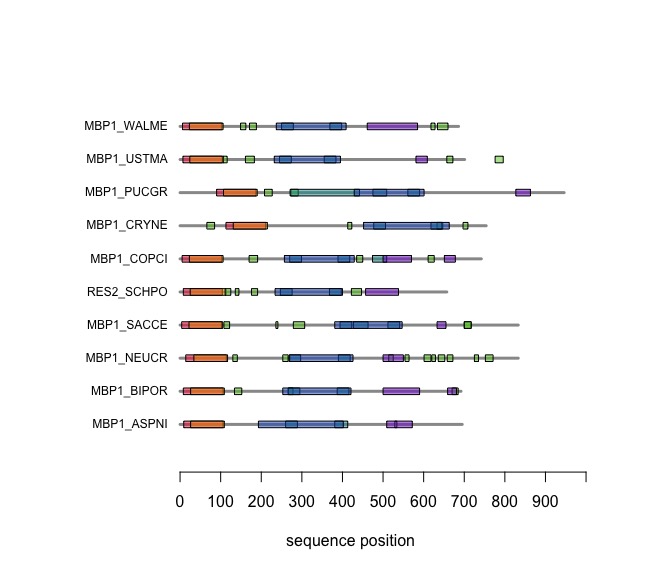
| − | + | [[Image:DomainAnnotations.jpg|frame|none|SMART domain annotations for Mbp1 proteins for the ten reference fungi. | |
| + | ]] | ||
| − | + | A note on the '''R''' code up to this point: You will find that we have been writing a lot of nested expressions for selections that join data from multiple tables of our data model. When I teach '''R''' workshops for graduate students, postdocs and research fellows, I find that the single greatest barrier in their actual research work is the preparation of data for analysis: filtering, selecting, cross-referencing, and integrating data from different sources. By now, I hope you will have acquired a somewhat robust sense for achieving this. You can imagine that there are ways to simplify those tasks with functions you write, or special resources from a variety of different packages you cab install. But the "pedestrian" approach we have been taking in our scripts has the advantage of working from a very small number of principles, with very few syntactic elements. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <!-- | |
| + | {{task|1= | ||
| − | + | ; Optional - care to share? | |
| − | |||
| − | |||
| − | + | # Copy one of the list definitions for Mbp1 domains and edit it with the appropriate values for your own annotations. | |
| − | + | # Test that you can add the YFO annotation to the plot. | |
| − | + | # Submit your validated code block to the [http://biochemistry.utoronto.ca/steipe/abc/students/index.php/BCH441_2014_Assignment_4_domain_annotations '''Student Wiki here''']. The goal is to compile an overview of all species we are studying in class. | |
| − | + | # If your working annotation block is in the Wiki before noontime on Wednesday, you will be awarded a 10% bonus on the quiz. | |
| − | + | }} | |
| − | + | --> | |
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | + | ==Multiple Sequence Alignment== | |
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | + | In order to perform a multiple sequence alignment, we obviously need a set of homologous sequences. This is not trivial. All interpretation of MSA results depends '''absolutely''' on how the input sequences were chosen. Should we include only orthologues, or paralogues as well? Should we include only species with fully sequenced genomes, or can we tolerate that some orthologous genes are possibly missing for a species? Should we include all sequences we can lay our hands on, or should we restrict the selection to a manageable number of ''representative'' sequences? All of these choices influence our interpretation: | |
| + | *orthologues are expected to be functionally and structurally conserved; | ||
| + | *paralogues may have divergent function but have similar structure; | ||
| + | *missing genes may make paralogs look like orthologs; and | ||
| + | *selection bias may weight our results toward sequences that are over-represented and do not provide a fair representation of evolutionary divergence. | ||
| − | + | <br> | |
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | + | ===Computing an MSA in R=== | |
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Let's use the Bioconductor msa package to align the sequences we have. Study and run the following code | |
| − | |||
| − | |||
| − | + | {{task|1 = | |
| − | + | * Return to your RStudio session. | |
| − | + | * Make sure you have saved <code>myDB</code> as instructed previously. | |
| − | + | * Bring code and data resources up to date: | |
| − | + | ** '''pull''' the most recent version of the project from GitHub | |
| − | + | ** type <code>init()</code> to lod the most recent files and functions | |
| − | + | ** re-merge your current <code>myDB</code> | |
| − | + | * Study and work through the code in the <code>Multiple sequence alignments</code> section of the <code>BCH441_A04.R</code> script. | |
| − | + | * Note that the final task asks you to print out some results and bring them to class for the next quiz. | |
| + | }} | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ===Sequence alignment editors=== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | + | Really excellent software tools have been written that help you visualize and manually curate multiple sequence alignments. If anything, I think they tend to do too much. Past versions of the course have used Jalview, but I have heard good things of AliView <small>(and if you are on a Mac [https://github.com/4ment/seqotron seqotron] might interest you, but I only cover software that is free and runs on all three major platforms)</small>. | |
| − | |||
| − | + | Right now, I am just mentioning the two alignment editors. If you have experience with comparing them, let us know. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * [[http://www.jalview.org/ '''Jalview''']] an integrated MSA editor and sequence annotation workbench from the Barton lab in Dundee. Lots of functions. | |
| − | + | * [[http://www.ormbunkar.se/aliview/ '''AliView''']] from Uppsala: fast, lean, looks to be very practical. | |
| − | |||
| − | |||
| − | |||
| − | |||
| + | {{Vspace}} | ||
| − | < | + | <!-- |
| − | ===Jalview: | + | ====Jalview: alignment editor==== |
| Line 1,432: | Line 865: | ||
| + | <!-- | ||
We will quickly install Jalview and explore its features in other assignments. | We will quickly install Jalview and explore its features in other assignments. | ||
| Line 1,446: | Line 880: | ||
| | ||
| − | + | --> | |
;That is all. | ;That is all. | ||
| − | + | {{Vspace}} | |
== Links and resources == | == Links and resources == | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | *{{#pmid: 15286655}} | ||
*{{PDFlink|[https://www.bioconductor.org/packages/release/bioc/vignettes/Biostrings/inst/doc/BiostringsQuickOverview.pdf Biostrings Quick Overview]| summary of Biostrings functions (PDF)}} | *{{PDFlink|[https://www.bioconductor.org/packages/release/bioc/vignettes/Biostrings/inst/doc/BiostringsQuickOverview.pdf Biostrings Quick Overview]| summary of Biostrings functions (PDF)}} | ||
| + | |||
| + | *[[Reference_species_for_fungi|Our 10 "reference" fungi]] | ||
<!-- {{#pmid: 19957275}} --> | <!-- {{#pmid: 19957275}} --> | ||
Latest revision as of 16:46, 7 August 2017
Assignment for Week 4
Sequence alignment
| < Assignment 3 | Assignment 5 > |
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz.
Contents
- 1 Introduction
- 2 Pairwise Alignments: Optimal
- 3 Heuristic pairwise alignments: BLAST
- 4 Heuristic profile-based alignment: PSI BLAST
- 5 Model Based Alignments: PSSMs and HMMs
- 6 SMART domain annotation
- 7 Multiple Sequence Alignment
- 8 Links and resources
- 9 Footnotes and references
- 10 Ask, if things don't work for you!
- Take care of things, and they will take care of you.
- Shunryu Suzuki
Introduction
Sequence alignment is a very large, and important topic.
One of the foundations of bioinformatics is the empirical observation that related sequences conserve structure, and often function. Much of what we know about a protein's physiological function is based on the conservation of that function as the species evolves. Indeed, conservation is a defining aspect of what can rightly be said to be a protein's "function" in the first place. Conservation - or its opposite: variation - is a consequence of selection under constraints: protein sequences change as a consequence of DNA mutations, this changes the protein's structure, this in turn changes functions and that has multiple effects on a species' reproductive fitness. Detrimental variants may be removed. Variation that is tolerated is largely neutral and therefore found only in positions that are neither structurally nor functionally critical. Conservation patterns can thus provide evidence for many different questions: structural conservation among proteins with similar 3D-structures, functional conservation among homologues with comparable roles, or amino acid propensities as predictors for protein engineering and design tasks.
We assess conservation by comparing sequences between related proteins. This is the basis on which we can make inferences from well-studied model organisms for species that have not been studied as deeply. The foundation is to measure protein sequence similarity. If two sequences are much more similar than we could expect from chance, we hypothesize that their similarity comes from shared ancestry plus conservation. The measurement of sequence similarity however requires sequence alignment[1].
A carefully done sequence alignment is a cornerstone for the annotation of the essential properties a gene or protein. It can already tell us a lot about which proteins we expect to have similar functions in different species.
Multiple sequence alignments (MSAs) are further useful to resolve ambiguities in the precise placement of "indels"[2] and to ensure that columns in alignments actually contain amino acids that evolve in a similar context. MSAs serve as input for
- functional annotation;
- protein homology modelling;
- phylogenetic analyses, and
- sensitive homology searches in databases.
In this assignment we will explore the essentials of
- optimal global and local pairwise alignment;
- Fast BLAST searches to determine best matches in large databases, and reciprocal best matches;
- PSI BLAST searches for exhaustive matches;
- Domain annotation by sequence alignment to statistical models; and
- Multiple sequence alignments.
As usual, the focus will be on practical, hands on approaches.
This is the scenario: you have previously identified a best match for a Mbp1 relative in YFO. Is this the most closely related protein? Is its DNA binding domain conserved? How can we identify all related genes in YFO? And, what can we learn from that collection of sequences?
Preparation: Updated Database Functions
The database contents and tables will change over time in this course. This means we need a mechanism to update the database, without throwing away previous work.
Task:
- Open the BCH441 project scripts in RStudio by selecting File → Recent Projects → BCH441_216
- Load the newest versions of scripts and data by pulling from the master file on GitHub.
- Study the code in the
Database maintenancesection of theBCH441_A04.Rscript
DotPlots and the Mutation Data Matrix
Before we start calculating alignments, we should get a better sense of the underlying sequence similarity. A Dotplot is a perfect tool for that, because it displays alignment-free similarity information. Let's make a dotplot that uses the BLOSUM62 Mutation Data Matrix to measure pairwise amino acid similarity. The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the BLOSUM62 matrix[3].
The NCBI makes its alignment matrices available by ftp. They are located at ftp://ftp.ncbi.nih.gov/blast/matrices - for example here is a link to the BLOSUM62 matrix[4].
Scoring matrices are also available in the Bioconductor Biostrings package.
BLOSUM62
A R N D C Q E G H I L K M F P S T W Y V B J Z X *
A 4 -1 -2 -2 0 -1 -1 0 -2 -1 -1 -1 -1 -2 -1 1 0 -3 -2 0 -2 -1 -1 -1 -4
R -1 5 0 -2 -3 1 0 -2 0 -3 -2 2 -1 -3 -2 -1 -1 -3 -2 -3 -1 -2 0 -1 -4
N -2 0 6 1 -3 0 0 0 1 -3 -3 0 -2 -3 -2 1 0 -4 -2 -3 4 -3 0 -1 -4
D -2 -2 1 6 -3 0 2 -1 -1 -3 -4 -1 -3 -3 -1 0 -1 -4 -3 -3 4 -3 1 -1 -4
C 0 -3 -3 -3 9 -3 -4 -3 -3 -1 -1 -3 -1 -2 -3 -1 -1 -2 -2 -1 -3 -1 -3 -1 -4
Q -1 1 0 0 -3 5 2 -2 0 -3 -2 1 0 -3 -1 0 -1 -2 -1 -2 0 -2 4 -1 -4
E -1 0 0 2 -4 2 5 -2 0 -3 -3 1 -2 -3 -1 0 -1 -3 -2 -2 1 -3 4 -1 -4
G 0 -2 0 -1 -3 -2 -2 6 -2 -4 -4 -2 -3 -3 -2 0 -2 -2 -3 -3 -1 -4 -2 -1 -4
H -2 0 1 -1 -3 0 0 -2 8 -3 -3 -1 -2 -1 -2 -1 -2 -2 2 -3 0 -3 0 -1 -4
I -1 -3 -3 -3 -1 -3 -3 -4 -3 4 2 -3 1 0 -3 -2 -1 -3 -1 3 -3 3 -3 -1 -4
L -1 -2 -3 -4 -1 -2 -3 -4 -3 2 4 -2 2 0 -3 -2 -1 -2 -1 1 -4 3 -3 -1 -4
K -1 2 0 -1 -3 1 1 -2 -1 -3 -2 5 -1 -3 -1 0 -1 -3 -2 -2 0 -3 1 -1 -4
M -1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5 0 -2 -1 -1 -1 -1 1 -3 2 -1 -1 -4
F -2 -3 -3 -3 -2 -3 -3 -3 -1 0 0 -3 0 6 -4 -2 -2 1 3 -1 -3 0 -3 -1 -4
P -1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7 -1 -1 -4 -3 -2 -2 -3 -1 -1 -4
S 1 -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4 1 -3 -2 -2 0 -2 0 -1 -4
T 0 -1 0 -1 -1 -1 -1 -2 -2 -1 -1 -1 -1 -2 -1 1 5 -2 -2 0 -1 -1 -1 -1 -4
W -3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 -4 -3 -2 11 2 -3 -4 -2 -2 -1 -4
Y -2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7 -1 -3 -1 -2 -1 -4
V 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4 -3 2 -2 -1 -4
B -2 -1 4 4 -3 0 1 -1 0 -3 -4 0 -3 -3 -2 0 -1 -4 -3 -3 4 -3 0 -1 -4
J -1 -2 -3 -3 -1 -2 -3 -4 -3 3 3 -3 2 0 -3 -2 -1 -2 -1 2 -3 3 -3 -1 -4
Z -1 0 0 1 -3 4 4 -2 0 -3 -3 1 -1 -3 -1 0 -1 -2 -2 -2 0 -3 4 -1 -4
X -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -1 -4
* -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 -4 1
Task:
- Study this and make sure you understand what this table is, how it can be used, and what a reasonable range of values for identities and pairscores for non-identical, similar and dissimilar residues is. Ask on the mailing list in case you have questions. This piece of data is the foundation of any sequence alignment. without it, no sensible alignment could be produced!
- Figure out the following values:
- Compare an identical match of histidine with an identical match of serine. What does this mean?
- How similar are lysine and leucine, as compared to leucine and isoleucine? Is this what you expect?
- PAM matrices are sensitive to an interesting artefact. Since W and R can be interchanged with a single point mutation, the probability of observing W→R and R→W exchanges in closely related sequences is much higher than one would expect from the two amino acid's biophysical properties. (Why?) PAM matrices were compiled from hypothetical point exchanges and then extrapolated. Therefore these matrices assign a relatively high degree of similarity to (W, R), that is not warranted considering what actually happens in nature. Do you see this problem in the BLOSUM matrix? If BLOSUM does not have this issue, why not?
Next, let's apply the scoring matrix for actual comparison:
Task:
- Return to your RStudio session.
- If you've been away from it for a while, it's probably a good idea to update to the newest versions of scripts and data by pulling from the master file on GitHub.
- Study and work through the code in the
Dotplot and MDMsection of theBCH441_A04.Rscript
Pairwise Alignments: Optimal
Optimal pairwise sequence alignment is the mainstay of sequence comparison. To consider such alignments in practice, we'll align the same sequences that we have just mapped in the dotplot exercise: Mbp1 and its YFO relative. For simplicity, I will call the two proteins MBP1_SACCE and MBP1_YFO through the remainder of the assignment. Your dotplots should have shown you two regions of similarity: a highly similar region focussed somewhere around the N-terminal 100 amino acids, and a more extended, but somewhat less similar region in the middle of the sequences. You can think of the sequence alignment algorithm as building the similarity matrix, and then discovering the best path along high-scoring diagonals.
Optimal Sequence Alignment: EMBOSS online tools
Online programs for optimal sequence alignment are part of the EMBOSS tools. The programs take FASTA files or raw text files as input.
Local optimal sequence alignment using "water"
Task:
- Fetch the sequences for
MBP1_SACCEandMBP1_YFOfrom your database. You can simply select them by name (if you have given your sequence the suggested name when you eneterd it into your database): paste the following into the console:
- to print the
MBP1_SACCEprotein sequence to the console
myDB$protein$sequence[myDB$protein$name == "MBP1_SACCE"]- to print the
MBP1_YFOprotein sequence to the console:
YFOseq <- paste("MBP1_", biCode(YFO), sep="")
myDB$protein$sequence[myDB$protein$name == YFOseq](If this didn't work, fix it. Did you give your sequence the right name?)
- Access the EMBOSS Explorer site (if you haven't done so yet, you might want to bookmark it.)
- Look for ALIGNMENT LOCAL, click on water, paste your sequences and run the program with default parameters.
- Study the results. You will probably find that the alignment extends over most of the protein, but does not include the termini.
- Considering the sequence identity cutoff we discussed in class (25% over the length of a domain), do you believe that the N-terminal domains (the APSES domains) are homologous?
- Change the Gap opening and Gap extension parameters to high values (e.g. 30 and 5). Then run the alignment again.
- Note what is different.
Global optimal sequence alignment using "needle"
Task:
- Look for ALIGNMENT GLOBAL, click on needle, paste the
MBP1_SACCEandMBP1_YFOsequences again and run the program with default parameters. - Study the results. You will find that the alignment extends over the entire protein, likely with long indels at the termini.
Optimal Sequence Alignment with R: Biostrings
Biostrings has extensive functions for sequence alignments. They are generally well written and tightly integrated with the rest of Bioconductor's functions. There are a few quirks however: for example alignments won't work with lower-case sequences[5].
Task:
- Return to your RStudio session.
- Once again, if you've been away from it for a while, it's always a good idea to update to pull updtaes from the master file on GitHub.
- Study and work through the code in the
Biostrings Pairwise Alignmentsection of theBCH441_A04.Rscript
Heuristic pairwise alignments: BLAST
BLAST is by a margin the most important computational tool of molecular biology. It is so important, that we have already used BLAST in Assignment 3 even before properly introducing the algorithm and the principles, to find the most similar sequence to MBP1_SACCE in YFO.
In this part of the assignment we will use BLAST to perform Reciprocal Best Matches.
One of the important questions of model-organism based inference is: which genes perform the same function in two different organisms. In the absence of other information, our best guess is that these are the two genes that are mutually most similar. The keyword here is mutually. If MBP1_SACCE from S. cerevisiae is the best match to RES2_SCHPO in S. pombe, the two proteins are only mutually most similar if RES2_SCHPO is more similar to MBP1_SACCE than to any other S. cerevisiae protein. We call this a Reciprocal Best Match, or "RBM"[6].
The argument is summarized in the figure on the right: genes that evolve under continuos selective pressure on their function have relatively lower mutation rates and are thus more similar to each other, than genes that undergo neo- or sub-functionalization after duplication.
However, there is a catch: proteins are often composed of multiple domains that implement distinct roles of their function. Under the assumptions above we could hypothesize:
- a gene in YFO that has the "same" function as the Mbp1 cell-cycle checkpoint switch in yeast should be an RBM to Mbp1;
- a gene that binds to the same DNA sites as Mbp1 should have a DNA-binding domain that is an RBM to the DNA binding domain of Mbp1.
Thus we'll compare RBMs in YFO for full-length Mbp1_SACCE and its DNA-binding domain, and see if the results are the same.
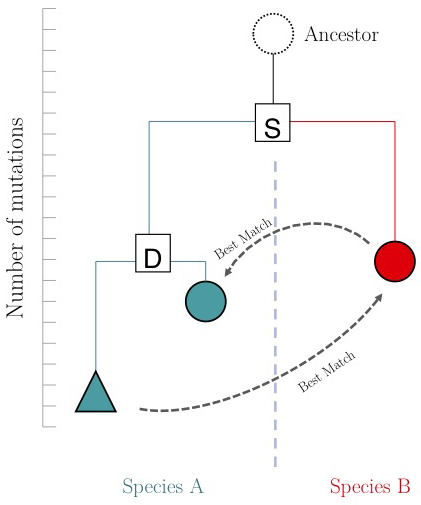
Full-length RBM
You have already performed the first half of the experiment: matching from S. cerevisiae to YFO. The backward match is simple.
Task:
- Access BLAST and follow the link to the protein blast program.
- Enter the RefSeq ID for
MBP1_YFOin the Query sequence field. - Select
refseq_proteinas the database to search in, and enterSaccharomyces cerevisiae (taxid:4932)to restrict the organism for which hits are reported. - Run BLAST. Examine the results.
If your top-hit is NP_010227, you have confirmed the RBM between Mbp1_SACCE and Mbp1_YFO. If it is not, let me know. I expect this to be the same and would like to verify your results if it is not[7].
RBM for the DNA binding domain
The DNA-binding domain of Mbp1_SACCE is called an APSES domain. If the RBM between Saccharomyces cerevisiae Mbp1 and YFO is truly an orthologue, we expect all of the protein's respective domains to have the RBM property as well. But let's not simply assume what we can easily test. We'll define the sequence of the APSES domain in MBP1_SACCE and YFO and see how these definitions reflect in a BLAST search.
Defining the range of the APSES domain annotation
The APSES domain is a well-defined type of DNA-binding domain that is ubiquitous in fungi and unique in that kingdom. Structurally it is a member of the Winged Helix-Turn-Helix family. Recently it was found that it is homologous to the somewhat shorter, prokaryotic KilA-N domain; thus the APSES domain was retired from pFam and instances were merged into the KilA-N family. However InterPro has a KilA-N entry but still recognizes the APSES domain.
KilA-N domain boundaries in Mbp1 can be derived from the results of a CDD search with the ID 1BM8_A (the Mbp1 DNA binding domain crystal structure). The KilA-N superfamily domain alignment is returned.
- (pfam 04383): KilA-N domain; The amino-terminal module of the D6R/N1R proteins defines a novel, conserved DNA-binding domain (the KilA-N domain) that is found in a wide range of proteins of large bacterial and eukaryotic DNA viruses. The KilA-N domain family also includes the previously defined APSES domain. The KilA-N and APSES domains may also share a common fold with the nucleic acid-binding modules of the LAGLIDADG nucleases and the amino-terminal domains of the tRNA endonuclease.
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
1BM8A 16 IHSTGSIMKRKKDDWVNATHILKAANFAKaKRTRILEKEVLKETHEKVQ---------------GGFGKYQGTWVPLNIA 80
Cdd:pfam04383 3 YNDFEIIIRRDKDGYINATKLCKAAGETK-RFRNWLRLESTKELIEELSeennvdkseiiigrkGKNGRLQGTYVHPDLA 81
90
....*....|....
1BM8A 81 KQLA----EKFSVY 90
Cdd:pfam04383 82 LAIAswisPEFALK 95
Note that CDD and SMART are not consistent in how they apply pFam 04383 to the Mbp1 sequence. See annotation below.
The CDD KilA-N domain definition begins at position 16 of the 1BM8 sequence. But virtually all fungal APSES domains have a longer, structurally defined, conserved N-terminus. Blindly applying the KilA-N domain definition to these proteins would lose important information. For most purposes we will prefer the sequence spanned by the 1BM8_A structure. The sequence is given below, the KilA-N domain is coloured dark green. By this definition the APSES domain is 99 amino acids long and comprises residues 4 to 102 of the NP_010227 sequence.
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
1BM8A 1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRILEKEVLKETHEKVQGGFGKYQGTWVPLNIA 80
90
....*....|....*....
1BM8A 81 KQLAEKFSVYDQLKPLFDF 99
- Yeast APSES domain sequence in FASTA format
>APSES_MBP1 Residues 4-102 of S. cerevisiae Mbp1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF
- Synopsis of ranges
| Domain | Link | Length | Boundary | Range (Mbp1) | Range (1BM8) |
| KilA-N: pfam04383 (CDD) | CDD alignment | 72 | STGSI ... KFSVY | 21 - 93 | 18 - 90 |
| KilA-N: pfam04383 (SMART) | Smart main page | 79 | IHSTG ... YDQLK | 19 - 97 | 16 - 94 |
| KilA-N: SM01252 (SMART) | Smart main page | 84 | TGSIM ... DFTQT | 22 - 105 | 19 - 99... |
| APSES: Interpro IPR003163 | (Interpro) | 130 | QIYSA ... IRSAS | 3 - 133 | 1 - 99... |
| APSES (1BM8) | – | 99 | QIYSA ... PLFDF | 4 - 102 | 1 - 99 |
Executing the forward search
Task:
- Access BLAST and follow the link to the protein blast program.
- Forward search:
- Paste only the APSES domain sequence for
MBP1_SACCEin the Query sequence field (copy the sequence from above). - Select
refseq_proteinas the database to search in, and enter the correct taxonomy ID for YFO in the Organism field. - Run BLAST. Examine the results.
- If the top hit is the same protein you have already seen, oK. If it's not add it to your protein database in RStudio.
- Paste only the APSES domain sequence for
With this we have confirmed the sequence with the most highly conserved APSES domain in YFO. Can we take the sequence for the reverse search from the alignment that BLAST returns? Actually, that is not a good idea. The BLAST alignment is not guaranteed to be optimal. We should do an optimal sequnece alignment instead. That is: we use two different tools here for two different purposes: we use BLAST to identify one protein as the most similar among many alternatives and we use optimal sequence alignment to determine the best alignment between two sequences. That best alignment is what we will annotate as the YFO APSES domain.
Alignment to define the YFO APSES domain for the reverse search
Task:
- Return to your RStudio session.
- Study and work through the code in the
APSES Domain annotation by alignmentsection of theBCH441_A04.Rscript
Executing the reverse search
Task:
- Paste the the APSES domain sequence for the YFO best-match and enter it into Query sequence field of the BLAST form.
- Select
refseq_proteinas the database to search in, and enterSaccharomyces cerevisiae (taxid:4932)to restrict the organism for which hits are reported. - Run BLAST. Examine the results.
- Select
If your top-hit is again NP_010227, you have confirmed the RBM between the APSES domain of Mbp1_SACCE and Mbp1_<YFO>. If it is not, let me know. There may be some organisms for which the full-length and APSES RBMs are different and I would like to discuss these cases.
Heuristic profile-based alignment: PSI BLAST
It is (deceptively) easy to perform BLAST searches via the Web interface, but to use such powerful computational tools to their greatest advantage takes a considerable amount of care, caution and consideration.
PSI-BLAST allows to perform very sensitive searches for homologues that have diverged so far that their pairwise sequence similarity has become insignificant. It achieves this by establishing a profile of sequences to align with the database, rather than searching with individual sequences. This deemphasizes parts of the sequence that are variable and inconsequential, and focusses on the parts of greater structural and functional importance. As a consequence, the signal to noise ratio is greatly enhanced.
In this part of the assignment, we will set ourselves the task to use PSI-BLAST and find all orthologs and paralogs of the APSES domain containing transcription factors in YFO. We will use these sequences for multiple alignments, calculation of conservation etc.
The first methodical problem we have to address is what sequence to search with. The full-length Mbp1 sequence from Saccharomyces cerevisiae or its RBM from YFO are not suitable: They contain multiple domains (in particular the ubiquitous Ankyrin domains) and would create broad, non-specific profiles. The APSES domain sequence by contrast is structurally well defined. The KilA-N domain, being shorter, is less likely to make a sensitive profile. Indeed one of the results of our analysis will be to find whether APSES domains in fungi all have the same length as the Mbp1 domain, or whether some are indeed much shorter, like the KILA-N domain, as suggested by the Pfam alignment.
The second methodical problem we must address is how to perform a sensitive PSI-BLAST search in one organism. We need to balance two conflicting objectives:
- If we restrict the PSI-BLAST search to YFO, PSI-BLAST has little chance of building a meaningful profile - the number of homologues that actually are in YFO is too small. Thus the search will not become very sensitive.
- If we don't restrict our search, but search in all species, the number of hits may become unwieldily large. It becomes increasingly difficult to closely check all hits as to whether they have good coverage. Also we need to evaluate the fringe cases of marginal E-value: should a new sequence be added to the profile, or should we hold off on it for one or two iterations, to see whether its E-value drops significantly. By all means, we need to avoid profile corruption.
Perhaps this is still be manageable when we are searching in fungi, but imagine you are working with a bacterial protein, or a protein that is conserved across the entire tree of life: your search may find tens of thousands of sequences. And by next year, thousands more will have been added.
Therefore we have to find a middle ground: add enough organisms (sequences) to compile a sensitive profile, but not so many that we can no longer individually assess the sequences that contribute to the profile. We need to define a broadly representative but manageable set of species - to exploit the transitivity of homology - even if we are interested only in matches in one species: YFO. Please reflect on this and make sure you understand why we include sequences in a PSI-BLAST search that we are not actually interested in.
We need a subset of species
- that represent as large a range as possible on the evolutionary tree;
- that are as well distributed as possible on the tree; and
- whose genomes are fully sequenced.
Selecting species for a PSI-BLAST search
To select species, we will use an approach that is conceptually simple: select a set of species according to their shared taxonomic rank in the tree of life. Biological classification provides a hierarchical system that describes evolutionary relatedness for all living entities. The levels of this hierarchy are so called taxonomic ranks. These ranks are defined in Codes of Nomenclature that are curated by the self-governed international associations of scientists working in the field. The number of ranks is not specified: there is a general consensus on seven principal ranks (see below, in bold) but many subcategories exist and may be newly introduced. It is desired–but not mandated–that ranks represent clades (a group of related species, or a "branch" of a phylogeny), and it is desired–but not madated–that the rank is sharply defined. The system is based on subjective dissimilarity. Needless to say that it is in flux.
If we follow a link to an entry in the NCBI's Taxonomy database, eg. Saccharomyces cerevisiae S228c, the strain from which the original "yeast genome" was sequenced in the late 1990s, we see the following specification of its taxonomic lineage:
cellular organisms; Eukaryota; Opisthokonta;
Fungi; Dikarya; Ascomycota; Saccharomyceta;
Saccharomycotina; Saccharomycetes;
Saccharomycetales; Saccharomycetaceae;
Saccharomyces; Saccharomyces cerevisiae
These names can be mapped into taxonomic ranks, since the suffixes of these names e.g. -mycotina, -mycetaceae are specific to defined ranks. (NCBI does not provide this mapping, but Wikipedia is helpful here.)
| Rank | Suffix | Example |
| Domain | Eukaryota (Eukarya) | |
| Subdomain | Opisthokonta | |
| Kingdom | Fungi | |
| Subkingdom | Dikarya | |
| Phylum | Ascomycota | |
| rankless taxon[8] | -myceta | Saccharomyceta |
| Subphylum | -mycotina | Saccharomycotina |
| Class | -mycetes | Saccharomycetes |
| Subclass | -mycetidae | |
| Order | -ales | Saccharomycetales |
| Family | -aceae | Saccharomycetaceae |
| Subfamily | -oideae | |
| Tribe | -eae | |
| Subtribe | -ineae | |
| Genus | Saccharomyces | |
| Species | Saccharomyces cerevisiae |
You can see that there is no common mapping between the yeast lineage listed at the NCBI and the commonly recognized categories - not all ranks are represented. Nor is this consistent across species in the taxonomic database: some have subfamily ranks and some don't. And the tree is in no way normalized - some of the ranks have thousands of members, and for some, only a single extant member may be known, or it may be a rank that only relates to the fossil record.
But the ranks do provide some guidance to evolutionary divergence. Say you want to choose four species across the tree of life for a study, you should choose one from each of the major domains of life: Eubacteria, Euryarchaeota, Crenarchaeota-Eocytes, and Eukaryotes. Or you want to study a gene that is specific to mammals. Then you could choose from the clades listed in the NCBI taxonomy database under Mammalia (a class rank, and depending how many species you would want to include, use the subclass-, order-, or family rank (hover over the names to see their taxonomic rank.)
I have chosen the 10 species below to define a well-distributed search-space for PSI-BLAST. Of course you must also include YFO in the selection (if YFO is not in this list already).
To enter these 10 species as an Entrez restriction, they need to be formatted as below. (One could also enter species one by one, by pressing the (+) button after the organism list)
"Wallemia mellicola"[organism] OR
"Puccinia Graminis"[organism] OR
"Ustilago maydis"[organism] OR
"Cryptococcus neoformans"[organism] OR
"Coprinopsis cinerea"[organism] OR
"Schizosaccharomyces pombe"[organism] OR
"Aspergillus nidulans"[organism] OR
"Neurospora crassa"[organism] OR
"Bipolaris oryzae"[organism] OR
"Saccharomyces cerevisiae"[organism]
Executing the PSI-BLAST search
We have a list of species. Good. Next up: how do we use it.
Task:
- Navigate to the BLAST homepage.
- Select protein BLAST.
- Paste the MBP1_SACCE APSES domain sequence into the search field:
>APSES_MBP1 Residues 4-102 of S. cerevisiae Mbp1 QIYSARYSGVDVYEFIHSTGSIMKRKKDDWVNATHILKAANFAKAKRTRI LEKEVLKETHEKVQGGFGKYQGTWVPLNIAKQLAEKFSVYDQLKPLFDF
- Select refseq as the database.
- Copy the Entrez restrictions from above and add the correct name for YFO to the list if it is not there already. (Obviously, you can't find sequences in YFO if YFO is not included among the genomes you are searching in.) Paste the list into the Entrez Query field.
- In the Algorithm section, select PSI-BLAST.
- Click on BLAST.
Evaluate the results carefully. Since we did not change the algorithm parameters, the threshold for inclusion was set at an E-value of 0.005 by default, and that may be a bit too lenient, i.e. it might include sequences that are not homologous. If you look at the table of your hits– in the Sequences producing significant alignments... section– there may also be a few sequences that have a low query coverage of less than 80%. Let's exclude these from the profile initially: not to worry, if they are true positives, the will come back with improved E-values and greater coverage in subsequent iterations. But if they were false positives, their E-values will rise and they will drop out of the profile and not contaminate it.
Task:
- In the header section, click on Formatting options and in the line "Format for..." set the with inclusion threshold to
0.001(This means E-values can't be above 10-03 for the sequence to be included.) - Click on the Reformat button (top right).
- In the table of sequence descriptions (not alignments!), click on Query cover to sort the table by coverage, not by score.
- Deselect the check mark next to these sequences in the second-to-rightmost column Select for PSI blast.
- Then scroll to Run PSI-BLAST iteration 2 ... and click on
Go.
This is now the "real" PSI-BLAST at work: it constructs a profile from all the full-length sequences and searches with the profile, not with any individual sequence. Note that we are controlling what goes into the profile in two ways:
- we are explicitly removing sequences with poor coverage; and
- we are requiring a more stringent minimum E-value for each sequence.
Task:
- Again, study the table of hits. Sequences highlighted in yellow have met the search criteria in the second iteration and are proposed for inclusion in the next iteration. Note that the coverage of (some) of the previously excluded sequences is now above 80%. These are the ones you need to check carefully: do you agree that they should be included? If there is any doubt, perhaps because of a really marginal E-value, poor coverage or a function annotation that is not compatible with your query, it is safer to exclude a sequence than to risk profile corruption. If the sequence is a true positive, it will return to the list in later iterations, usually with a better E-value as the profile improves. It's a good idea to note such sequences in your journal so you can keep track of how their E-values change.
- Let's exclude partial matches one more time. Again, deselect all sequences with less than 80% coverage. Then run the third iteration.
- Iterate the search in this way, successively relaxing the coverage threshold, until no more "New" sequences are added to the profile. The search has converged. Obviously the result depends on your data, but it would be unusual if the search had not converged after 6 iterations or so, and there is probably a mistake if there are more than 70 hits or so.
- Now look at the list of excluded hits (if any), the hits that are reasonable but didn't quite make the cut. Are there any from YFO that seem like they should actually be included? Perhaps their E-value is only marginally above the threshold? If that's the case, try returning the E-value threshold to the default 0.005 and see what happens...
Once no "new" sequences have been added, we would always get the same result on additional iterations because there are no more changes to the profile. We say that the search has converged. Time to harvest.
Task:
- In the header section of the BLAST report, click on Taxonomy reports and find YFO in the Organism Report section. These are your APSES domain homologs. All of them. There is a link to the alignment, the BLAST score, the E-value, and a link to the entry in RefSeq.
- From the report copy the sequence identifiers from YFO, with E-values above your defined threshold to your notebook.
For example, the list of Saccharomyces genes contains the following information:
Saccharomyces cerevisiae S288c [ascomycetes] taxid 559292
4e-37 ref|NP_010227.1| Mbp1p [Saccharomyces cerevisiae S288c]
2e-30 ref|NP_011036.1| Swi4p [Saccharomyces cerevisiae S288c]
4e-27 ref|NP_012881.1| Phd1p [Saccharomyces cerevisiae S288c]
4e-27 ref|NP_013729.1| Sok2p [Saccharomyces cerevisiae S288c]
7e-06 ref|NP_012165.1| Xbp1p [Saccharomyces cerevisiae S288c]
Xbp1 is a special case. It has only very low coverage, but that is because it has a long domain insertion and the N-terminal match often is not recognized by alignment because the gap scores for long indels are unrealistically large. For now, I keep that sequence with the others.
Task:
- To add the sequences to your database, open each of the links to the RefSeq record for YFO organism into a separate tab.
- Find the UniProt IDs
- Go through the (short) section
add PSI BLAST resultsin the Assignment 04 R-script.
So much for using PSI-BLAST. The last step seems a bit tedious, adding all this information by hand. There's got to be a better way, right?
But for now, we'll have a look at what the sequences tell us.
Model Based Alignments: PSSMs and HMMs
- Position Specific Scoring Matrices (PSSMs)
The sensitivity of PSI-BLAST is based on the alignment of profiles of related sequences. The profiles are represented as position specific scoring matrices compiled from the alignment of hits, first to the original sequence and then to the profile. Incidentally, this process can also be turned around, and a collection of pre-compiled PSSMs can be used to annotate protein sequence: this is the principle employed by RPS-BLAST, the tool that identifies conserved domains at the beginning of every BLAST search, and has been used to build the CDD database of conserved domains (for a very informative help-page on CDD see here.
- Hidden Markov Models (HMMs)
An approach to represent such profile information that is more general than PSSMs is a Hidden Markov model (HMM) and the standard tool to use HMMs in Bioinformatics is HMMER, written by Sean Eddy. HMMER has allowed to represent the entirety of protein sequences as a collection of profiles, stored in databases such as Pfam, Interpro, and SMART. While the details are slightly different, all of these services allow to scan sequences for the presence of domains. Importantly thus, the alignment results are not collections of full-length protein families, but annotate to domain families, i.e. full length proteins are decomposed into their homologous domains. This is a very powerful approach towards the functional annotation of unknown sequences.
In this section, we will annotate the YFO sequence with the domains it contains, using the database of domain HMMs curated by SMART in Heidelberg and Pfam at the EMBL. We will then compare these annotations with those determined for the orthologues in the reference species. In this way we can enhance the information about one protein by determining how its features are conserved.
SMART domain annotation
The SMART database at the EMBL in Heidelberg integrates a number of feature detection tools including Pfam domain annotation and its own, HMM based SMART domain database. You can search by sequence, or by accession number and retrieve domain annotations and more.
SMART search
Task:
- Access the SMART database at http://smart.embl-heidelberg.de/
- Click the lick to access SMART in the normal mode.
- Paste the YFO Mbp1 UniProtKB Accession number into the Sequence ID or ACC field. If you were not able to find a UniProt ID, paste the sequence instead.
- Check all the boxes for:
- outlier homologues (also including homologues in the PDB structure database)
- PFAM domains (domains defined by sequence similarity in the PFAM database)
- signal peptides (using the Gunnar von Heijne's SignalP 4.0 server at the Technical University in Lyngby, Denmark)
- internal repeats (using the programs ariadne and prospero at the Wellcome Trust Centre for Human Genetics at Oxford University, England)
- Click on Sequence SMART to run the search and annotation. (In case you get an error like: "Sorry, your entry seems to have no SMART domain ...", try again with the actual sequence instead of the accession number.)
Study the results.
- Note down the following information so you can enter the annotation in the protein database for YFO:
- From the section on "Confidently predicted domains ..."
- The start and end coordinates of the KilA-N domain (...according to SMART, not Pfam, in case the two differ).
- All start and end coordinates of low complexity segments
- All start and end coordinates of ANK (Ankyrin) domains
- Start and end coordinates of coiled coil domain(s) I expect only one.
- Start and end coordinates of AT hook domain(s) I expect at most one - not all Mbp1 orthologues have one.
- From the section on "Features NOT shown ..."
- All start and end coordinates of low complexity segments for which the Reason is "overlap".
- Any start and end coordinates of overlapping coiled coil segments.
- I expect all other annotations - besides the overlapping KilA-N domain defined by Pfam - to arise from the succession of ankyrin domains that the proteins have, both Pfam_ANK.. domains, as well as internal repeats. However, if there are other features I have not mentioned here, feel encouraged to let me know.
- From the section on "Outlier homologues ..."
- Start and end coordinates of a PDB:1SW6|B annotation (if you have one): this is a region of sequence similarity to a protein for which the 3D structural coordinate are known.
- Of course there should also be annotations to the structure of 1BM8 / 1MB1 and/or 1L3G - all of which are structures of the Mbp1 APSES domain that we have already annotated as an"APSES fold" feature previously. And there will be BLAST annotations to Ankyrin domains. We will not annotate these separately either.
- From the section on "Confidently predicted domains ..."
- Follow the links to the database entries for the information so you know what these domains and features are.
Next we'll enter the features into our database, so we can compare them with the annotations that I have prepared from SMART annotations of Mbp1 orthologues from the ten reference fungi.
Visual comparison of domain annotations in R
The versatile plotting functions of R allow us to compare domain annotations. The distribution of segments that are annotated as "low-complexity, presumably disordered, is particularly interesting: these are functional features that are often not associated with sequence similarity but may have arisen from convergent evolution. Those would not be detectable through sequence alignment - which is after all based on amino acid pair scores and therefore context independent.
In the following code tutorial, we create a plot similar to the CDD and SMART displays. It is based on the SMART domain annotations of the six-fungal reference species for the course.
Task:
- Return to your RStudio session.
- Make sure you have saved
myDBas instructed previously. Then quit the program, restart, and re-open the project via the File → Recent projects ... menu. This is to clear out-of-date assignments and functions from the workspace. - Do not type
init()yet, but pull the most recent version of files from github. Then typeinit(). - Study and work through the code in the
SMART domain annotationssection of theBCH441_A04.Rscript. This includes entering your domain and other feature annotations into the database. - At the end of the script, print out your plot of the domain annotations for MB1_YFO and the reference proteins. Bring this plot with you for the next quiz.
- Can this plot be improved? What would you do differently to maximize its utility from an information-design point of view?
When you execute the code, your plot should look similar to this one:
A note on the R code up to this point: You will find that we have been writing a lot of nested expressions for selections that join data from multiple tables of our data model. When I teach R workshops for graduate students, postdocs and research fellows, I find that the single greatest barrier in their actual research work is the preparation of data for analysis: filtering, selecting, cross-referencing, and integrating data from different sources. By now, I hope you will have acquired a somewhat robust sense for achieving this. You can imagine that there are ways to simplify those tasks with functions you write, or special resources from a variety of different packages you cab install. But the "pedestrian" approach we have been taking in our scripts has the advantage of working from a very small number of principles, with very few syntactic elements.
Multiple Sequence Alignment
In order to perform a multiple sequence alignment, we obviously need a set of homologous sequences. This is not trivial. All interpretation of MSA results depends absolutely on how the input sequences were chosen. Should we include only orthologues, or paralogues as well? Should we include only species with fully sequenced genomes, or can we tolerate that some orthologous genes are possibly missing for a species? Should we include all sequences we can lay our hands on, or should we restrict the selection to a manageable number of representative sequences? All of these choices influence our interpretation:
- orthologues are expected to be functionally and structurally conserved;
- paralogues may have divergent function but have similar structure;
- missing genes may make paralogs look like orthologs; and
- selection bias may weight our results toward sequences that are over-represented and do not provide a fair representation of evolutionary divergence.
Computing an MSA in R
Let's use the Bioconductor msa package to align the sequences we have. Study and run the following code
Task:
- Return to your RStudio session.
- Make sure you have saved
myDBas instructed previously. - Bring code and data resources up to date:
- pull the most recent version of the project from GitHub
- type
init()to lod the most recent files and functions - re-merge your current
myDB
- Study and work through the code in the
Multiple sequence alignmentssection of theBCH441_A04.Rscript. - Note that the final task asks you to print out some results and bring them to class for the next quiz.
Sequence alignment editors
Really excellent software tools have been written that help you visualize and manually curate multiple sequence alignments. If anything, I think they tend to do too much. Past versions of the course have used Jalview, but I have heard good things of AliView (and if you are on a Mac seqotron might interest you, but I only cover software that is free and runs on all three major platforms).
Right now, I am just mentioning the two alignment editors. If you have experience with comparing them, let us know.
- [Jalview] an integrated MSA editor and sequence annotation workbench from the Barton lab in Dundee. Lots of functions.
- [AliView] from Uppsala: fast, lean, looks to be very practical.
- That is all.
Links and resources
Eddy (2004) Where did the BLOSUM62 alignment score matrix come from?. Nat Biotechnol 22:1035-6. (pmid: 15286655) [ PubMed ] [ DOI ] Many sequence alignment programs use the BLOSUM62 score matrix to score pairs of aligned residues. Where did BLOSUM62 come from?
- Biostrings Quick Overview ( summary of Biostrings functions (PDF))
Footnotes and references
- ↑ This is not strictly true in all cases: some algorithms measure similarity through an alignment-free approach, for example by comparing structural features, or domain annotations. These methods are less sensitive, but important when sequences are so highly diverged that no meaningful sequence alignment can be produced.
- ↑ "indel": insertion / deletion – a difference in sequence length between two aligned sequences that is accommodated by gaps in the alignment. Since we can't tell from the comparison of two sequences whether such a change was introduced by insertion into or deletion from the ancestral sequence, we join both into a portmanteau.
- ↑ That directory also contains sourcecode to generate the PAM matrices. This may be of interest if you ever want to produce scoring matrices from your own datasets.
- ↑ That directory also contains sourcecode to generate the PAM matrices. This may be of interest if you ever want to produce scoring matrices from your own datasets.
- ↑ While this seems like an unnecessary limitation, given that we could easily write such code to transform to-upper when looking up values in the MDM, perhaps it is meant as an additional sanity check that we haven't inadvertently included text in the sequence that does not belong there, such as the FASTA header line perhaps.
- ↑ Note that RBMs are usually orthologues, but the definition of orthologue and RBM is not the same. Most importantly, many orthologues are not RBMs. We will explore this more when we discuss phylogenetic inference.
- ↑ One such case we encountered involved a protein that has a corrupted annotation for the DNA binding domain. It appears to be the correct orthologue, but it only has the second highest BLAST score.
- ↑ The -myceta are well supported groups above the Class rank. See Leotiomyceta for details and references.
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.
< Assignment 3 Assignment 5 >