Difference between revisions of "BIO Assignment Week 3"
m (→Search input) |
m |
||
| (54 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
<span style="font-size: 70%">Sequence Analysis</span> | <span style="font-size: 70%">Sequence Analysis</span> | ||
</div> | </div> | ||
| + | <table style="width:100%;"><tr> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:left; font-size:80%;">[[BIO_Assignment_Week_2|< Assignment 2]]</td> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:right; font-size:80%;">[[BIO_Assignment_Week_4|Assignment 4 >]]</td> | ||
| + | </tr></table> | ||
{{Template:Inactive}} | {{Template:Inactive}} | ||
| Line 16: | Line 20: | ||
==Introduction== | ==Introduction== | ||
| + | ==Model organism and YFO== | ||
| − | In this assignment we will | + | In this assignment we will set out on our exploration of the system that regulates the G1/S transition by focussing first on the Mbp1 protein in selected species from the {{WP|Kingdom_(biology)|kingdom}} of fungi, whose genome has been completely sequenced; our quest is thus also an exercise in ''model-organism reasoning'': the transfer of knowledge from one, well-studied organism to others. It's reasonable to hypothesize that such central control machinery is conserved in most if not all fungi. But we don't know. Many of the species that we will be working with have not been characterized in great detail, and some of them are new to our class this year. And while we know a fair bit about Mbp1, we probably don't know very much at all about the related genes in other organisms: whether they exist, whether they have similar functional features and whether they might contribute to the ''G1/S checkpoint system'' in a similar way. Thus we might discover things that are new and interesting. This is a quest of discovery. |
| − | * | + | |
| − | + | {{Vspace}} | |
| + | |||
| + | ==The BCH441 RStudio Project== | ||
| + | |||
| + | I have begun to unify '''R'''-scripts and other resources in an RStudio project that will make it easy to update and distribute code. Conveniently, if I push update material to the github repository, all you need to do is to ''pull'' the updated project to receive all updates and new files on your computer. Version control is really good for this. However, there is one limitation to this approach that you need to be aware of. If you create your own, local files and then '''commit''' them, ''git'' will complain that it would be overwriting such local material. As long as you don't '''commit''' your files then all should be fine. This means you'll need to do your own "versioning" by saving your own scripts under a different name from time to time. Once again: in this context: | ||
| + | *'''saving''' your own files is fine; | ||
| + | *'''committing''' your own files to version control will cause problems; | ||
| + | * changes you make to course material files and save '''under the same filename''' (like adding comments and notes) will not persist, these changes will be overwritten with the next update. You need to "Save As..." with a new filename (e.g. just prefix the original name with "<code>my</code>"). | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | {{task|1= | ||
| + | * Open RStudio and create a '''New Project...''' cloned from a git version control directory. The repository URL is <code><nowiki>https://github.com/hyginn/BCH441_2016</nowiki></code>. Create this in the same way as you did for the [[R_tutorial#Git_Version_control|'''R'''-tutorial]]. | ||
| + | * As requested on the console, type <code>init()</code>. This will create a file called <code>.myProfile.R</code> and ask you for your UofT eMail address and Student ID. You need to enter the correct values because other scripts will make assumptions about that these variables exist and are valid. | ||
| + | * Work through the task: <code>"local script"</code> in the <code>BCH441_2016.R</code> script. | ||
| + | * Then load the script for the current assignment as instructed. | ||
| + | |||
| + | }} | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ==Choosing YFO (Your Favourite Organism)== | ||
| + | |||
| + | Since we were trying to find related proteins in a different species, our next task is to find suitable species. | ||
| + | |||
| + | For this purpose we create a lottery to assign species at (pseudo) random to students, so that everyone has a good chance to be working on their own species. The technical details are in the R scripts that implement the species search and distribution. In brief, we define a function that picks one species from a long list at random - but to make sure this process is reproducible, we'll set a seed for the random number generator. Obviously, everyone has to use a different seed, or else everyone would end up getting the same species assigned. Thus we'll use your Student Number as the seed. This is an integer, so it can be used as an argument to '''R''''s <code>set.seed()</code> function, and it's unique to each of you. The choice is then random, reproducible and unique. | ||
| + | |||
| + | <small>You may notice that this process does not guarantee that everyone gets a different species, and that all species are chosen at least once. I don't think doing that is possible in a "stateless" way (i.e. I don't want to have to remember who chose what species), given that I don't know all of your student numbers. But if anyone can think of a better solution, that would be neat.</small> | ||
| + | |||
| + | Is it possible that all of you end up working on the same species anyway? Indeed. That's the problem with randomness. But it is not very likely. | ||
| − | |||
| − | + | What about the "suitable species" though? Where do they come from? For the purposes of the course "quest", we need species | |
| + | * that actually have transcription factors that are related to Mbp1; | ||
| + | * whose genomes have been sequenced; and | ||
| + | * for which the sequences have been deposited in the RefSeq database, NCBI's unique sequence collection. | ||
| − | + | {{task| | |
| + | |||
| + | * Work through PART ONE: YFO Species of the BCH441_A03.R script. | ||
| + | * There are some deep "rabbitholes" that you are encouraged to follow to explore the code that went into generating the YFO species list. The minimal required result however is that you have picked an '''YFO''', and that it's name got written into your personalized profile file. | ||
| + | * Note down the species name and its five letter label on your Student Wiki user page. '''Use this species whenever this or future assignments refer to YFO'''. | ||
| + | }} | ||
| − | |||
| − | |||
| + | {{Vspace}} | ||
| − | + | ===Selecting the YFO "Mbp1"=== | |
| + | {{Vspace}} | ||
{{task|1= | {{task|1= | ||
| − | |||
| − | |||
| − | |||
| − | |||
| + | # Back at the [http://www.ncbi.nlm.nih.gov/protein/NP_010227 Mbp1 protein page] follow the link to [http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins&PROGRAM=blastp&BLAST_PROGRAMS=blastp&QUERY=NP_010227.1&LINK_LOC=protein&PAGE_TYPE=BlastSearch Run BLAST...] under "Analyze this sequence". | ||
| + | # This allows you to perform a sequence similarity search. You need to set two parameters: | ||
| + | ## As '''Database''', select '''Reference proteins (refseq_protein)''' from the drop down menu; | ||
| + | ## In the '''Organism''' field, type the species you have selected as YFO and select the corresponding taxonomy ID. | ||
| + | # Click on '''Run BLAST''' to start the search. This should find a handful of genes, all of them in YFO. If you find none, or hundreds, or they are not all in the same species, you did something wrong. Ask on the mailing list and make sure to fix the problem. | ||
| + | # Look at the top "hit" in the '''Descriptions''' section. The rightmost column contains sequence IDs unter the '''Accession''' heading. The alignment and alignment score are shown in the '''Alignments''' section a bit further down the page. Look at the result. | ||
| + | # In the header information for each hit is a link to its database entry, right next to '''Sequence ID'''. It says something like <code>ref|NP_123456789.1</code> or <code>ref|XP_123456789</code> ... follow that link. | ||
| + | # Note the RefSeq ID, and the search results %ID, E-value, whether one or more similar regions were found etc. in your Journal. We will refer to this sequence as "''YFO'' Mbp1" or similar in the future. | ||
| + | # Finally access the [http://www.uniprot.org/uploadlists/ UniProt ID mapping service] to retrieve the UniProt ID for the protein. Paste the RefSeq ID and choose '''RefSeq Protein''' as the '''From:''' option and '''UniProtKB''' as the '''To:''' option. | ||
| − | + | :<small>If the mapping works, the UniProt ID will be in the '''Entry:''' column of the table that is being returned. Click the link and have a look at the UniProt entry page while you're there.</small> | |
| + | <!-- What could go wrong? Sometimes the mapping does not work: | ||
| + | I don't know why the mapping for some sequences is not available. | ||
| + | If this happens, you can work around the problem as follows. | ||
| − | + | 1. Load the RefSeq protein page | |
| − | + | 2. View the protein as FASTA and copy the sequence. | |
| − | + | 3. Open the UniProt BLAST page http://www.uniprot.org/blast/ | |
| − | + | (Yes, UniProt runs its own BLAST version, and that searches UniProt databases, not Genbank) | |
| − | + | 4. Paste the sequence into the search form and run BLAST. | |
| − | + | ... if the sequence is in UniProt, you will get the top hit with 100% sequence identity. In your case it is: | |
| + | H1VQK3 ( http://www.uniprot.org/uniprot/H1VQK3 ) | ||
| + | I.e. UniProt contains the sequence, but the mapping service does not know. | ||
| + | --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | }} | |
| − | + | {{Vspace}} | |
| − | + | ||
| − | + | ==Store Data== | |
| − | + | ||
| − | + | {{Vspace}} | |
| − | + | ||
| − | + | ===The ''System'' datamodel=== | |
| − | + | ||
| + | {{Vspace}} | ||
| + | |||
| + | [[Image:SystemDB_Datamodel.png|frame|Entity-Relationship Diagram (ERD) for a data model that stores data for a systems project. Entity names are at the top of each box, attributes are listed below. If you think of the entity as a table, its attributes are the column names and each row stores the values for one particular instance. Semantically related entities are shaded in similar colours; this helps to make the design-principles visible, but one must be careful not to overdo the use of colour. As with all graphical elements in information design: "less is more". All relationships link to unique primary keys and are thus 1 to (0, n): i.e. as attributes, the relationship does not have to exist but there could be many, as the primary key, exactly one key must exist. The diagram was drawn as a "Google presentation" slide and you can [https://docs.google.com/presentation/d/1_nYWiwQc-9Z4anUAwOeVqWXgXIvM1Zl_AffMTM_No2Q/edit?usp=sharing view it on the Web] and make a copy for your own purposes.]] | ||
| + | |||
| + | Here is a first version of a systems data model, based on what we discussed in class: | ||
| + | * The '''<code>feature</code> table''' is at the centre. This was not intentional, but emerged from iterating the model through a number of revisions. It emphasizes that the main purpose of this model is to integrate and annotate data from various sources. ''Feature'' in the way we understand it here is an abstraction of quite disparate categories of information items. This includes domain annotations, system functions, literature references, and cross-references to other databases. The ''type'' attribute will require some thought: this attribute really needs a "controlled vocabulary" to ensure that the same category is described consistently with the same string ("PubMed"? "Lit."? "reference"?). There are a number of ways to achieve this, the best way<ref>Relational databases like MySQL, PostgresQL, and MariaDB offer the datatype "Enum" for this purpose but this is an inferior solution. Enum types need to be fixed at the time the schema is created, they can't store information about their semantics, i.e. how the keywords are defined and when they should be used, and they can't be used in more than one table, since they are metadata of one particular column.</ref> is to store these types in their own "reference" table '''<code>type</code>''' - and link to that table via a ''foreign key''. | ||
| + | * The '''<code>protein</code> table''' is at the centre. Its ''primary key'' is a unique integer. We store the NCBI RefSeq ID and the Uniprot ID in the table. We would not call these "foreign keys", since the information they reference is not in our schema but at the NCBI resp. EBI. For example, we can't guarantee that they are unique keys. | ||
| + | * The '''<code>taxonomy</code> table''' holds information about species. We use the NCBI taxonomy ID as its ''primary key''. The same key appears in the protein table as the ''foreign key'' that links the protein with its proper taxonomy information. This is an instance where the data model actually does not describe reality well. The problem is that particular proteins that we might retrieve from database searches will often be annotated to a specific ''strain'' of a ''species'' – and there is no easy way to reference strains to species. We'll see whether this turns out to be a problem in practice. But it may be that an additional table may be required that stores parent/child relationships of the taxonomic tree of life. | ||
| + | * The '''<code>protein_feature</code> table''' links a protein with all the features that we annotate it with. ''start'' and ''end'' coordinates identify the region of the sequence we have annotated. | ||
| + | * The '''<code>...Annotation</code> tables''' link feature-information with annotated entities. | ||
| + | * Should the '''<code>system</code> table''' have its own taxonomy attribute? Or should the species in which the system is observed be inferred from the component protein's '''<code>taxonomy.ID</code>'''? What do you think? I decided not to add a taxonomy attribute to that table. How would you argue for or against this decision? | ||
| + | * The '''<code>component</code> table''' links proteins that collaborate together as a system. There is an implicit assumption in this model that only proteins are system components (and not e.g. RNA), and that components are atomic (i.e. we can't link to subsystems). How would you change the model to accommodate more realistic biological complexity? | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ===Implementing the Data Model in R=== | ||
| − | + | {{Vspace}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | </ | + | To actually implement the data model in '''R''' we will create the tables as data frames, and we will collect them in a list. We don't '''have''' to keep the tables in a list - we could also keep them as independent objects, but with a table named "protein" we should be worried of inadvertently overwriting the table. A list is neater, more flexible (we might want to have several of these), it reflects our intent about the model better, and doesn't require very much more typing. For now, to keep things simple, we will implement only two tables: '''<code>protein</code>''' and '''<code>taxonomy</code>'''. We'll add the rest when we actually need them. |
| − | </ | ||
| + | {{task|1 = | ||
| − | + | * study the code in the <code>Creating two tables</code> section of the '''R''' script | |
}} | }} | ||
| + | {{Vspace}} | ||
| − | + | As you see we can edit any component of our data model directly by assigning new values to the element. But in general that's a really bad idea, since it is far too easy to bring the whole model into an inconsistent state. It's much better to write functions that ''get'' and ''set'' data elements, not only to keep the data consistent, but also because it is much easier, if the model ever changes, to simply edit the function code, rather than having to repeat every single data entry by hand. | |
| + | |||
| + | What would an <code>setData</code> function have to look like? It should | ||
| + | *create a new entry if the requested row of a table does not exist yet; | ||
| + | *update data if the protein exists; | ||
| + | *perform consistency checks (i.e. check that the data has the correct type); | ||
| + | *perform sanity checks (i.e. check that data values fall into the expected range); | ||
| + | *perform completeness checks (i.e. handle incomplete data) | ||
| + | |||
| + | Let's start simple, and create a '''set-''' function to | ||
| + | add new values to existing sequence data. Also, for clarity, we'll forgo many checks. The first thing we should do is to add the actual sequence. | ||
| + | |||
| + | We only entered a placeholder for the sequence field. | ||
| + | Sequences come in many different flavours when we copy them from a Webpage: | ||
| + | there can be whitespace, carriage returns, numbers, they can be upper-case, lower-case | ||
| + | mixed-case ... | ||
| + | |||
| + | What we want in our sequence data field is one string | ||
| + | that contains the entire sequence, and nothing but | ||
| + | upper-case, amino-acid code letters. | ||
| + | |||
| + | We'll need to look at how we work with strings in '''R''', how we identify and work with patterns in strings. This is a great time to introduce regular expressions. | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ====A Brief First Encounter of Regular Expressions==== | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ;Regular expressions are a concise description language to define patterns for pattern-matching in strings. | ||
| + | |||
| + | Truth be told, many programmers have a love-hate relationship with regular expressions. The syntax of regular expressions is very powerful and expressive, but also terse, not always intuitive, and sometimes hard to understand. I'll introduce you to a few principles here that are quite straightforward and they will probably cover 99% of the cases you will encounter. | ||
| + | |||
| + | Here is our test-case: the sequence of Mbp1, copied from the [https://www.ncbi.nlm.nih.gov/protein/NP_010227 NCBI Protein database page for yeast Mbp1]. | ||
| + | |||
| + | 1 msnqiysary sgvdvyefih stgsimkrkk ddwvnathil kaanfakakr trilekevlk | ||
| + | 61 ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf dftqtdgsas pppapkhhha | ||
| + | 121 skvdrkkair sastsaimet krnnkkaeen qfqsskilgn ptaaprkrgr pvgstrgsrr | ||
| + | 181 klgvnlqrsq sdmgfprpai pnssisttql psirstmgpq sptlgileee rhdsrqqqpq | ||
| + | 241 qnnsaqfkei dledglssdv epsqqlqqvf nqntgfvpqq qssliqtqqt esmatsvsss | ||
| + | 301 pslptspgdf adsnpfeerf pgggtspiis miprypvtsr pqtsdindkv nkylsklvdy | ||
| + | 361 fisnemksnk slpqvllhpp phsapyidap idpelhtafh wacsmgnlpi aealyeagts | ||
| + | 421 irstnsqgqt plmrsslfhn sytrrtfpri fqllhetvfd idsqsqtvih hivkrksttp | ||
| + | 481 savyyldvvl skikdfspqy rielllntqd kngdtalhia skngdvvffn tlvkmgaltt | ||
| + | 541 isnkegltan eimnqqyeqm miqngtnqhv nssntdlnih vntnnietkn dvnsmvimsp | ||
| + | 601 vspsdyityp sqiatnisrn ipnvvnsmkq masiyndlhe qhdneikslq ktlksisktk | ||
| + | 661 iqvslktlev lkesskdeng eaqtnddfei lsrlqeqntk klrkrliryk rlikqkleyr | ||
| + | 721 qtvllnklie detqattnnt vekdnntler lelaqeltml qlqrknklss lvkkfednak | ||
| + | 781 ihkyrriire gtemnieevd ssldvilqtl iannnknkga eqiitisnan sha | ||
| + | // | ||
{{task|1= | {{task|1= | ||
| − | + | ||
| − | + | Navigate to http://regexpal.com and paste the sequence into the '''lower''' box. This site is one of a number of online regular expression testers; their immediate, visual feedback is invaluable when you are developing regular expression patterns. | |
| − | < | + | |
| − | < | + | Lets try some expressions: |
| − | + | ||
| − | + | ;Most characters are matched literally. | |
| − | + | :Type "<code>a</code>" in to the '''upper''' box and you will see all "<code>a</code>" characters matched. Then replace <code>a</code> with <code>q</code>. | |
| − | < | + | : Now type "<code>aa</code>" instead. Then <code>krnnkk</code>. ''Sequences'' of characters are also matched literally. |
| − | + | ||
| − | </ | + | ;The pipe character {{pipe}} that symbolizes logical OR can be used to define that more than one character should match: |
| + | :<code>i(s{{pipe}}m{{pipe}}q)n</code> matches <code>isn</code> OR <code>imn</code> OR <code>iqn</code>. Note how we can group with parentheses, and try what would happen without them. | ||
| + | |||
| + | ;We can more conveniently specify more than one character to match if we place it in square brackets. | ||
| + | :<code>[lq]</code> matches <code>l</code> OR <code>q</code>. <code>[familyvw]</code> matches hydrophobic amino acids. | ||
| + | |||
| + | ;Within square brackets, we can specify "ranges". | ||
| + | :<code>[1-5]</code> matches digits from 1 to 5. | ||
| + | |||
| + | ;Within square brackets, we can specify characters that should NOT be matched, with the caret, <code>^</code>. | ||
| + | :<code>[^0-9]</code> matches everything EXCEPT digits. <code>[^a-z]</code> matches everything that is not a lower-case letter. That's what we need (try it). | ||
| + | |||
| + | }} | ||
| + | |||
| + | One of the '''R''' functions that uses regular expressions is the function <code>gsub()</code>. It replaces characters that match a "regex" with other characters. That is useful for our purpose: we can | ||
| + | #match all characters that are NOT a letter, and | ||
| + | #replace them by - nothing: the empty string <code>""</code>. | ||
| + | This deletes them. | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | {{task|1 = | ||
| + | * study the code in the <code>An excursion into regular expressions</code> section of the '''R''' script | ||
| + | }} | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | ===Updating the database=== | ||
| + | |||
| + | {{Vspace}} | ||
| + | |||
| + | {{task|1 = | ||
| + | * study the code in the <code>Updating the database</code> section of the '''R''' script | ||
}} | }} | ||
| + | {{Vspace}} | ||
| − | + | ===Add "your" YFO Sequence=== | |
| + | {{Vspace}} | ||
{{task|1= | {{task|1= | ||
| − | + | ||
| − | + | ;Add the YFO Mbp1 protein data to the database: | |
| − | + | ||
| − | + | # Copy the '''code template''' to add a new protein and its taxonomy entry into the script file <code>myCode.R</code> that you created at the very beginning. | |
| − | + | # Add your protein to the database by editing a copy of the code template in your script file. Ask on the mailing list if you don't know how (but be specific) or if you don't know how to find particular information items. | |
| − | # | + | # Add the taxonomy entry to the taxonomy table, again simply modifying a copy of the code template in your own script file. |
| − | # | + | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | # | ||
| − | |||
}} | }} | ||
| − | + | {{Vspace}} | |
| − | == | + | ==Analyze== |
| − | Let us perform a few simple sequence | + | Let us perform a few simple sequence analyses using the online EMBOSS tools. EMBOSS (the European Molecular Biology laboratory Open Software Suite) combines a large number of simple but fundamental sequence analysis tools. The tools can be [http://emboss.sourceforge.net/download/ installed locally on your own machine], or run via a public Web interface. Google for [http://www.google.ca/search?q=EMBOSS+explorer EMBOSS explorer], public access points include http://emboss.bioinformatics.nl/ . |
| − | Access | + | Access an EMBOSS Explorer service and explore some of the tools: |
| Line 147: | Line 271: | ||
;Local composition | ;Local composition | ||
# Find <code>pepinfo</code> under the '''PROTEIN COMPOSITION''' heading. | # Find <code>pepinfo</code> under the '''PROTEIN COMPOSITION''' heading. | ||
| − | # | + | # Retrieve the YFO Mbp1 related sequence from your '''R''' database, e.g. with something like <br /><code> cat(db$protein[db$protein$name == "UMAG_1122"), "sequence"]</code> |
| + | # Copy and paste the sequence into the input field. | ||
# Run with default parameters. | # Run with default parameters. | ||
| + | # Scroll to the figures all the way at the bottom. | ||
# Do the same in a separate window for yeast Mbp1. | # Do the same in a separate window for yeast Mbp1. | ||
| − | # Try to compare ... kind of hard without reference, overlay and expectation, isn't it? | + | # Try to compare ... <small>(kind of hard without reference, overlay and expectation, isn't it?)</small> |
}} | }} | ||
| Line 157: | Line 283: | ||
;Global composition | ;Global composition | ||
# Find <code>pepstats</code> under the '''PROTEIN COMPOSITION''' heading. | # Find <code>pepstats</code> under the '''PROTEIN COMPOSITION''' heading. | ||
| − | # Paste the YFO Mbp1 sequence | + | # Paste the YFO Mbp1 sequence into the input field. |
# Run with default parameters. | # Run with default parameters. | ||
# Do the same in a separate window for yeast Mbp1. | # Do the same in a separate window for yeast Mbp1. | ||
| Line 166: | Line 292: | ||
{{task|1= | {{task|1= | ||
;Motifs | ;Motifs | ||
| − | # Find <code> | + | # Find <code>pepcoil</code>, an algorithm to detect {{WP|coiled coil}} motifs. |
| − | # Run | + | # Run this with the YFO Mbp1 sequence and yeast Mbp1. |
| − | # Try to compare ... do both sequences have | + | # Try to compare ... do both sequences have coiled-coil motif predictions? Are they annotated in approximately comparable regions of the respective sequence? |
}} | }} | ||
| Line 175: | Line 301: | ||
;Transmembrane sequences | ;Transmembrane sequences | ||
# Find <code>tmap</code>. Also find <code>shuffleseq</code>. | # Find <code>tmap</code>. Also find <code>shuffleseq</code>. | ||
| − | # Use your YFO | + | # Use your YFO sequence to annotate transmembrane helices for your protein and for a few shuffled sequences. The YFO is not expected to have TM helices, nor are the shuffled sequences expected to have any. If you '''do''' find some, these are most likely "''false positives''". |
| − | # Also compare the following positive | + | |
| − | + | # Also compare the following positive control: Gef1 - a yeast chloride channel with 10 trans-membrane helices and outward localized N-terminus: | |
| − | Gef1 - a yeast chloride channel with 10 trans-membrane helices and outward localized N-terminus: | ||
| − | |||
<source lang="text"> | <source lang="text"> | ||
>gi|6322500|ref|NP_012574.1| Gef1p [Saccharomyces cerevisiae S288c] | >gi|6322500|ref|NP_012574.1| Gef1p [Saccharomyces cerevisiae S288c] | ||
| Line 195: | Line 319: | ||
TTNRNGNVI | TTNRNGNVI | ||
</source> | </source> | ||
| − | + | ||
| − | + | # Evaluate the output: does the algorithm (wrongly) predict TM-helices in your protein? In the shuffled sequences? Does it find all ten TM-helices in Gef1? | |
| − | # Evaluate the output: does the algorithm (wrongly) predict | ||
}} | }} | ||
| Line 203: | Line 326: | ||
Try to familiarize yourself with the offerings in the EMBOSS package. I find some of the nucleic acid tools indispensable in the lab, such as restriction-site mapping tools, and I frequently use the alignment tools <code>Needle</code> and <code>Water</code>, but by and large the utility of many of the components–while fast, efficient and straightforward to use– suffers from lack of reference and comparison and from terse output. The routines show their conceptual origin in the 1970s and 1980s. We will encounter alternatives in later assignments. | Try to familiarize yourself with the offerings in the EMBOSS package. I find some of the nucleic acid tools indispensable in the lab, such as restriction-site mapping tools, and I frequently use the alignment tools <code>Needle</code> and <code>Water</code>, but by and large the utility of many of the components–while fast, efficient and straightforward to use– suffers from lack of reference and comparison and from terse output. The routines show their conceptual origin in the 1970s and 1980s. We will encounter alternatives in later assignments. | ||
| + | {{Vspace}} | ||
| − | == | + | =='''R''' Sequence Analysis Tools== |
| + | It's interesting to see this collection of tools that were carefully designed some twenty years ago, as an open source replacement for a set of software tools - the GCG package - that was indispensable for molecular biology labs in the 80s and 90s, but whose cost had become prohibitive. Fundamentally this is a building block approach, and the field has turned to programming solutions instead. | ||
| − | + | As for functionality, much more sophisticated functions are available on the Web: do take a few minutes and browse the [http://bioinformatics.ca/links_directory/tag/protein-sequence-analysis curated Web services directory of bioinformatics.ca]. | |
| − | + | As for versatility, '''R''' certainly has the edge. Let's explore some of the functions available in the <code>seqinr</code> package that you already encountered in the introductory [[R tutorial]]. They are comparatively basic - but it is easy to prepare our own analysis. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Vspace}} | |
| − | |||
| − | + | {{task|1 = | |
| − | + | * Study the code in the <code>Sequence Analysis</code> section of the '''R''' script | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
| − | + | {{Vspace}} | |
;That is all. | ;That is all. | ||
| + | {{Vspace}} | ||
| − | + | == Links and resources == | |
| − | + | * [http://regexpal.com RegEx Pal] - a tool for testing and developing regular expressions. | |
<!-- {{#pmid: 19957275}} --> | <!-- {{#pmid: 19957275}} --> | ||
| Line 361: | Line 361: | ||
{{#lst:BIO_Assignment_Week_1|assignment_footer}} | {{#lst:BIO_Assignment_Week_1|assignment_footer}} | ||
| + | <table style="width:100%;"><tr> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:left; font-size:80%;">[[BIO_Assignment_Week_2|< Assignment 2]]</td> | ||
| + | <td style="height:30px; vertical-align:middle; text-align:right; font-size:80%;">[[BIO_Assignment_Week_4|Assignment 4 >]]</td> | ||
| + | </tr></table> | ||
| | ||
[[Category:Bioinformatics]] | [[Category:Bioinformatics]] | ||
</div> | </div> | ||
Latest revision as of 09:10, 5 October 2016
Assignment for Week 3
Sequence Analysis
| < Assignment 2 | Assignment 4 > |
Note! This assignment is currently inactive. Major and minor unannounced changes may be made at any time.
Concepts and activities (and reading, if applicable) for this assignment will be topics on next week's quiz.
Contents
Introduction
Model organism and YFO
In this assignment we will set out on our exploration of the system that regulates the G1/S transition by focussing first on the Mbp1 protein in selected species from the kingdom of fungi, whose genome has been completely sequenced; our quest is thus also an exercise in model-organism reasoning: the transfer of knowledge from one, well-studied organism to others. It's reasonable to hypothesize that such central control machinery is conserved in most if not all fungi. But we don't know. Many of the species that we will be working with have not been characterized in great detail, and some of them are new to our class this year. And while we know a fair bit about Mbp1, we probably don't know very much at all about the related genes in other organisms: whether they exist, whether they have similar functional features and whether they might contribute to the G1/S checkpoint system in a similar way. Thus we might discover things that are new and interesting. This is a quest of discovery.
The BCH441 RStudio Project
I have begun to unify R-scripts and other resources in an RStudio project that will make it easy to update and distribute code. Conveniently, if I push update material to the github repository, all you need to do is to pull the updated project to receive all updates and new files on your computer. Version control is really good for this. However, there is one limitation to this approach that you need to be aware of. If you create your own, local files and then commit them, git will complain that it would be overwriting such local material. As long as you don't commit your files then all should be fine. This means you'll need to do your own "versioning" by saving your own scripts under a different name from time to time. Once again: in this context:
- saving your own files is fine;
- committing your own files to version control will cause problems;
- changes you make to course material files and save under the same filename (like adding comments and notes) will not persist, these changes will be overwritten with the next update. You need to "Save As..." with a new filename (e.g. just prefix the original name with "
my").
Task:
- Open RStudio and create a New Project... cloned from a git version control directory. The repository URL is
https://github.com/hyginn/BCH441_2016. Create this in the same way as you did for the R-tutorial. - As requested on the console, type
init(). This will create a file called.myProfile.Rand ask you for your UofT eMail address and Student ID. You need to enter the correct values because other scripts will make assumptions about that these variables exist and are valid. - Work through the task:
"local script"in theBCH441_2016.Rscript. - Then load the script for the current assignment as instructed.
Choosing YFO (Your Favourite Organism)
Since we were trying to find related proteins in a different species, our next task is to find suitable species.
For this purpose we create a lottery to assign species at (pseudo) random to students, so that everyone has a good chance to be working on their own species. The technical details are in the R scripts that implement the species search and distribution. In brief, we define a function that picks one species from a long list at random - but to make sure this process is reproducible, we'll set a seed for the random number generator. Obviously, everyone has to use a different seed, or else everyone would end up getting the same species assigned. Thus we'll use your Student Number as the seed. This is an integer, so it can be used as an argument to R's set.seed() function, and it's unique to each of you. The choice is then random, reproducible and unique.
You may notice that this process does not guarantee that everyone gets a different species, and that all species are chosen at least once. I don't think doing that is possible in a "stateless" way (i.e. I don't want to have to remember who chose what species), given that I don't know all of your student numbers. But if anyone can think of a better solution, that would be neat.
Is it possible that all of you end up working on the same species anyway? Indeed. That's the problem with randomness. But it is not very likely.
What about the "suitable species" though? Where do they come from? For the purposes of the course "quest", we need species
- that actually have transcription factors that are related to Mbp1;
- whose genomes have been sequenced; and
- for which the sequences have been deposited in the RefSeq database, NCBI's unique sequence collection.
Task:
- Work through PART ONE: YFO Species of the BCH441_A03.R script.
- There are some deep "rabbitholes" that you are encouraged to follow to explore the code that went into generating the YFO species list. The minimal required result however is that you have picked an YFO, and that it's name got written into your personalized profile file.
- Note down the species name and its five letter label on your Student Wiki user page. Use this species whenever this or future assignments refer to YFO.
Selecting the YFO "Mbp1"
Task:
- Back at the Mbp1 protein page follow the link to Run BLAST... under "Analyze this sequence".
- This allows you to perform a sequence similarity search. You need to set two parameters:
- As Database, select Reference proteins (refseq_protein) from the drop down menu;
- In the Organism field, type the species you have selected as YFO and select the corresponding taxonomy ID.
- Click on Run BLAST to start the search. This should find a handful of genes, all of them in YFO. If you find none, or hundreds, or they are not all in the same species, you did something wrong. Ask on the mailing list and make sure to fix the problem.
- Look at the top "hit" in the Descriptions section. The rightmost column contains sequence IDs unter the Accession heading. The alignment and alignment score are shown in the Alignments section a bit further down the page. Look at the result.
- In the header information for each hit is a link to its database entry, right next to Sequence ID. It says something like
ref|NP_123456789.1orref|XP_123456789... follow that link. - Note the RefSeq ID, and the search results %ID, E-value, whether one or more similar regions were found etc. in your Journal. We will refer to this sequence as "YFO Mbp1" or similar in the future.
- Finally access the UniProt ID mapping service to retrieve the UniProt ID for the protein. Paste the RefSeq ID and choose RefSeq Protein as the From: option and UniProtKB as the To: option.
- If the mapping works, the UniProt ID will be in the Entry: column of the table that is being returned. Click the link and have a look at the UniProt entry page while you're there.
Store Data
The System datamodel
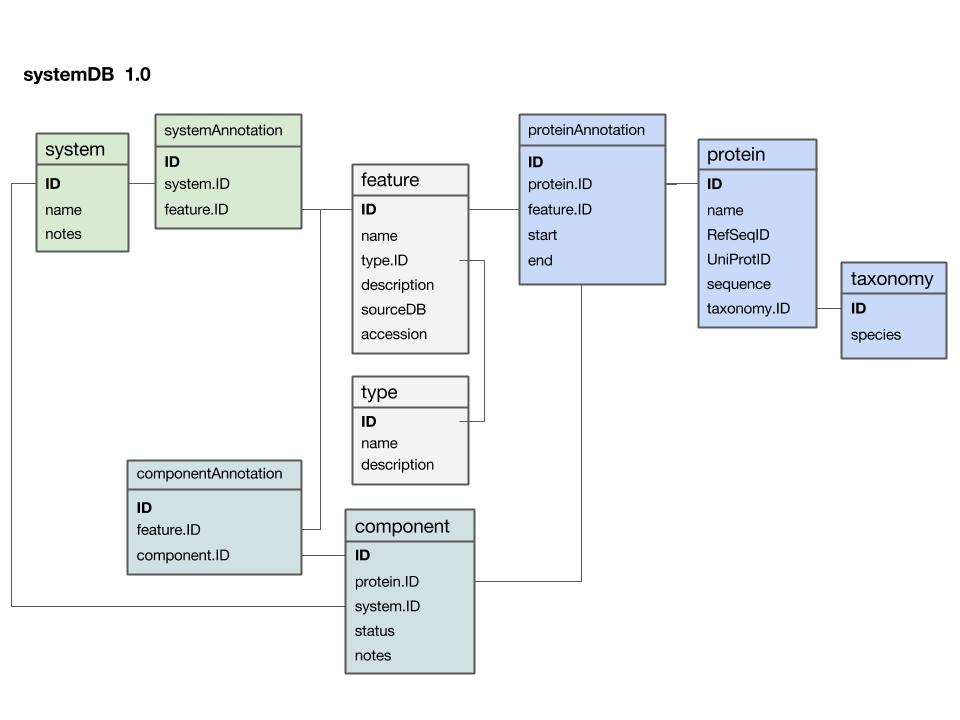
Here is a first version of a systems data model, based on what we discussed in class:
- The
featuretable is at the centre. This was not intentional, but emerged from iterating the model through a number of revisions. It emphasizes that the main purpose of this model is to integrate and annotate data from various sources. Feature in the way we understand it here is an abstraction of quite disparate categories of information items. This includes domain annotations, system functions, literature references, and cross-references to other databases. The type attribute will require some thought: this attribute really needs a "controlled vocabulary" to ensure that the same category is described consistently with the same string ("PubMed"? "Lit."? "reference"?). There are a number of ways to achieve this, the best way[1] is to store these types in their own "reference" tabletype- and link to that table via a foreign key. - The
proteintable is at the centre. Its primary key is a unique integer. We store the NCBI RefSeq ID and the Uniprot ID in the table. We would not call these "foreign keys", since the information they reference is not in our schema but at the NCBI resp. EBI. For example, we can't guarantee that they are unique keys. - The
taxonomytable holds information about species. We use the NCBI taxonomy ID as its primary key. The same key appears in the protein table as the foreign key that links the protein with its proper taxonomy information. This is an instance where the data model actually does not describe reality well. The problem is that particular proteins that we might retrieve from database searches will often be annotated to a specific strain of a species – and there is no easy way to reference strains to species. We'll see whether this turns out to be a problem in practice. But it may be that an additional table may be required that stores parent/child relationships of the taxonomic tree of life. - The
protein_featuretable links a protein with all the features that we annotate it with. start and end coordinates identify the region of the sequence we have annotated. - The
...Annotationtables link feature-information with annotated entities. - Should the
systemtable have its own taxonomy attribute? Or should the species in which the system is observed be inferred from the component protein'staxonomy.ID? What do you think? I decided not to add a taxonomy attribute to that table. How would you argue for or against this decision? - The
componenttable links proteins that collaborate together as a system. There is an implicit assumption in this model that only proteins are system components (and not e.g. RNA), and that components are atomic (i.e. we can't link to subsystems). How would you change the model to accommodate more realistic biological complexity?
Implementing the Data Model in R
To actually implement the data model in R we will create the tables as data frames, and we will collect them in a list. We don't have to keep the tables in a list - we could also keep them as independent objects, but with a table named "protein" we should be worried of inadvertently overwriting the table. A list is neater, more flexible (we might want to have several of these), it reflects our intent about the model better, and doesn't require very much more typing. For now, to keep things simple, we will implement only two tables: protein and taxonomy. We'll add the rest when we actually need them.
Task:
- study the code in the
Creating two tablessection of the R script
As you see we can edit any component of our data model directly by assigning new values to the element. But in general that's a really bad idea, since it is far too easy to bring the whole model into an inconsistent state. It's much better to write functions that get and set data elements, not only to keep the data consistent, but also because it is much easier, if the model ever changes, to simply edit the function code, rather than having to repeat every single data entry by hand.
What would an setData function have to look like? It should
- create a new entry if the requested row of a table does not exist yet;
- update data if the protein exists;
- perform consistency checks (i.e. check that the data has the correct type);
- perform sanity checks (i.e. check that data values fall into the expected range);
- perform completeness checks (i.e. handle incomplete data)
Let's start simple, and create a set- function to add new values to existing sequence data. Also, for clarity, we'll forgo many checks. The first thing we should do is to add the actual sequence.
We only entered a placeholder for the sequence field. Sequences come in many different flavours when we copy them from a Webpage: there can be whitespace, carriage returns, numbers, they can be upper-case, lower-case mixed-case ...
What we want in our sequence data field is one string that contains the entire sequence, and nothing but upper-case, amino-acid code letters.
We'll need to look at how we work with strings in R, how we identify and work with patterns in strings. This is a great time to introduce regular expressions.
A Brief First Encounter of Regular Expressions
- Regular expressions are a concise description language to define patterns for pattern-matching in strings.
Truth be told, many programmers have a love-hate relationship with regular expressions. The syntax of regular expressions is very powerful and expressive, but also terse, not always intuitive, and sometimes hard to understand. I'll introduce you to a few principles here that are quite straightforward and they will probably cover 99% of the cases you will encounter.
Here is our test-case: the sequence of Mbp1, copied from the NCBI Protein database page for yeast Mbp1.
1 msnqiysary sgvdvyefih stgsimkrkk ddwvnathil kaanfakakr trilekevlk
61 ethekvqggf gkyqgtwvpl niakqlaekf svydqlkplf dftqtdgsas pppapkhhha
121 skvdrkkair sastsaimet krnnkkaeen qfqsskilgn ptaaprkrgr pvgstrgsrr
181 klgvnlqrsq sdmgfprpai pnssisttql psirstmgpq sptlgileee rhdsrqqqpq
241 qnnsaqfkei dledglssdv epsqqlqqvf nqntgfvpqq qssliqtqqt esmatsvsss
301 pslptspgdf adsnpfeerf pgggtspiis miprypvtsr pqtsdindkv nkylsklvdy
361 fisnemksnk slpqvllhpp phsapyidap idpelhtafh wacsmgnlpi aealyeagts
421 irstnsqgqt plmrsslfhn sytrrtfpri fqllhetvfd idsqsqtvih hivkrksttp
481 savyyldvvl skikdfspqy rielllntqd kngdtalhia skngdvvffn tlvkmgaltt
541 isnkegltan eimnqqyeqm miqngtnqhv nssntdlnih vntnnietkn dvnsmvimsp
601 vspsdyityp sqiatnisrn ipnvvnsmkq masiyndlhe qhdneikslq ktlksisktk
661 iqvslktlev lkesskdeng eaqtnddfei lsrlqeqntk klrkrliryk rlikqkleyr
721 qtvllnklie detqattnnt vekdnntler lelaqeltml qlqrknklss lvkkfednak
781 ihkyrriire gtemnieevd ssldvilqtl iannnknkga eqiitisnan sha
//
Task:
Navigate to http://regexpal.com and paste the sequence into the lower box. This site is one of a number of online regular expression testers; their immediate, visual feedback is invaluable when you are developing regular expression patterns.
Lets try some expressions:
- Most characters are matched literally.
- Type "
a" in to the upper box and you will see all "a" characters matched. Then replaceawithq. - Now type "
aa" instead. Thenkrnnkk. Sequences of characters are also matched literally.
- The pipe character | that symbolizes logical OR can be used to define that more than one character should match
i(s|m|q)nmatchesisnORimnORiqn. Note how we can group with parentheses, and try what would happen without them.
- We can more conveniently specify more than one character to match if we place it in square brackets.
[lq]matcheslORq.[familyvw]matches hydrophobic amino acids.
- Within square brackets, we can specify "ranges".
[1-5]matches digits from 1 to 5.
- Within square brackets, we can specify characters that should NOT be matched, with the caret,
^. [^0-9]matches everything EXCEPT digits.[^a-z]matches everything that is not a lower-case letter. That's what we need (try it).
One of the R functions that uses regular expressions is the function gsub(). It replaces characters that match a "regex" with other characters. That is useful for our purpose: we can
- match all characters that are NOT a letter, and
- replace them by - nothing: the empty string
"".
This deletes them.
Task:
- study the code in the
An excursion into regular expressionssection of the R script
Updating the database
Task:
- study the code in the
Updating the databasesection of the R script
Add "your" YFO Sequence
Task:
- Add the YFO Mbp1 protein data to the database
- Copy the code template to add a new protein and its taxonomy entry into the script file
myCode.Rthat you created at the very beginning. - Add your protein to the database by editing a copy of the code template in your script file. Ask on the mailing list if you don't know how (but be specific) or if you don't know how to find particular information items.
- Add the taxonomy entry to the taxonomy table, again simply modifying a copy of the code template in your own script file.
Analyze
Let us perform a few simple sequence analyses using the online EMBOSS tools. EMBOSS (the European Molecular Biology laboratory Open Software Suite) combines a large number of simple but fundamental sequence analysis tools. The tools can be installed locally on your own machine, or run via a public Web interface. Google for EMBOSS explorer, public access points include http://emboss.bioinformatics.nl/ .
Access an EMBOSS Explorer service and explore some of the tools:
Task:
- Local composition
- Find
pepinfounder the PROTEIN COMPOSITION heading. - Retrieve the YFO Mbp1 related sequence from your R database, e.g. with something like
cat(db$protein[db$protein$name == "UMAG_1122"), "sequence"] - Copy and paste the sequence into the input field.
- Run with default parameters.
- Scroll to the figures all the way at the bottom.
- Do the same in a separate window for yeast Mbp1.
- Try to compare ... (kind of hard without reference, overlay and expectation, isn't it?)
Task:
- Global composition
- Find
pepstatsunder the PROTEIN COMPOSITION heading. - Paste the YFO Mbp1 sequence into the input field.
- Run with default parameters.
- Do the same in a separate window for yeast Mbp1.
- Try to compare ... are there significant and unexpected differences?
Task:
- Motifs
- Find
pepcoil, an algorithm to detect coiled coil motifs. - Run this with the YFO Mbp1 sequence and yeast Mbp1.
- Try to compare ... do both sequences have coiled-coil motif predictions? Are they annotated in approximately comparable regions of the respective sequence?
Task:
- Transmembrane sequences
- Find
tmap. Also findshuffleseq. - Use your YFO sequence to annotate transmembrane helices for your protein and for a few shuffled sequences. The YFO is not expected to have TM helices, nor are the shuffled sequences expected to have any. If you do find some, these are most likely "false positives".
- Also compare the following positive control: Gef1 - a yeast chloride channel with 10 trans-membrane helices and outward localized N-terminus:
>gi|6322500|ref|NP_012574.1| Gef1p [Saccharomyces cerevisiae S288c]
MPTTYVPINQPIGDGEDVIDTNRFTNIPETQNFDQFVTIDKIAEENRPLSVDSDREFLNSKYRHYREVIW
DRAKTFITLSSTAIVIGCIAGFLQVFTETLVNWKTGHCQRNWLLNKSFCCNGVVNEVTSTSNLLLKRQEF
ECEAQGLWIAWKGHVSPFIIFMLLSVLFALISTLLVKYVAPMATGSGISEIKVWVSGFEYNKEFLGFLTL
VIKSVALPLAISSGLSVGKEGPSVHYATCCGYLLTKWLLRDTLTYSSQYEYITAASGAGVAVAFGAPIGG
VLFGLEEIASANRFNSSTLWKSYYVALVAITTLKYIDPFRNGRVILFNVTYDRDWKVQEIPIFIALGIFG
GLYGKYISKWNINFIHFRKMYLSSWPVQEVLFLATLTALISYFNEFLKLDMTESMGILFHECVKNDNTST
FSHRLCQLDENTHAFEFLKIFTSLCFATVIRALLVVVSYGARVPAGIFVPSMAVGATFGRAVSLLVERFI
SGPSVITPGAYAFLGAAATLSGITNLTLTVVVIMFELTGAFMYIIPLMIVVAITRIILSTSGISGGIADQ
MIMVNGFPYLEDEQDEEEEETLEKYTAEQLMSSKLITINETIYLSELESLLYDSASEYSVHGFPITKDED
KFEKEKRCIGYVLKRHLASKIMMQSVNSTKAQTTLVYFNKSNEELGHRENCIGFKDIMNESPISVKKAVP
VTLLFRMFKELGCKTIIVEESGILKGLVTAKDILRFKRIKYREVHGAKFTYNEALDRRCWSVIHFIIKRF
TTNRNGNVI- Evaluate the output: does the algorithm (wrongly) predict TM-helices in your protein? In the shuffled sequences? Does it find all ten TM-helices in Gef1?
Try to familiarize yourself with the offerings in the EMBOSS package. I find some of the nucleic acid tools indispensable in the lab, such as restriction-site mapping tools, and I frequently use the alignment tools Needle and Water, but by and large the utility of many of the components–while fast, efficient and straightforward to use– suffers from lack of reference and comparison and from terse output. The routines show their conceptual origin in the 1970s and 1980s. We will encounter alternatives in later assignments.
R Sequence Analysis Tools
It's interesting to see this collection of tools that were carefully designed some twenty years ago, as an open source replacement for a set of software tools - the GCG package - that was indispensable for molecular biology labs in the 80s and 90s, but whose cost had become prohibitive. Fundamentally this is a building block approach, and the field has turned to programming solutions instead.
As for functionality, much more sophisticated functions are available on the Web: do take a few minutes and browse the curated Web services directory of bioinformatics.ca.
As for versatility, R certainly has the edge. Let's explore some of the functions available in the seqinr package that you already encountered in the introductory R tutorial. They are comparatively basic - but it is easy to prepare our own analysis.
Task:
- Study the code in the
Sequence Analysissection of the R script
- That is all.
Links and resources
- RegEx Pal - a tool for testing and developing regular expressions.
Footnotes and references
- ↑ Relational databases like MySQL, PostgresQL, and MariaDB offer the datatype "Enum" for this purpose but this is an inferior solution. Enum types need to be fixed at the time the schema is created, they can't store information about their semantics, i.e. how the keywords are defined and when they should be used, and they can't be used in more than one table, since they are metadata of one particular column.
Ask, if things don't work for you!
- If anything about the assignment is not clear to you, please ask on the mailing list. You can be certain that others will have had similar problems. Success comes from joining the conversation.
- Do consider how to ask your questions so that a meaningful answer is possible:
- How to create a Minimal, Complete, and Verifiable example on stackoverflow and ...
- How to make a great R reproducible example are required reading.
| < Assignment 2 | Assignment 4 > |